Abstract
Described here is a method for facile generation of markerless gene deletion mutants of Actinomyces oris. Homologous integration of a nonreplicative vector carrying a gene exchange cassette into the bacterial chromosome was selected for by using mCherry fluorescence and resistance to kanamycin. Completion of allelic replacement was counterselected for by using loss of fluorescence.
Actinomyces oris (formerly Actinomyces naeslundii [3]) is Gram positive, facultatively anaerobic, and commonly found in the human oral cavity and plays a major role in the formation of oral biofilm or dental plaque. It is thought that adherence of A. oris to the tooth surface and its coaggregation with oral streptococci create an adhesive platform for subsequent colonization of bacteria in the plaque community (4). A. oris surface molecules such as fimbriae and pili have been shown previously to be required for the bacterial interactions with host tissues and other oral bacteria (7). However, the roles of fimbrial molecules or other surface proteins involved in these processes and their molecular assembly on the cell surface remain elusive. Lack of a facile gene disruption technology is the main reason for this obscurity.
Conventional methods of genetic manipulation employing nonreplicative plasmids as delivery vectors in A. oris have been used to create gene disruption by allelic exchange, which allows insertion of a selectable marker (1, 8, 9). Often, this strategy generates polar mutations that affect downstream genes, and it is inadequate for multigene deletion because antibiotic markers for Actinomyces are scarce. To circumvent this problem, we successfully developed a method that utilizes a pUC19 derivative (namely, pHTT177) to generate a nonpolar, in-frame deletion of the sortase gene srtC2 (5). However, this system proved extremely laborious because the second homologous recombination (double-crossover) event leading to chromosomal excision and loss of the plasmid could not be efficiently selected for. Consequently, we explored fluorescence as a positive selection marker for A. oris, as described below.
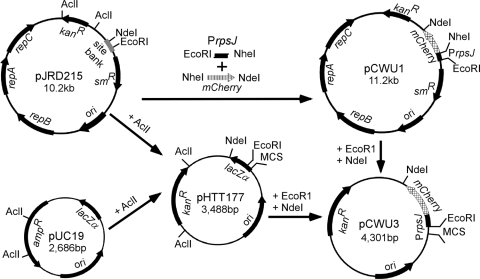
To generate a nonreplicative delivery vector for gene replacement with a counterselectable marker, we cloned the gene encoding the red fluorescent protein mCherry under the control of the constitutive promoter PrpsJ into pHTT177 by using EcoRI and NdeI sites (5) (Fig. 1). Initially, the mCherry sequence was amplified from plasmid pRSET-B-mCherry DNA (6) by using primers P1 (5′-GGCGGCTAGCATGGTGAGCAAGGGCGAGGAG-3′) and P2 (5′-GGCGCATATGCTACTACTTGTACAGCTCGTCCATG-3′), which contain NheI and NdeI sites (underlined), respectively. Primers P3 (5′-GGCGGAATTCCGCCCGAGCGCGGGGACCAGT-3′) and P4 (5′-GGCGGCTAGCGGCGCCTAACCTCTCTTGTACTTG-3′), containing EcoRI and NheI sites, were used to amplify the untranslated region of rpsJ from A. oris MG-1 chromosomal DNA (see gene identification no. ANA_0026 in the A. oris database at www.oralgen.lanl.gov). Both fragments were subcloned into pJRD215 at EcoRI and NdeI sites (2). The resulting vector, pCWU3, has a multiple-cloning site (MCS) containing EcoRI, SacI, KpnI, BamHI, XbaI, SalI, and HindIII sites for cloning purposes (Fig. 1).
FIG. 1.
Construction of the nonreplicative delivery vector with red fluorescent mCherry protein as a counterselectable marker. The mCherry gene under the control of the A. oris rpsJ promoter was subcloned into the Escherichia coli/Actinomyces shuttle vector pJRD215 before being cloned into pHTT177, which is a derivative of pUC19. The resulting plasmid, pCWU3, has a kanamycin resistance cassette (kanR) and an MCS containing EcoRI, SacI, KpnI, BamHI, XbaI, SalI, and HindIII sites.
As a proof of concept, we utilized the vector pCWU3, created as described above, to generate an in-frame deletion of acaA (see gene identification no. ANA_0196 in the database at www.oralgen.lanl.gov), encoding a putative cell wall anchor protein (called Aca for actinomyces cell wall anchor). Primer sets P5/P6 (5′-GGCGGAATTCGCCGGAGGCGCCGTCGGGGAAG-3′/5′-GGCGGGTACCAGGATCTCCGTTAGACACGG-3′) and P7/P8 (5′-GGCGGGTACCCAGCGAGACTGCGACCAGCAG-3′/5′-GGCGTCTAGAGGTGGGCGTACTTCTGGTCCAT-3′) were used to amplify ∼1.0-kb sequences upstream and downstream, respectively, of acaA from A. oris MG-1 chromosomal DNA. The upstream DNA fragment was digested with EcoRI and KpnI, while the downstream fragment was digested with KpnI and XbaI (restriction enzyme sites are underlined); both fragments were ligated into pCWU3, which had been precut with EcoRI and XbaI. A. oris MG-1 was transformed with the resulting plasmid by electroporation (5), and kanamycin-resistant colonies representing integration of the plasmid into the bacterial chromosome were selected for their ability to grow on heart infusion (HI; Difco) agar plates supplemented with 50-μg ml−1 kanamycin. These colonies were also examined for their fluorescence by using an Olympus XI71 inverted microscope equipped with a Hamamatsu charge-coupled device camera and a tetramethyl rhodamine isothiocyanate (TRITC) filter set (Fig. 2 A).
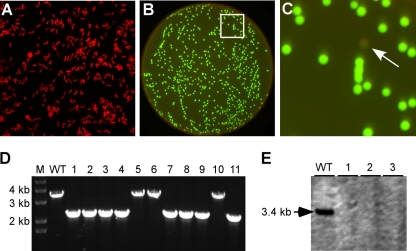
FIG. 2.
Analysis of a gene deletion in A. oris. (A) A. oris cells expressing mCherry under the control of the rpsJ promoter were viewed with an Olympus inverted microscope using a TRITC filter. (B and C) Selection of A. oris acaA deletion mutants was performed with a FluorChem Q imaging system (Alpha Innotech, CA). Fluorescent cells appeared green (pseudocolored) with the Cy3 setting, while nonfluorescent cells (potential mutants, indicated by the white arrow in panel C) were gray. An enlarged area (indicated by the white box in panel B) is shown in panel C. (D and E) Nonfluorescent bacteria were further examined for the absence of the acaA gene by PCR amplification (D) and Southern blot analysis (E). Bands of approximately 2.2 kb indicate acaA deletion, whereas bands of approximately 3.3 kb indicate a wild-type (WT) genotype (D). Samples from the parent strain MG-1 (WT) and size markers (M) are indicated, and the black arrow marks a 3.4-kb hybridized fragment.
To select Actinomyces clones that had undergone the double-crossover event leading to chromosomal excision and loss of the plasmid, we inoculated a sample of bacteria carrying the integrated plasmid into HI broth overnight at 37°C. The bacterial culture was then serially passaged seven times with a 1:40 dilution in HI broth without antibiotics. Forty-microliter aliquots of the 10,000-fold-diluted final culture were plated onto HI agar plates. After 3 days of growth at 37°C, plates were screened for nonfluorescent colonies by using a FluorChem Q imaging system (Alpha Innotech, CA) with a Cy3 filter. The Cy3 filter was chosen because it produced brighter images than those produced by the Cy5 filter (data not shown) and the images were given with false green coloring (Fig. 2B). Of approximately 16,000 colonies that were screened (a procedure taking less than 30 min), 11 showed no fluorescence (an example indicated by an arrow is shown in Fig. 2C), corresponding to a frequency of ∼7.0 × 10−4. This is consistent with the low frequency of homologous recombination in A. oris (5). Nonfluorescent colonies were also confirmed to be sensitive to kanamycin (data not shown), and their genomic DNA was extracted for PCR and Southern blot analyses. For PCR analysis, primers P5 and P8 were used. As shown in Fig. 2D, 8 of the 11 isolates generated amplicons of approximately 2.2 kb, which is indicative of acaA deletion, while the remaining 3 isolates generated the expected wild-type amplicons of approximately 3.3 kb. For further confirmation, DNA samples from three acaA mutants and the wild-type strain MG-1 were analyzed by Southern blotting using a 550-bp probe generated by primers 5′-AGTCTCCAACGCATCCGTCTC-3′ and 5′-GTGTCCCGAGACATTGGCCGTG-3′. Based on sequence analysis of acaA and surrounding genes, digestion by PstI will generate a 3.4-kb fragment for the wild type. As expected, the probe hybridized to the 3.4-kb DNA fragment, which was missing from the three mutant samples (Fig. 2E). Thus, the lack of a hybridization signal for the three mutants further confirmed the absence of acaA in these mutants.
In summary, we have developed a facile allelic exchange system for A. oris that reduces the laborious step of screening for a double-crossover event to less than 30 min. To our knowledge, this is the first report of an application that employs a fluorescent protein as a positive selectable marker for gene disruption in bacteria. Conceptually, this strategy can be applied for gene disruption in any system.
Acknowledgments
We thank Roger Y. Tsien (UCSD) for pRSET-B-mCherry and Timothy Fothergill, I-Hsiu Huang, and Elizabeth Rogers (UTHSC—Houston) and members of our laboratory for critical review of the manuscript and discussion.
This work was supported by NIH grant DE017382 from the National Institute of Dental and Craniofacial Research to H.T.-T.
Footnotes
Published ahead of print on 2 July 2010.
REFERENCES
- 1.Chen, P., J. O. Cisar, S. Hess, J. T. Ho, and K. P. Leung. 2007. Amended description of the genes for synthesis of Actinomyces naeslundii T14V type 1 fimbriae and associated adhesin. Infect. Immun. 75:4181-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davison, J., M. Heusterspreute, N. Chevalier, V. Ha-Thi, and F. Brunel. 1987. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene 51:275-280. [DOI] [PubMed] [Google Scholar]
- 3.Do, T., U. Henssge, S. C. Gilbert, D. Clark, and D. Beighton. 2008. Evidence for recombination between a sialidase (nanH) of Actinomyces naeslundii and Actinomyces oris, previously named ‘Actinomyces naeslundii genospecies 1 and 2.’ FEMS Microbiol. Lett. 288:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolenbrander, P. E., R. J. Palmer, Jr., A. H. Rickard, N. S. Jakubovics, N. I. Chalmers, and P. I. Diaz. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42:47-79. [DOI] [PubMed] [Google Scholar]
- 5.Mishra, A., A. Das, J. O. Cisar, and H. Ton-That. 2007. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J. Bacteriol. 189:3156-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 7.Yeung, M. K. 1999. Molecular and genetic analyses of Actinomyces spp. Crit. Rev. Oral Biol. Med. 10:120-138. [DOI] [PubMed] [Google Scholar]
- 8.Yeung, M. K., J. A. Donkersloot, J. O. Cisar, and P. A. Ragsdale. 1998. Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. Infect. Immun. 66:1482-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeung, M. K., and P. A. Ragsdale. 1997. Synthesis and function of Actinomyces naeslundii T14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect. Immun. 65:2629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]