Abstract
Shiga toxin-converting bacteriophages (Stx phages) are involved in the pathogenicity of some enteric bacteria, such as Escherichia coli O157:H7. Stx phages are released from their bacterial hosts after lytic induction and remain free in the environment. Samples were analyzed for the presence of free Stx phages by an experimental approach based on the use of real-time quantitative PCR (qPCR), which enables stx to be detected in the DNA from the viral fraction of each sample. A total of 150 samples, including urban raw sewage samples, wastewater samples with fecal contamination from cattle, pigs, and poultry, and fecal samples from humans and diverse animals, were used in this study. Stx phages were detected in 70.0% of urban sewage samples (10 to 103 gene copies [GC] per ml) and in 94.0% of animal wastewater samples of several origins (10 to 1010 GC per ml). Eighty-nine percent of cattle fecal samples were positive for Stx phages (10 to 105 GC per g of sample), as were 31.8% of other fecal samples of various origins (10 to 104 GC per g of sample). The stx2 genes and stx2 variants were detected in the viral fraction of some of the samples after sequencing of stx2 fragments amplified by conventional PCR. The occurrence and abundance of Stx phages in the extraintestinal environment confirm the role of Stx phages as a reservoir of stx in the environment.
Shiga toxin-producing Escherichia coli (STEC) is associated with diarrhea, hemorrhagic enterocolitis, and hemolytic-uremic syndrome in humans (46). Escherichia coli serotype O157:H7 is the main cause of these diseases, although other serotypes of E. coli and other enterobacteria species have been described (36). These E. coli serotypes produce at least two immunologically distinct Shiga toxins, called Stx1 and Stx2. In addition to these, several variations of these toxins have been reported in recent years, showing differences in virulence and distribution in the host populations examined (48, 51). Shiga toxin genes are carried by temperate bacteriophages (19, 35). Stx-encoding bacteriophages investigated to date consist of double-stranded DNA and have lambdoid genetic structures (19, 27, 32, 37, 47). The induction and regulation of these phages are directly involved in the production of toxin and, therefore, in the pathogenicity of the strains (8, 50). Stx phages are efficient vectors for the transfer of toxin genes, being able to convert nonpathogenic bacterial hosts into Stx-producing strains by transduction of stx, as has been demonstrated under various conditions (1, 4, 27, 28, 41, 49).
Most of the reported outbreaks of STEC infections are associated with cattle products (10, 17), with the consumption of contaminated foods (10, 34), and with several waterborne infections (30). Stx phages are present within fecally contaminated aquatic environments (9, 28, 30, 32, 45). Moreover, a high percentage of STEC strains present in extraintestinal environments carry inducible Stx phages (14, 30).
As individuals infected with STEC strains shed large quantities of Stx phages in feces, Stx phages should be prevalent in the environment, as are other viruses transmitted by the fecal-oral route (5, 11) or bacteriophages infecting bacteria present in the intestinal tract (16, 23). Moreover, those STEC strains isolated from food and animals carry inducible Stx phages (24, 27, 42). The virulence profiles of STEC strains isolated from food also suggest the presence of inducible Stx phages (10).
Stx phages in sewage have been detected by nested PCR (28, 29, 31). However, to quantify them, the most probable number (MPN) method was applied, which allows only a rough estimate of the amount of Stx phages present in the sample. To assess the number of Stx phages accurately, real-time quantitative PCR (qPCR) technology is a useful tool. This technology is both sensitive and specific, and it gives accurate quantitative results (25). Comparison with a standard enables the number of copies of stx to be quantified, which can then be translated into the number of Stx phage particles.
Little is known about the prevalence of phages carrying stx in fecal samples. The data available on the numbers of these phages in fecally contaminated water samples were only roughly estimated. The first step to evaluate the role of Stx phages in the environment as lateral gene transfer vectors is to know the extent of these viruses in the environment. The aim of this study is to report quantitative data on the abundance of Stx phages in urban sewage samples, in wastewater samples from cattle, pigs, and poultry, and in diverse fecal samples, calculated by means of a methodology based on qPCR.
MATERIALS AND METHODS
Samples. (i) Urban sewage.
This study was performed with 50 sewage samples collected from the influent raw urban sewage at two wastewater treatment plants. Treatment plant 1 serves a large urban area, consisting of a number of cities and towns, of approximately 500,000 inhabitants; treatment plant 2 receives urban sewage from a population of about 5,000 inhabitants. There are no other noteworthy differences between the two plants. Samples were regularly collected approximately every 20 days for a 2.5-year period. No incidence of enterocolitis caused by STEC was reported during the study period (2007-2009) in the areas (15). Fifty milliliters of each sample was analyzed. Microbiological parameters measured for all the samples are summarized in Table 1.
TABLE 1.
Bacterial and viral indicators detected in urban sewage, animal wastewater, and fecal samples
| Sample | No. of samples | Detection of indicated bacterial or viral indicator (no. of log10 CFU·ml−1 or g−1)a |
Samples positive for stx detection in phage DNA (%) | ||
|---|---|---|---|---|---|
| Fecal coliform | E. coli | Somatic coliphage | |||
| Sewage from plant 1 | 30 | 5.08 (0.31) | 4.70 (0.39) | 4.46 (0.32) | 90.0 |
| Sewage from plant 2 | 20 | 5.05 (0.58) | 4.25 (0.85) | 4.07 (0.93) | 40.0 |
| Cattle wastewater | 14 | 4.81 (0.58) | 4.52 (0.60) | 4.41 (0.98) | 85.7 |
| Pig wastewater | 8 | 5.77 (0.42) | 5.66 (0.43) | 5.49 (0.50) | 100.0 |
| Poultry wastewater | 14 | 5.15 (0.92) | 4.89 (0.82) | 4.49 (0.80) | 100.0 |
| Cattle feces | 38 | 3.87 (1.50) | 3.62 (1.32) | 3.74 (0.41) | 89.5 |
| Human feces | 5 | NA | NA | 4.74 (0.30) | 20.0 |
| Pig feces | 4 | NA | NA | 5.84 (0.24) | 75.0 |
| Rabbit feces | 4 | NA | NA | 3.85 (1.20) | 0 |
| Cat feces | 3 | NA | NA | 5.44 (0.67) | 0 |
| Sheep feces | 2 | NA | NA | 4.35 (0.49) | 100.0 |
| Dog feces | 2 | NA | NA | 5.33 (0.53) | 50.0 |
| Mouse feces | 1 | NA | NA | 3.48 | 0 |
| Bird feces | 1 | NA | NA | 4.43 | 0 |
Bacterial indicators include fecal coliforms and E. coli strains, and viral indicators include somatic coliphages. Results are the average values of the number of samples tested, with SD shown in parentheses. NA, not analyzed.
(ii) Animal wastewater.
Thirty-six samples containing exclusively fecal contaminants of a single animal origin (cattle, swine, or poultry) were collected from slaughterhouse wastewater effluents (Table 1). Cattle and poultry wastewater samples were regularly collected approximately every 2 months during a 2.5-year period, and pig samples were collected during a 1.5-year period. No human fecal contamination was expected in these samples. Fifty milliliters of each sample was analyzed.
(iii) Feces.
Five samples were collected from humans (healthy individuals). The animal fecal samples were collected from farms and domestic animals. Fecal samples were also obtained from 38 dairy cows, 4 farm pigs, 4 farm rabbits, 3 domestic cats, 2 sheep, 2 domestic dogs (Alsatians), 2 domestic mice, and 2 wild birds (sparrows). Portions of each fecal sample (2.5 g) were homogenized in 50 ml of phosphate-buffered saline (PBS; 137 mM NaCl, 10 mM phosphate, 2.7 mM KCl at pH 7.4), and the whole volume was processed for Stx phage detection and somatic coliphage quantification. Values were then referred to 1 g of sample. Fecal samples were collected in a single sampling campaign.
Determination of microbial indicators.
Fecal coliforms (FCs) and E. coli strains were enumerated as indicators of bacterial fecal pollution, and somatic coliphages were enumerated as indicators of viral fecal pollution. Each analysis was performed in duplicate. FCs and E. coli strains were counted by membrane filtration, according to previously standardized methods (2). mFC agar (Difco, France) and Chromocult coliform agar (Merck, Darmstadt, Germany) were used for fecal coliform and E. coli strains determination, respectively. The number of somatic coliphages, indicators of viral fecal pollution, was counted using the International Organization for Standardization (ISO) double-agar-layer method (3). E. coli strain WG5 (ATCC 700078) (3) was used as a host for detection of somatic coliphages, as described below. Modified Scholten's broth or modified Scholten's agar was used for the detection of phages infecting E. coli WG5 (3).
Purification of bacteriophages.
Bacteriophage 933W was used in the experiments as a positive control. Phage 933W (35) was induced from lysogenic E. coli strain C600(933W) (35) and purified, and the number of these phages was counted as previously described (22).
To purify bacteriophages from the samples, all the environmental samples were passed through low-protein-binding 0.22-μm-pore-size membrane filters (Millex-GP; Millipore, Bedford, MA). When necessary, several filter units were used to filter the whole volume. Partially purified bacteriophages were then 100-fold concentrated by protein concentrators (100-kDa Amicon Ultra centrifugal filter units; Millipore, Bedford, MA), following the manufacturer's instructions. The total volume of the filtered sample was placed in the units and centrifuged at 4,000 × g for the time necessary to reduce the volume to 0.5 ml. The amount of centrifugation time varied depending on the sample and ranged from 10 to 90 min. The concentrate was recovered from the tube, and the volume was adjusted to 2 ml with double-distilled sterile water.
The bacteriophage concentrates were then treated with DNase (100 units·ml−1 of the phage lysate) to eliminate free DNA outside the phage particles. An aliquot of the phage lysate at this stage was amplified by qPCR to confirm that free DNA containing stx had been removed from the sample.
Nucleic acid extraction.
DNA from all the Stx phages was isolated from phage lysates by proteinase K digestion and phenol-chloroform (1:1, vol/vol) treatment (39). The phenol-chloroform/phage lysate mixture was added to Phase Lock Gel tubes (5 Prime; VWR International, Madrid, Spain) and centrifuged by following the manufacturer's instructions. The DNA from the supernatant was precipitated with 100% ethanol and 3 M sodium acetate, and the volume was adjusted to 250 μl. DNA was further purified by means of Microcon YM-100 centrifugal filter units (Millipore, Bedford, MA) by following the manufacturer's instructions. Purified DNA was eluted in a final volume of 50 μl and evaluated by agarose (0.8%) gel electrophoresis, and bands were viewed by ethidium bromide staining. The concentration and purity of phage DNA extracted was determined by the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Scientific Instruments, Wilmington, DE).
Standard PCR procedures.
PCR amplifications were performed with GeneAmp PCR system 2400 (PerkinElmer, PE Applied Biosystems, Barcelona, Spain). A 378-bp fragment of the stx2 A subunit was amplified with primers UP378/LP378 (Table 2). The complete stx2 A subunit was amplified with primers S2Aup/S2Alp (Table 2).
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence | Gene region/use | Size (bp) | Reference or source |
|---|---|---|---|---|
| UP378 | GCGTTTTGACCATCTTCGT | 378-bp fragment of stx2 A subunit | 378 | 27 |
| LP378 | ACAGGAGCAGTTTCAGACAG | |||
| S2Aup | ATGAAGTGTATATTATTTA | stx2 A subunit | 960 | 26 |
| S2Alp | TTCTTCATGCTTAACTCCT | |||
| pBADf | ATGCCATAGCATTTTTATCC | Binds pBAD vector upstream of the cloning site | Invitrogen | |
| pBADr | GATTTAATCTGTATCAGG | Binds pBAD vector downstream of the cloning site | Invitrogen | |
| stxANY-f | ACGGACAGCAGTTATACCACTCT | qPCR for detection of a fragment of the stx2 A subunit | 65 | 21 |
| stxANY-r | CTGATTTGCATTCCGGAACGT | |||
| stxANY-M | FAM-CCAGCGCTGCGACACG-NFQ |
qPCR procedures. (i) Preparation of standard curves.
For the generation of standards to use in qPCR assays, a plasmid construction was employed. A 378-bp fragment obtained by conventional PCR, as described above, was cloned with a pBAD-TOPO vector for insertion of PCR products by following the manufacturer's instructions (Invitrogen Corporation, Barcelona, Spain). The construct was transformed by electroporation into competent cells.
Electroporation-competent cells were prepared from 10 ml of cultures of E. coli strain DH5α in LB medium and were concentrated by centrifugation at 3,000 × g for 5 min. They were then washed in 1 ml of ice-cold double-distilled water. After four washing steps, the cells were suspended in 100 μl of ice-cold double-distilled water. Cells were mixed with 10 μl of plasmid DNA in an ice-cold microcentrifuge tube and transferred to a 0.2-cm electroporation cuvette (Bio-Rad Inc., Barcelona, Spain). Cells were electroporated at 2.5 kV, 25 F capacitance, and 200 Ω resistance. Immediately after electroporation, 1 ml of SOC medium (39) was added to the cuvette. The cells were transferred to a 17- by 100-mm polypropylene tube and recovered in SOC medium for 1 to 4 h at 37°C, without shaking. A total of 100 μl of culture was incubated on LB agar with ampicillin (100 μg·ml−1). Colonies were selected and screened by conventional PCR to evaluate the presence of the vector containing the insert.
The vector was purified from the positive colonies using the Qiagen plasmid purification midikit (Qiagen Inc., Valencia, CA). The presence of the insert in the vector and its orientation were assessed by conventional PCR and sequencing, as described above, using primers UP378/LP378 and pBADf/pBADr (Invitrogen, Barcelona, Spain) (Table 2). The concentration of the vector construct was quantified by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Scientific Instruments, Wilmington, DE). The reaction product was linearized by digestion with EcoRV restriction endonuclease (Promega Co., Madison, WI). The restricted product was purified and quantified again.
To calculate the number of construct gene copies (GC), the following formula was used: [concentration of pBAD-TOPO::378stx (ng·μl−1)/molecular weight (ng·mol−1)] × 6.022 × 1023 molecules·mol−1 = number of molecules of pBAD-TOPO::378stx/μl−1 (22). Since each molecule of construct contains 1 copy of the stx fragment, it was calculated that 1 μl of our stock contained 5.89 × 1010 GC.
Serial decimal dilutions of this stock (5.89 to 5.89 × 109 GC·μl−1) were done in double-distilled water to prepare the standard curve for qPCR. The standard dilutions were then aliquoted and stored at −80°C until use. Three replicates of each dilution were added to each qPCR.
(ii) Quantification by qPCR.
A custom TaqMan (Applied Biosystems, Spain) set of primers and probe was designed (Table 2). The forward stxANY-f and reverse stxANY-r primers, amplifying a 65-bp fragment of the stx2 A subunit, and a minor groove binding probe stxANY-M with a FAM (6-carboxyfluorescein) reporter and an NFQ (nonfluorescent quencher) (Table 2) were used under standard conditions in a StepOne real-time PCR system (Applied Biosystems, Spain). This qPCR set amplifies stx2 and variants stx2c, stx2d, stx2e, and stx2g but not stx2f (22). The stx genes were amplified in a 20-μl reaction mixture with the TaqMan environmental real-time PCR mastermix 2.0 (Applied Biosystems, Spain). The reaction mixture contained 2 μl of the DNA sample or quantified plasmid DNA. Thermal cycler conditions used were an initial setup of 10 min at 95°C, 40 cycles of 15-s denaturing phase at 95°C, and 1 min of annealing/extending at 60°C. All samples were run in triplicate, including the standards—positive and negative controls. As a positive control, a 1:10,000 dilution of phage 933W DNA was used. As negative controls, double-distilled sterile water and the aliquots taken after DNase treatment during the DNA extraction procedure were used. The number of GC was defined as the average of the data in triplicate obtained. Since Stx phages are known to carry 1 stx copy only, the stx GC values can be extrapolated to the number of Stx phages in each sample.
(iii) Sequencing of the amplicons obtained by PCR.
Further confirmation of the sequence of the amplified DNA from 22 samples was achieved by sequencing. For this, amplicons of stx were generated by conventional PCR, using a combination of primers UP378, LP378, S2Aup, and S2Alp (Table 2). Amplicons were electrophoretically analyzed in a 1% agarose gel, and bands were viewed by ethidium bromide staining. The bands were excised from the agarose gel and purified using a QIAquick gel extraction kit (Qiagen Inc., Valencia, CA) by following the manufacturer's instructions. The purified amplicons were used as a template for sequencing. Sequencing was performed with the ABI Prism BigDye 3.1 Terminator cycle sequencing ready reaction kit (Applied Biosystems, Spain) in an ABI Prism 3730 DNA analyzer (Applied Biosystems, Spain), according to the manufacturer's instructions. All sequencing was performed in duplicate.
Nucleotide sequence analysis searches for homologous DNA sequences in the EMBL and GenBank database libraries were carried out with the Wisconsin Package version 10.2, Genetics Computer Group (GCG; Madison, WI). BLAST analyses were performed with the tools available on the National Institutes of Health (NIH) website (http://www.ncbi.nlm.nih.gov). Sequences were assembled with the MultAlin program available on the MultAlin website (http://bioinfo.genotoul.fr/multalin/multalin.html) (7).
(iv) Statistical analyses.
Data and statistical tests were computed with Statistical Package for Social Science software (SPSS). One-way analysis of variance (ANOVA) tests with a P value of 0.05 were used to evaluate the differences between samples obtained from urban sewage plants 1 and 2 and between bacterial and viral indicators. Pearson's correlation coefficients (r) between bacterial and viral indicators and between those indicators and log10 stx GC·ml−1 values were calculated. The box plot graph used to compare the summarized values of samples obtained from the same origin positive for stx was composed with Excel software (Microsoft Excel 2000). Calculations performed to generate the box plot graph included mean, standard deviation, median, quartile, minimum, and maximum values for each group of samples.
RESULTS
Numbers of bacterial and viral indicators in urban sewage and animal wastewater samples.
The numbers of FCs, E. coli strains, and somatic coliphages in urban sewage and animal wastewater samples were quite homogeneous across all the samples tested. Each sample value was the mean result of the two plates assayed. The values given in Table 1 are the average values of the number of samples tested from each origin (Table 1). The values of fecal coliforms significantly (P < 0.05) exceeded those of E. coli strains by 0.10 to 0.80 log10 units and exceeded those of somatic coliphages by 0.13 to 0.98 log10 units. However, FC values correlated with E. coli values in all groups of samples (Pearson's r = 0.91 to 0.99). Negative or very low correlation coefficients were found when comparing bacterial indicators (E. coli strains or FCs) with somatic coliphages (r = −0.10 to 0.41). In general, the values of all three indicators in the human samples were lower than those in the animal samples, with the exception of those in cattle samples. Rabbit and mouse feces also contained lower numbers, but the values were too low to draw more conclusions. Bacterial indicators detected in cattle fecal samples showed the highest variation among animals, as observed by the standard deviations (SD) calculated. The same samples showed less variability for somatic coliphages. The rest of the fecal samples were analyzed only for somatic coliphages because of the limited amount of the sample available. In these samples, correlations between indicators were not calculated.
Evaluation of the procedure.
Using phage 933W, we estimated the minimal number of phages detected by qPCR. After qPCR data acquisition, the cycle threshold (CT) value was calculated by determining the point at which fluorescence exceeded an arbitrary threshold signal. Standard curves were prepared by plotting CT versus log10 of the number of GC·μl−1. Amplification efficiency (E) was calculated by the formula E = (10−1/slope) − 1 (18). The slope was calculated using the regression line obtained with the points of the standard curve included (CT value versus the number of GC) for each run. Efficiencies for all reactions showing a slope of −3.32 were taken as 100%. Efficiencies of our reactions ranged from 94 to 100%. The detection limit of the qPCR, evaluated with the plasmid construct and with the stock of phage 933W, was calculated as 5.29 stx copies.
The amplifications of 933W phage lysates indicated a certain variability in the quantitative results obtained with a low density of phages (below 9 GC per ml, which corresponds to 9 Stx phage particles), hampering precise quantification at that level because of methodological limits. The limit of detection of the qPCR was calculated as 5.29 GC·ml−1, and the limit of quantification was 9 GC·ml−1. Those samples with values between the two limits were positive, although accurate quantification of the number of Stx phages was not possible.
To evaluate the recovery of the Stx phages in the different samples, phage 933W was inoculated in urban sewage samples, in cattle and pig fecal samples, and in poultry wastewater samples. Two phage concentrations, 1.7 × 108 and 8.0 × 104 phage particles·ml−1 or 8.24 and 4.90 log10 phages·ml−1, as shown in Table 3, were used. Phage 933W was obtained from a lysate of a known concentration, prepared, and evaluated by electron microscopy, as described elsewhere (22). The number of Stx phages was quantified by qPCR before and after inoculation of the samples, and the results were compared (Table 3). Each assay was performed with three independent samples from each origin. Results for the different samples showed few differences in regard to the number of Stx phages detected in the inoculated sample. A lower recovery percentage was observed for samples of cattle and pig feces when a high phage concentration (8.24 log10 phages·ml−1) was used. Nevertheless, even at the lowest recovery level, the reduction in log10 units did not exceed 0.4 and, in the other samples, was below 0.1 (Table 3). These results suggest that this is a suitable method for the extraction of Stx phages.
TABLE 3.
Representative experiment showing the recovery of phage 933W detected by qPCR from stock and after being inoculated in samples
| Sample | 933W (no. of log10 GC·ml−1) detected in: |
Log10 reductionc | |
|---|---|---|---|
| Phage lysatea | Sampleb | ||
| Urban sewage | 8.24 | 8.19 | 0.05 |
| 4.90 | 4.84 | 0.06 | |
| Cattle feces | 8.24 | 7.84 | 0.40 |
| 4.90 | 4.70 | 0.20 | |
| Pig feces | 8.24 | 7.85 | 0.40 |
| 4.90 | 4.86 | 0.05 | |
| Poultry wastewater | 8.24 | 8.23 | 0.01 |
| 4.90 | 4.86 | 0.04 | |
Number of log10 stx copies (GC) evaluated from the phage lysates used to inoculate the samples.
Number of log10 stx copies (GC) detected in the inoculated samples.
Values are the difference between the number of log10 stx copies (GC) of 933W found in the phage lysates and the number of log10 stx copies detected in the samples.
In addition, to rule out the interference of inhibitors potentially present in the samples, 2 μl of DNA extract for each sample was mixed with 2 μl of the standard containing 106 GC·μl−1, and the mixture was processed by qPCR. A reduction in the number of GC for the standard in these mixtures would reveal the presence of inhibitors. Of the 150 samples assayed, 5 showed more GC·μl−1 than the standard, since the values of Stx phages exceeded the number of GC in the standard; 102 samples showed no reduction, and 41 showed a reduction in the number of GC·μl−1 of the standard of less than 0.01 log10 GC. Those samples were considered free of inhibitors. Only two samples of cattle feces showed a reduction in the GC values of the standard of more than 0.5 log10 GC·μl−1. These two samples were analyzed by diluting the DNA 1/10 in order to minimize the effect of inhibitors. Both samples showed positive results following dilution.
Stx phages in urban sewage and animal wastewater samples.
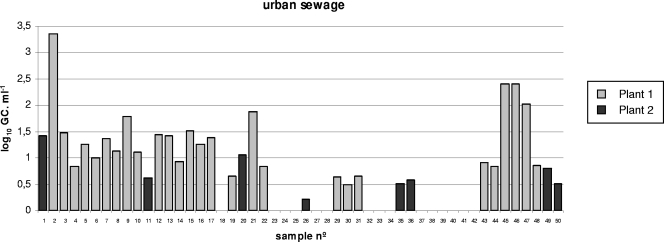
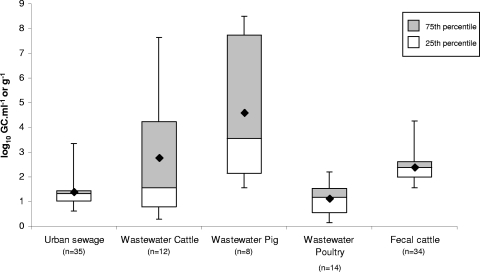
The number of stx GC detected per ml of urban sewage and animal wastewater samples are shown in Fig. 1 and 2 (Fig. 1 gives results from samples of urban sewage from plants 1 and 2, and Fig. 2 gives results from cattle, pig, and poultry wastewater samples). Seventy percent of the sewage samples (plant 1 and 2 taken together) were positive for the presence of Stx phages (Table 1 and Fig. 1). Results indicated a significant difference in the CT values obtained when the two plants were compared (ANOVA; P < 0.05). When considering values of positive samples from both plants, the mean value of the Stx phage counts for urban sewage samples was 1.37 log10 GC·ml−1 (Fig. 3). Urban sewage samples had less variability than samples with other origins (Fig. 3). The levels of bacterial (FC and E. coli) and viral (somatic coliphage) fecal indicators for each sample do not correlate with the densities of Stx phages in the same sample. Pearson's r gave values of −0.1 to 0.23 for FCs and E. coli strains and −0.1 to 0.4 for somatic coliphages, indicating that there is no correlation between Stx phages and the fecal input.
FIG. 1.
Values of Stx phages (the number of log10 GC·ml−1) in urban raw sewage samples evaluated by qPCR. n°, number.
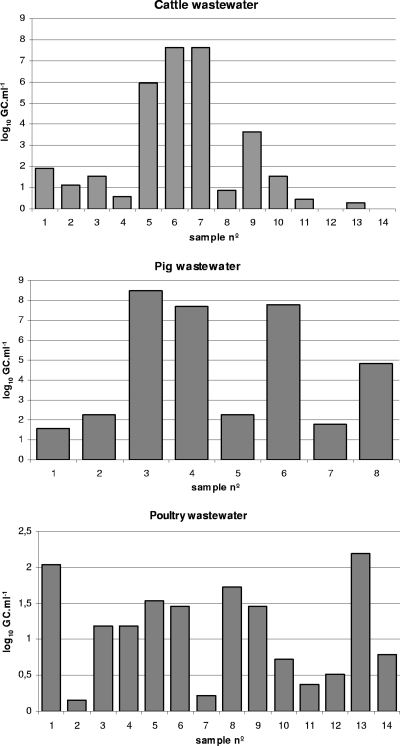
FIG. 2.
Values of Stx phages (the number of log10 GC·ml−1) in samples of wastewater with fecal pollution obtained from cattle, pigs, and poultry evaluated by qPCR.
FIG. 3.
Variability of the numbers of Stx phages in each group of samples. Each box plot indicates the counts obtained from samples of the same origin that were positive for stx. The crosspieces of each box plot represent (from top to bottom) the maximum (top black line), upper quartile (gray box), median (middle black line), lower quartile (white box), and minimum (bottom black line) values. The gray boxes include samples showing values within the 75th percentile, and the white boxes include samples showing values within the 25th percentile. The black diamonds show the mean values.
Wastewater samples from cattle, pigs, and poultry (Fig. 2) showed a high percentage of positive samples and, in general, higher values of Stx phages than those found in urban sewage samples (Fig. 3). In cattle wastewater samples, the average density of Stx phages was 2.77 log10 GC·ml−1 for positive samples (Fig. 3). All pig and poultry wastewater samples were positive. The average value in pig wastewater samples was 4.59 log10 GC·ml−1. Pig wastewater samples showed higher densities of Stx phages than samples with other origins (Fig. 2 and 3). Cattle and pig wastewater samples showed greater variability than other samples (Fig. 3). In poultry, the average value of positive samples (1.11 log10 GC·ml−1) was lower than that of samples with other origins (Fig. 3).
Controls of sterile water and the samples processed after DNase treatment and before proteinase K digestion were negative in all cases.
Stx phages in individual fecal samples.
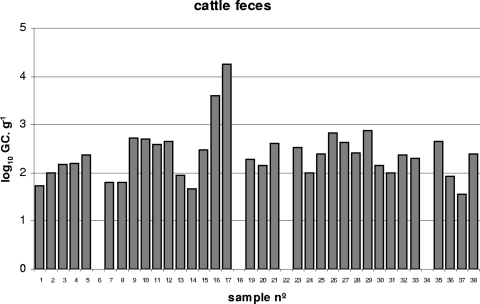
Thirty-seven fecal samples collected from individual cows were analyzed in this study (Fig. 4). Of these, 89.5% showed the presence of Stx phages (Fig. 4). The average value of Stx phages in cattle fecal samples, when only positive samples were considered, was 2.32 log10 GC·g−1. Cattle fecal samples were less variable than wastewater samples, with most of the values close to the median (Fig. 3).
FIG. 4.
Values of Stx phages (the number of log10 GC·g−1) in samples of feces obtained from 38 cows evaluated by qPCR.
Twenty-two fecal samples from healthy humans and domestic animals were also analyzed. Seven samples (three from pigs, one from a human, two from sheep, and one from a dog) were positive for Stx phages. Of these, the value for Stx phages in the human sample was 2.90 log10 GC·g−1. Values for Stx phages in the three pig samples were 2.15, 2.40, and 2.94 log10 GC·g−1. One sheep sample showed a value of 2.47 log10 GC·g−1, while the other one showed 1.19 log10 GC·g−1. The dog fecal sample showed a value of 1.45 log10 GC·g−1.
Sequences of the amplified DNAs.
Those samples showing the largest concentrations of Stx phages were selected. The viral DNA fraction of the sample was used as a template to amplify the complete stx2 sequence by conventional PCR, as described above. Not all the samples allowed amplification of the whole stx sequence; some allowed only amplification of a shorter fragment. Wherever it was possible to obtain stx amplicons from those samples, these were sequenced. Twenty-two sequences were obtained and compared with the sequences available in the databases (Table 4). A total of 99 to 100% similarities between our amplicons and the published sequences were considered. Most of the sequences corresponded to that of the stx2 variant described previously for phage 933W (GenBank accession no. AF125520) (37), while in some samples, variants were detected (Table 4).
TABLE 4.
Comparison of the stx2 fragment sequence amplified from phage DNA isolated from 22 samples and previously described sequences
| Sample | Sample no. | Fragment size (bp) | stx2 variant(s) | Sequence homologue(s) | GenBank accession no. |
|---|---|---|---|---|---|
| Urban sewage | 1 | 700 | stx2v, stx2d, stx2c, stx2, and stx2g | E. coli stx2v genes, strain TS17/08, serotype O113:H21 | FM998851 |
| E. coli stx2d genes, strain TS06/08, serotype OR:H29 | FM998848 | ||||
| E. coli stx2 genes, strain N2688, serotype O88:H38 | GQ429163 | ||||
| E. coli stx2c genes, strain 5021/96, serotype Orauh:H− | AJ567994 | ||||
| E. coli stx2g genes, serotype O28ab:H28 | AY095209 | ||||
| 5, 7, 15, 21, and 47 | 700 | stx2 | 933W phage | AF125520 | |
| 10 | 500 | stx2 | E. coli stx2 genes, serotype O157:H− | EU526759 | |
| 12 and 20 | 500 | stx2, stx2v, stx2d, and stx2d1 | E. coli stx2 genes, strain N5545, serotype ONT:H7 | GQ429167 | |
| E. coli stx2v genes, strain TS27/08, serotype OR:NM | FM998860 | ||||
| E. coli stx2d genes, isolate EC782, serotype ONT:NM | AF500193 | ||||
| E. coli stx2d1 genes, serotype O91:H21 | AF479828.1 | ||||
| Pig wastewater | 1 | 500 | stx2e | E. coli stx2e genes, strain TS29/08, serotype ONT:NM | FN182286 |
| E. coli stx2e genes, strain TS16/08, serotype O8:NM | FN182285 | ||||
| E. coli stx2e genes, strain TS13/08, serotype O8:H9 | FN182284 | ||||
| Bacteriophage P27 complete genome | AJ298298 | ||||
| 2, 3, 4, and 6 | 1,100 | stx2 | 933W phage | AF125520 | |
| Cattle wastewater | 5, 6, and 7 | 1,100 | stx2 | 933W phage | AF125520 |
| Poultry wastewater | 1 | 1,100 | stx2 and stx2v | E. coli stx2 genes, strain N2688, serotype O88:H38 | GQ429163 |
| E. coli stx2v genes, strain TS07/07, serotype O130:H11 | FM998861 | ||||
| 8 | 1,100 | stx2 | E. cloacae stx2 genes | Z50754.1 | |
| stx2vh-d | E. coli stx2vh-d genes, serotype O157:H7 | AB071845 | |||
| 13 | 1,100 | stx2 | 933W phage | AF125520 | |
| Cattle feces | 16 and 17 | 1,100 | stx2 | 933W phage | AF125520 |
DISCUSSION
It is possible that phage-mediated transduction of virulence genes into environmental bacteria could cause the emergence of new pathogenic E. coli strains. Some reports suggest that human pathogenic STEC strains have evolved from other nonhuman serotypes by incorporation of new virulence genes in their genomes (26), with some of them carried by bacteriophages, like stx. This hypothesis was also put forward in the study of an outbreak caused by E. coli strain O103:H25 in Norway (42). In this outbreak, STEC O103 strains carried an Stx phage that was similar to the Stx phages found in O103 strains isolated from previous cases. It was suggested that the Stx2 phage, present in the environment either as a free phage particle or within a limited pool of Stx-producing E. coli O103 strains, had infected or integrated into the Stx-negative E. coli O103:H25 isolates from the Norwegian outbreak, generating new STEC strains.
Stx phages are present in urban sewage, wastewater, and river water (9, 28, 29, 45). Environmental Stx phages were characterized (32) and found to persist in the water environment in a way similar to that of lytic E. coli-infecting phages and better than E. coli O157 (9, 31). Nevertheless, the numbers of Stx phages in fecally contaminated samples detected in these previous studies were only roughly estimated (at least 1 Stx phage particle in 10 ml of urban sewage and from 1 to 10 infectious Stx phages·ml−1 of urban sewage) (28). The density of Stx phages in urban sewage samples in the present study is of the same order or even higher than previously estimated (28).
The approach used yielded a high recovery of Stx phages, with a log10 reduction of Stx phages ranging from 0.01 to 0.4 log10 units. The worst recoveries obtained from pig and cattle fecal samples could be attributed to the matrix itself, which affects the phage purification procedure. Inhibitors did not seem to be the cause, since higher recoveries were observed in the same samples spiked with lower densities of phages (4.90 log10GC·ml−1). The presence of inhibitors in the phage DNA samples was ruled out in most of the samples, since the 150 samples were spiked with standard dilutions and no reduction in the number of stx copies in the standard was seen. This approach was preferred to the use of the internal PCR inhibition control (IC) of DNA nonrelated with the test sample in the same reaction tube. The latter option implies the design of a duplex qPCR that interferes and competes with the Stx qPCR used and suggests that differential amplification favors one of the templates more than the other (38, 43). There is also competition between target DNA and the IC. Therefore, the amount of internal control is critical to the detection limit in low-template DNA samples. This can reduce the amplification efficiency of the target gene, producing false-negative results (20). Similarly, the IC can also be inhibited by an excess in the target gene. Finally, there are no guarantees that inhibitors affect the sample and IC in the same way, reducing IC usefulness (21). The approach used here allows detection of inhibition and avoids some of these handicaps.
In animal wastewater samples, the percentage of positive samples indicates the widespread existence of Stx phages among fecal matter of different origins. The variability observed in fecal samples could be attributable to differences in the release of Stx phages among individuals. These differences have been reported in cattle for those animals that excrete more Escherichia coli O157 than others (6, 10). Those animals that excrete more STEC strains might also be expected to excrete more Stx phages than others. As with bacteria, these high-shedding animals might also increase contamination with Stx phages, although our results do not allow us to confirm this hypothesis. Together with cattle wastewater samples, a larger number of cattle fecal samples were analyzed, and results confirmed the abundance of Stx phages in cattle feces, which was expected to be highly heterogeneous, in line with previous descriptions of Stx phages from cattle (27). Values of 2.6 log10 units·ml−1 of STEC strains were previously reported from cattle slaughterhouses in the same geographical area (12).
Pig wastewater samples showed higher densities of Stx phages than samples with other origins (up to 8 log10 GC·ml−1). The number of Stx-producing bacteria in pig slaughterhouses in the same area was 103 MPN·ml−1 (12). There are fewer descriptions of the impact of Stx phages isolated from pigs than those of the impact of Stx phages isolated from cows. Although it has not been proven that human strains can be converted with Stx phages induced from pig isolates, the release of Stx phages from swine fecal samples should be taken into account. The variant stx2e was found in one of the pig samples, and an Stx2e phage induced from a pig STEC strain was described elsewhere (33). Poultry wastewater samples showed the lowest levels of Stx phages. The primers used allow for amplification of all Stx2 variants described so far, except the Stx2f variant, which was isolated from pigeons (40). It is possible that, with the primer set used, the real amount of Stx phages carrying Stx2 variants, either Stx2f or others not described and typically with poultry as reservoir, was underestimated.
Since each fecal sample was collected from a single animal, the chance of detecting an Stx phage shedder is lower than the chance of detecting one in animal wastewater or urban sewage samples, which comprise a mixture from numerous animals/individuals. Although the number of samples from each species was limited, and few of them (pigs, humans, and dogs) were positive, this is, to our knowledge, the first description of Stx phages quantified in fecal samples of animal origin.
The stx sequences obtained from human samples indicate that the Stx phages carry mostly stx2 but also carry some stx2 variants. Many prophages expressing the stx2 variant show a high level of spontaneous induction (8), which could explain why this variant is predominantly found in those samples with a large amount of Stx phages. The stx2 variant is associated mainly with the most pathogenic human-derived strains belonging to seropathotype A (O157:H7 strains) (8, 51), although there are no recent outbreak reports in our geographical area. Some other human samples showed equal identity among variants Stx2d, Stx2c, and Stx2g, which have also been described in phages (13, 44, 47). Among the variants obtained from animal samples, the stx2 variant was predominant. stx2e was detected in only one pig sample.
Despite the abundance of Stx phages, there were no STEC outbreaks in our geographical area during the period of the study (15). Nevertheless, not only stx but also other factors contribute to the virulence of pathogenic STEC strains. Besides, detection of Stx phages by qPCR does not determine whether these phages are infectious and able to transduce the toxin. It has previously been reported that the ratio of infectious Stx phages to Stx phages detected by qPCR could range from 1/10 to 1/1,000, depending on the Stx phage and the host strain used (22). If this calculation is correct for all Stx phages, this indicates that not all samples carry infectious Stx phages.
The results reported in this study demonstrate that Stx phages are widely distributed in fecally polluted environments. Besides, the data of fecal indicators (especially somatic coliphages) indicate that certain numbers of Stx phages do not correlate directly with larger inputs of fecal pollutants. The possibility of another stx environmental reservoir, located in bacteria or most probably in phages, could be considered. The abundance of Stx phages supports their contribution to the gene flux between bacteria in the extraintestinal environment. This gene flux can be important both in bacterial evolution and in the movement of genes that are relevant to the emergence and reemergence of human and animal pathogens and consequently have significant implications for public health. More studies on the occurrence and abundance of phages carrying virulence genes may help to modify present-day practices of urban sewage and slurry treatment and disposal, and of food management, in order to minimize the spread of virulence factors.
Acknowledgments
This study was supported by the Generalitat de Catalunya (grant 2009SGR1043), by the Spanish Ministry of Education and Science (grants AGL200601566/ALI and AGL2009-07576), and by the Xarxa de Referència en Biotecnologia (XeRBa). L. Imamovic is a recipient of a grant from the Spanish Ministry of Education and Science (grant FPI 20060054361).
Footnotes
Published ahead of print on 9 July 2010.
REFERENCES
- 1.Acheson, D. W. K., J. Reidl, X. Zahang, G. T. Keusch, J. J. Mekelanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, American Works Association, and Water Environmental Federation, Washington, DC.
- 3.Anonymous. 2000. ISO 10705-2: water quality. Detection and enumeration of bacteriophages. Part 2: enumeration of somatic coliphages. International Organization for Standardization, Geneva, Switzerland.
- 4.Beutin, L., E. Strauch, and I. Fischer. 1999. Isolation of Shigella sonnei lysogenic for a bacteriophage encoding gene for production of Shiga toxin. Lancet 353:1498. [DOI] [PubMed] [Google Scholar]
- 5.Bosch, A. 1998. Human enteric viruses in the water environment: a minireview. Int. Microbiol. 1:191-196. [PubMed] [Google Scholar]
- 6.Chase-Topping, M., D. Gally, C. Low, L. Matthews, and M. Woolhouse. 2008. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 12:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Sablet, T., Y. Bertin, M. Vareille, J. P. Girardeau, A. Garrivier, A. P. Gobert, and C. Martin. 2008. Differential expression of stx2 variants in Shiga toxin-producing Escherichia coli belonging to seropathotypes A and C. Microbiology 154(Pt. 1):176-186. [DOI] [PubMed] [Google Scholar]
- 9.Dumke, R., U. Schroter-Bobsin, E. Jacobs, and I. Roske. 2006. Detection of phages carrying the Shiga toxin 1 and 2 genes in waste water and river water samples. Lett. Appl. Microbiol. 42:48-53. [DOI] [PubMed] [Google Scholar]
- 10.Erickson, M. C., and M. P. Doyle. 2007. Food as a vehicle for transmission of Shiga toxin-producing Escherichia coli. J. Food Prot. 70:2426-2449. [DOI] [PubMed] [Google Scholar]
- 11.Fong, T. T., and E. K. Lipp. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 69:357-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Aljaro, C., M. Muniesa, J. Jofre, and A. R. Blanch. 2004. Prevalence of the stx2 gene in coliform populations from aquatic environments. Appl. Environ. Microbiol. 70:3535-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Aljaro, C., M. Muniesa, J. Jofre, and A. R. Blanch. 2006. Newly identified bacteriophages carrying the stx2g Shiga toxin gene isolated from Escherichia coli strains in polluted waters. FEMS Microbiol. Lett. 258:127-135. [DOI] [PubMed] [Google Scholar]
- 14.García-Aljaro, C., M. Muniesa, J. Jofre, and A. R. Blanch. 2009. Genotypic and phenotypic diversity among induced, stx2-carrying bacteriophages from environmental Escherichia coli strains. Appl. Environ. Microbiol. 75:329-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Generalitat de Catalunya, Departament de Salut. 2009. Brot de gastroenteritis per E. coli O157:H7 en diferents escoles de Catalunya. Bol. Epidemiol. Cataluña 5:90-95. [Google Scholar]
- 16.Grabow, W. O. K., T. E. Neubrech, C. S Holtzhausen, and J. Jofre. 1995. Bacteroides fragilis and Escherichia coli bacteriophages. Excretion by humans and animals. Water Sci. Technol. 31:223-230. [Google Scholar]
- 17.Gyles, C. L. 2007. Shiga toxin-producing E. coli: an overview. J. Anim. Sci. 85:45-62. [DOI] [PubMed] [Google Scholar]
- 18.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 19.Herold, S., H. Karch, and H. Schmidt. 2004. Shiga toxin-encoding bacteriophages—genomes in motion. Int. J. Med. Microbiol. 294:115-121. [DOI] [PubMed] [Google Scholar]
- 20.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huggett, J. F., T. Novak, J. A. Garson, C. Green, S. D. Morris-Jones, R. F. Miller, and A. Zumla. 2008. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res. Notes 1:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamovic, L., R. Serra-Moreno, J. Jofre, and M. Muniesa. 2010. Quantification of Shiga toxin 2-encoding bacteriophages by real-time PCR and correlation with phage infectivity. J. Appl. Microbiol. 108:1105-1114. [DOI] [PubMed] [Google Scholar]
- 23.Jofre, J. 2002. Bacteriophage as indicators, p. 354-363. In G. Bitton (ed.), Encyclopedia of environmental microbiology, vol. 1. John Wiley & Sons, New York, NY. [Google Scholar]
- 24.Koitabashi, T., S. Cui, M. Kamruzzaman, and M. Nishibuchi. 2008. Isolation and characterization of the Shiga toxin gene (stx)-bearing Escherichia coli O157 and non-O157 from retail meats in Shandong Province, China, and characterization of the O157-derived stx2 phages. J. Food Prot. 71:706-713. [DOI] [PubMed] [Google Scholar]
- 25.Mackay, I. M. 2004. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 10:190-212. [DOI] [PubMed] [Google Scholar]
- 26.Mellmann, A., S. Lu, H. Karch, J. G. Xu, D. Harmsen, M. A. Schmidt, and M. Bielaszewska. 2008. Recycling of Shiga toxin 2 genes in sorbitol fermenting enterohemorrhagic Escherichia coli O157:NM. Appl. Environ. Microbiol. 74:67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muniesa, M., J. E. Blanco, M. De Simón, R. Serra-Moreno, A. R. Blanch, and J. Jofre. 2004. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 150:2959-2971. [DOI] [PubMed] [Google Scholar]
- 28.Muniesa, M., and J. Jofre. 1998. Abundance in sewage of bacteriophages that infect Escherichia coli O157:H7 and that carry the Shiga toxin 2 gene. Appl. Environ. Microbiol. 64:2443-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muniesa, M., and J. Jofre. 2000. Occurrence of phages infecting Escherichia coli O157:H7 carrying the Shiga toxin 2 gene in sewage from different countries. FEMS Microbiol. Lett. 183:197-200. [DOI] [PubMed] [Google Scholar]
- 30.Muniesa, M., J. Jofre, C. García-Aljaro, and A. R. Blanch. 2006. Occurrence of Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli in the environment. Environ. Sci. Technol. 40:7141-7149. [DOI] [PubMed] [Google Scholar]
- 31.Muniesa, M., F. Lucena, and J. Jofre. 1999. Comparative survival of free Shiga toxin 2-encoding phages and Escherichia coli strains outside the gut. Appl. Environ. Microbiol. 65:5615-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muniesa, M., R. Serra-Moreno, and J. Jofre. 2004. Free Shiga toxin bacteriophages isolated from sewage showed diversity although the stx genes appeared conserved. Environ. Microbiol. 6:716-725. [DOI] [PubMed] [Google Scholar]
- 33.Muniesa, M., J. Recktenwald, M. Bielaszewska, H. Karch, and H. Schmidt. 2000. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect. Immun. 68:4850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from E. coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 36.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage Φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sekse, C., M. Muniesa, and Y. Wasteson. 2008. Conserved Stx2 phages from Escherichia coli O103:H25 isolated from patients suffering from hemolytic uremic syndrome. Foodborne Pathog. Dis. 5:801-810. [DOI] [PubMed] [Google Scholar]
- 43.Shanks, O. C., E. Atikovic, A. D. Blackwood, J. Lu, R. T Noble, J. S. Domingo, S. Seifring, M. Sivaganesan, and R. A. Haugland. 2008. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 74:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauch, E., J. A. Hammerl, A. Konietzny, S. Schneiker-Bekel, W. Arnold, A. Goesmann, A. Pühler, and L. Beutin. 2008. Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7. Infect. Immun. 76:5466-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanji, Y., K. Mizoguchi, M. Yoichi, M. Morita, N. Kijima, H. Kator, and H. Unno. 2003. Seasonal change and fate of coliphages infected to Escherichia coli O157:H7 in a waste water treatment plant. Water Res. 37:1136-1142. [DOI] [PubMed] [Google Scholar]
- 46.Tarr, P. I. 1995. Escherichia coli O157:H7: clinical, diagnostic and epidemiological aspects of human infection. Clin. Infect. Dis. 20:1-8. [DOI] [PubMed] [Google Scholar]
- 47.Teel, L. D., A. R. Melton-Celsa, C. K. Schmitt, and A. D. O'Brien. 2002. One of two copies of the gene for the activatable Shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect. Immun. 70:4282-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorpe, C. M., J. M. Ritchie, and D. W. K. Acheson. 2002. Enterohemorrhagic and other Shiga toxin-producing E. coli, p. 119-141. In M. Donnenberg (ed.), E. coli: virulence mechanisms of a versatile pathogen. Academic Press, New York, NY.
- 49.Toth, I., H. Schmidt, M. Dow, A. Malik, E. Oswald, and B. Nagy. 2003. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a Shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 69:7242-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, P. L., D. W Acheson, and M. K. Waldor. 1999. Isogenic lysogens of diverse Shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng, J., S. Cui, L. D. Teel, S. Zhao, R. Singh, A. D. O'Brien, and J. Meng. 2008. Identification and characterization of Shiga toxin type 2 variants in Escherichia coli isolates from animals, food, and humans. Appl. Environ. Microbiol. 74:5645-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]