Abstract
Here, we report an efficient method for extracting high-quality mRNA from soil. Key steps in the isolation of total RNA were low-pH extraction (pH 5.0) and Q-Sepharose chromatography. The removal efficiency of humic acids was 94 to 98% for all soils tested. To enrich mRNA, subtractive hybridization of rRNA was most efficient. Subtractive hybridization may be followed by exonuclease treatment if the focus is on the analysis of unprocessed mRNA. The total extraction method can be completed within 8 h, resulting in enriched mRNA ranging from 200 bp to 4 kb in size.
Over the last decade, several methods have been reported for the extraction of environmental RNA (e.g., 11, 13, 32). These methods have been widely used to study the compositions and dynamics of microbial communities at the rRNA level (e.g., 19, 33). However, rRNA surveys provide no or only indirect information on the functional status of a microbial community. Therefore, monitoring the environmental expression of key genes of particular metabolic pathways, such as the genes for nitrogen fixation (nifH), nitrite reduction (nirS, nirK), ammonia oxidation (amoA), or methane oxidation (pmoA), received increasing attention (3, 5, 8, 16, 17, 25). Most recently, new sequencing platforms, such as 454 pyrosequencing, allowed the metatranscriptome analysis of complex microbial communities (9, 10, 23, 31). However, it remains challenging to extract mRNA of high quality from soil for use in metatranscriptomics, due to the coextraction of humic acids and other organic compounds. These contaminants inhibit downstream analyses such as RNA amplification and reverse transcription-PCR, thus calling for the need of their quantitative removal. The low content of mRNA in total RNA extracts (1 to 5%) and their greater susceptibility to degradation by RNases than rRNA also hamper the efficient extraction of intact mRNA from soil (1, 2).
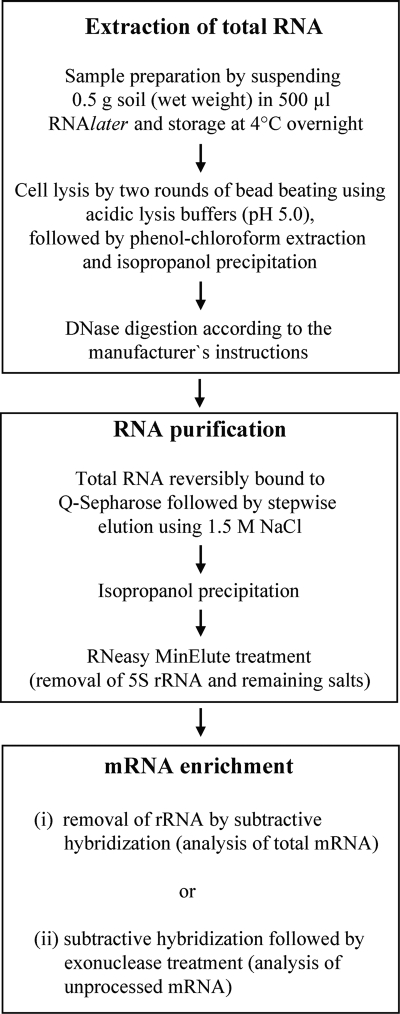
Here, we report an efficient method for the extraction of total RNA and enrichment of mRNA from soils differing in their amounts and compositions of humic acids, including (i) rice paddy, (ii) grassland, (iii) agricultural, and (iv) forest soils. Assessments were made with regard to the (i) quantitative removal of humic acids, (ii) yield and integrity of total RNA, and (iii) size distribution of enriched mRNA. The optimized method allows the extraction of total RNA and enrichment of mRNA from multiple samples within 8 h. The procedural steps are described in their order of application (Fig. 1). Additional information on the procedural steps is given in the supplemental material.
FIG. 1.
Flow chart showing the procedural steps in the extraction of total RNA and enriched mRNA from soil.
Extraction of total RNA.
Samples from all four different soil types were processed in the same way. Fresh soil (0.5 g, wet weight) was suspended in 500 μl of RNAlater (Ambion, Germany) and stored at 4°C overnight. Soil samples were pelleted at 20,000 × g for 1 min, and the supernatants were discarded. No rRNA or other nucleic acids could be detected in the supernatants, indicating that the RNAlater treatment and subsequent centrifugation do not lead to the loss of RNA.
The pellets were mixed with equal volumes of glass beads (0.17 to 0.18 mm in diameter) and resuspended in 700 μl of precooled TPM buffer (50 mM Tris-HCl [pH 5.0], 1.7% [wt/vol] polyvinylpyrrolidone, 20 mM MgCl2). Subsequently, the mixture was shaken in a bead beater (FastPrep FP120; Qbiogene) at 6.0 ms−1 for 35 s. Harsh lysis was chosen to ensure a fast and complete disruption of all microorganisms (6, 27). Soil and cell debris were pelleted by centrifugation at 20,000 × g for 1 min at 4°C, and the supernatant was transferred to a fresh tube. The pellet was suspended in 700 μl of PBL buffer (5 mM Tris-HCl [pH 5.0], 5 mM Na2EDTA, 0.1% [wt/vol] sodium dodecyl sulfate, and 6% [vol/vol] water-saturated phenol), and the lysis procedure was repeated as described above. The supernatants of the two lysis treatments were pooled.
The pooled supernatant was extracted first with water-saturated phenol (pH 4.5), second with phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol], pH 4.5), and third with chloroform-isoamyl alcohol (24:1 [vol/vol], pH 5.5), each time using 500 μl of extractant. The resulting aqueous phase was mixed with 0.1 volume of 3 M sodium acetate (pH 5.7) and 0.7 volume of isopropanol, incubated at room temperature for 5 min, and centrifuged for 30 min at 20,000 × g and 4°C. The nucleic acid pellet was washed with 70% ethanol, air dried, and resuspended in 50 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). Subsequently, the sample was adjusted to 1× DNase buffer, and 5 U DNase and 10 U RNasin (Promega, Germany) were added, followed by incubation for 60 min at 37°C and denaturation of the DNase at 65°C for 10 min. The final volume of the DNase-treated RNA sample was 60 μl.
Humic acids consist of various components whose capabilities to dissolve in aqueous solutions and organic solvents vary with pH (15, 34). Therefore, we assessed the correspondence between pH and coextraction of humic acids. The pH of the lysis buffer was stepwise changed from neutral pH, as recommended in previous reports (e.g., references 13 and 32), to pH 4.5, in steps of 0.5 pH. The sequential use of low-pH lysis buffers (pH 5.0) and organic solvents (pH 4.5) was found to be most effective in minimizing the coextraction of humic acids. Another benefit of low-pH extraction is the increased stability of RNA (21).
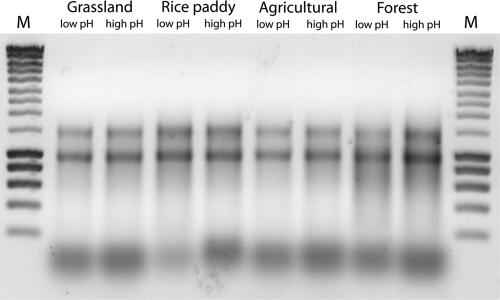
Contrary to a previous report (32), no decline in the RNA recovery was observed when low-pH extraction conditions were used, as deduced from agarose gel electrophoresis (Fig. 2). Low-pH extraction involved the use of lysis buffer (pH 5.0), water-saturated phenol (pH 4.5), and phenol-chloroform-isoamyl alcohol (pH 4.5), while high-pH extraction was defined by the use of lysis buffer (pH 7.0), water-saturated phenol (pH 8.0), and phenol-chloroform-isoamyl alcohol (pH 8.0). The removal of DNA never failed, suggesting that the DNase activity is not strongly inhibited by humic acids (Fig. 2).
FIG. 2.
Raw extracts of DNase-treated RNA. The extracts were obtained from grassland, rice paddy, agricultural, and forest soils using either low- or high-pH extraction conditions. Equivalent amounts of the raw extracts were loaded onto the gel. SmartLadder (Eurogentec) was used as the molecular size standard (lanes M).
The quantification of total RNA in the raw extracts was not possible using the RiboGreen RNA quantitation kit (Invitrogen, Germany), presumably due to dual effects: (i) RiboGreen also binds to humic acids (22) and (ii) humic acids absorb at the excitation and emission wavelengths of the RNA-dye complex. Similarly, NanoDrop ND-1000 spectrophotometry (NanoDrop Technologies, United States) could not be used to quantify the RNA content, due to the overlapping absorption spectra of humic acids and RNA.
Determination of humic acid content.
In order to establish a quantitative assay for monitoring the efficiency of humic acid removal, serial dilutions of commercially available humic acids (Carl Roth, Germany) and forest soil-derived humic acids were prepared in distilled water. The extraction of humic acids from forest soil was done by suspending soil in 0.1 M NaOH on a stirrer for 3 h (29). The suspension was centrifuged at 2,500 × g for 10 min. The supernatant was transferred to a new tube, acidified with 6 M HCl to a pH of 1.0, and incubated for 13 h at room temperature, finally resulting in a precipitation of humic acids. The precipitate was pelleted at 2,500 × g for 10 min and air dried overnight.
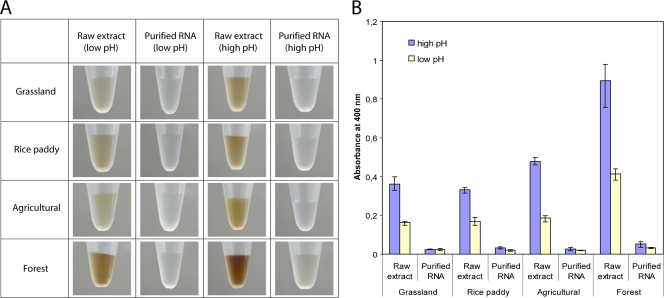
Using NanoDrop ND-1000 spectrophotometry, a linear relationship between absorbance at 400 nm (A400) and the concentration of humic acids was determined for both commercial and forest soil-derived humic acids (see Fig. S1 in the supplemental material), thus showing that a reliable quantitation of humic acids in RNA extracts is possible. The amounts of coextracted humic acids in raw extracts prepared under low-pH conditions were much lower than those in raw extracts prepared under high-pH conditions (Fig. 3).
FIG. 3.
Correspondence between the efficiency of humic acid removal and the pH of the extraction buffer, as assessed by eye (A) and NanoDrop ND-1000 spectrophotometry at 400 nm (B). Total RNA was extracted under either high-pH or low-pH conditions and was subsequently purified by Q-Sepharose column chromatography.
No additive effects between the absorption of humic acids and RNA were observed at A400, as assessed by spiking commercially available humic acids with E. coli RNA (see Fig. S2 in the supplemental material). Previous studies used a wavelength of 320 nm (A320) or a ratio of 465 to 665 nm (E4:E6) for quantitative measurement of humic acids (18, 20, 32). A320 measurements result in higher absorbance values for given amounts of humic acids than those at A400. However, the A320 measurements are close to the curves' intersection point, where the absorbance spectra of RNA and humic acids start to overlap strongly. The sensitivity of A400 measurements was sufficient to quantitatively monitor the humic acid removal in the raw and purified extracts, thereby avoiding incorrect measurements due to overlapping absorbances.
RNA purification.
Various methods were tested for their efficiency in purifying total RNA, including Q-Sepharose chromatography (24), hydrophobic interaction with polyvinylpolypyrrolidone (12), Sephadex G-50 column chromatography (30), and pretreatment with aluminum sulfate (7). Raw extracts from forest soil, due to their high humic acid content, were used for comparative assessment of the purification methods (Fig. 3). Although never used in previous studies for the purification of RNA, Q-Sepharose chromatography was found to be the most efficient method to remove humic acids. However, the Q-Sepharose treatment that we initially used had been developed to purify environmental DNA (24), leading to a small recovery of total RNA. Therefore, the Q-Sepharose treatment was successively modified in order to improve RNA yield. The quantitative removal of humic acids and the purity of total RNA were assessed spectrophotometrically (Table 1).
TABLE 1.
Efficiency of RNA purification measured by NanoDrop ND-1000 spectrophotometry
| Purification methoda | % reduction in humic acidsb |
A400c |
A260/A230d | A260/A280e | Reference | |
|---|---|---|---|---|---|---|
| Before | After | |||||
| Q-Sepharose | 96.0 ± 2.1 | 0.721 | 0.029 | 1.45 | 1.79 | This work |
| PVPP | 79.1 ± 5.3 | 0.721 | 0.152 | 1.09 | 1.45 | 12 |
| Sephadex G-50 | 69.2 ± 5.6 | 0.721 | 0.176 | 1.01 | 1.31 | 30 |
| Al2(SO4)3f | 0.225 | 0.77 | 1.06 | 7 | ||
The same raw extract of nucleic acids was used for assessing the different purification methods except for the treatment with Al2(SO4)3. The purification treatments were carried out according to the referenced literature.
Percentage reduction in humic acid content as calculated from the absorbance measurements at 400 nm (A400) before and after RNA purification.
A400 measurements of the raw versus purified RNA extracts.
Absorbance ratio between RNA (260 nm) and both humic acids and salts (230 nm).
Absorbance ratio between RNA (260 nm) and both humic acids and proteins (280 nm).
Aliquots of the same soil sample were used for the Al2(SO4)3 treatment and the other three purification methods. However, unlike with the other three methods, the Al2(SO4)3 treatment was performed prior to cell lysis and recovery of the raw extract. Therefore, a calculation of the percent reduction in humic acids by the Al2(SO4)3 pretreatment was not possible.
Q-Sepharose Fast Flow (Sigma Aldrich, Germany) was delivered in 20% ethanol. A 600-μl aliquot was washed three times with 3 ml of diethyl pyrocarbonate (DEPC) water, involving mixing, centrifuging at 650 × g for 10 s, and discarding of the supernatant after each washing step. The washed Q-Sepharose was finally suspended in 300 μl of DEPC water, poured into an empty Illustra Autoseq G-50 column (GE Healthcare, Germany), and packed by centrifugation at 650 × g for 10 s. The DNase-treated RNA sample (60 μl) was then loaded onto the resin and centrifuged at 400 × g for 7 s, immediately followed by the RNA elution steps (compare with Fig. 1). The RNA was eluted by loading 80 μl of 1.5 M NaCl in DEPC water (pH 5.5) onto the resin and spinning at 400 × g for 7 s. The 1.5 M NaCl elution step was repeated five times. The eluate was collected in a single 2-ml tube and subjected to isopropanol precipitation as described above. The precipitate was dissolved in 100 μl RNase-free TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5; U.S. Biochemicals [USB], Cleveland, OH) and subjected to an RNeasy MinElute cleanup treatment to remove 5S rRNA and remaining salts. The treatment was carried out according to the manufacturer's instructions (Qiagen, Germany), finally resulting in highly purified total RNA dissolved in 20 μl of RNase-free TE buffer (USB).
Several eluents were tested for optimal recovery of total RNA by Q-Sepharose chromatography, including (i) DEPC water, (ii) Tris-HCl, and (iii) sodium and potassium phosphate buffers with different counter-ion concentrations (1.0 M, 1.3 M, 1.5 M, 1.8 M, 2.0 M) and pH values (pHs 5.0 to 8.0 in pH 0.5 steps). The optimal elution conditions were found to be 1.5 M NaCl and pH 5.5. Under these conditions in Q-Sepharose chromatography, the humic acid content in the raw extracts of all four different soil types was reduced by an average of 96% (Fig. 3B). This level of purification was more than sufficient for downstream applications. We assume that the residual humic acids were less negatively charged than the bulk of humic acids, thereby resulting in a weak binding to the positively charged resin and, as a consequence, coelution at 1.5 M NaCl. The purity of the RNA extracts could be improved by lowering the NaCl concentration, but concentrations below 1.5 M NaCl reduced RNA recovery (data not shown). The optimized Q-Sepharose purification procedure increased the yield from initially 0.5 ± 0.1 μg to 1.6 ± 0.4 μg of total RNA per 0.5 g of soil (wet weight), regardless of the soil type tested (three replicates each).
mRNA enrichment.
We tested two commercially available kits for rRNA removal. The MICROBExpress bacterial mRNA enrichment kit (Ambion) is based on rRNA capture oligonucleotides hybridized with probes linked to magnetic beads, while the mRNA-ONLY prokaryotic mRNA isolation kit (Epicentre Biotechnologies, United States) uses a 5′-monophosphate-dependent exonuclease to degrade the rRNA. Both methods were previously used to enrich mRNA for global transcriptome analysis (10, 23, 26). In order to compare the efficiencies of the two methods, total RNA was divided into two aliquots, which were processed by either subtractive hybridization or exonuclease treatment. Enriched mRNA was purified and concentrated using the RNeasy MinElute kit (Qiagen). All kits, including MICROBExpress, mRNA-ONLY, and RNeasy MinElute, were used according to the manufacturers' instructions. The quantities of total RNA and enriched mRNA as well as the quantitative removal of rRNA and the size distribution of enriched mRNA were checked by RNA picochip electrophoresis using an Agilent 2100 bioanalyzer (Agilent Technology, England). The picochip analysis was carried out according to the manufacturer's protocol.
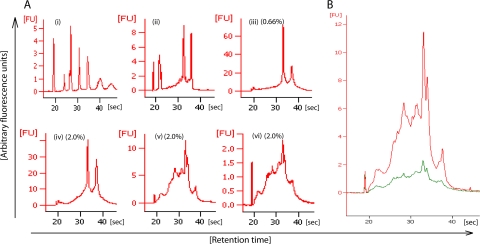
Subtractive hybridization was more efficient in depleting the rRNA content than the exonuclease treatment (Fig. 4A). The rRNA removal efficiency of subtractive hybridization was 94 to 97%, as determined by comparing the fluorescence intensities of the 16S and 23S rRNA signals after picochip electrophoresis of enriched mRNA and total RNA. These rRNA removal efficiencies matched the manufacturer's specifications. As previously reported (10, 23, 26), this approach fails to completely remove rRNA due to the insufficient compatibility of the capture probes. These probes capture rRNA from most members of the domain Bacteria, except rRNA of species such as Chloroflexus aurantiacus and Propionibacterium freudenreichii (a complete list is available on Ambion's website). In addition, archaeal and eukaryotic rRNAs are not captured.
FIG. 4.
Electropherograms of total RNA and enriched mRNA extracted from forest soil. The mRNA was enriched by exonuclease treatment, subtractive hybridization, or a combination of these two methods. RNA 6000 picochip electrophoresis was carried out on an Agilent 2100 bioanalyzer in the mRNA Pico series II mode. (A, i) Ladder (sizes of fragments are 25, 200, 500, 1,000, 2,000, 4,000, and 6,000 nucleotides); (ii) E. coli RNA standard (2 ng/μl); (iii to vi) environmental extracts of total RNA (iii), RNA treated with exonuclease (iv), RNA treated by subtractive hybridization (v), and RNA treated by subtractive hybridization and then with exonuclease (vi). The amounts (percentages) of total extract loaded onto the chip are given in parentheses for total RNA (iii) and enriched mRNA (iv to vi). (B) Overlay of mRNA samples enriched by either subtractive hybridization (red) or the combined treatment of subtractive hybridization and exonuclease digestion (green). The x axis shows the retention times of size-separated fragments in seconds, and the y axis shows the signal intensities in arbitrary fluorescence units (FU). Note that the fluorescence signal at 19 s represents a methods-inherent marker.
The efficiency of the exonuclease treatment in removing rRNA was in the range of 80 to 86% for the four soil types tested, while a depletion efficiency of 94% was observed for pure-culture rRNA of E. coli (data not shown). The reduced efficiency of the exonuclease treatment when applied to the environmental extracts may be due to the slight amount of residual humic acid impurities (Fig. 3). Therefore, we performed a second round of purification using size exclusion chromatography (G-50 columns) according to the manufacturer's instructions. However, the additional purification treatment had no significant effect on the residual humic acid content or the rRNA removal efficiency of the subsequent exonuclease treatment.
In order to achieve the most quantitative depletion of rRNA, we combined subtractive hybridization with exonuclease digestion. However, the additional exonuclease treatment had no further enrichment effect on the mRNA, and the RNA yield decreased by 80% compared to that obtained when only subtractive hybridization was used (Fig. 4B). The activity of the exonuclease accounted for approximately half of the RNA loss, while the loss of the other half was due to the need of an additional round of RNeasy MinElute purification, as concluded from control experiments with E. coli RNA extracts (data not shown).
At first glance, it was unexpected that the exonuclease activity did not further enrich mRNA relative to rRNA. Most likely, however, a considerable fraction of the mRNA transcripts were 5′-monophosphorylated (14) and therefore sensitive to exonuclease degradation. Using a newly developed technique, Celesnik and colleagues recently showed that E. coli cells in the stationary phase contain a major fraction of 5′-monophosphorylated mRNAs. This fraction accounted for 30% of mRNA transcripts, representing a particular state in the mRNA decay (4). Therefore, it is reasonable to assume that a considerable portion of the soil metatranscriptome is 5′-monophosphorylated rather than triphosphorylated. The latter is a characteristic of newly transcribed, unprocessed mRNAs.
There were no differences in the yields of enriched mRNA between grassland, rice paddy, agricultural, and forest soils. The yields of the enriched mRNA fractions were 220 ± 60 ng (subtractive hybridization), 420 ± 110 ng (exonuclease), and 50 ± 20 ng (sequential approach of subtractive hybridization and exonuclease) per 0.5 g of wet soil. The size distribution of the enriched mRNA was between 200 bp and 4 kb, with the bulk of mRNA ranging in size from 500 bp to 3 kb (Fig. 4; see also Fig. S3 in the supplemental material). The enriched mRNA was used for RNA amplification and cDNA production. Using random hexamers (Fermentans, Germany), cDNA was amplified by PCR for TA cloning (see Fig. S4 in the supplemental material). Slightly more than half of the cDNA clones were derived from mRNA, while the source of the other cDNA clones was rRNA. This agrees well with our previous finding that the use of MICROBExpress depleted the rRNA content of the extracts by 94 to 97%. The residual rRNA in the extracts approximately equals the amount of mRNA, considering that 1 to 5% of total RNA in bacterial cells is mRNA (1, 2). Notably, nearly all rRNA-cDNA clones were derived from bacterial 23S rRNA.
Concluding remarks.
As shown in Fig. 1, we developed a simple and efficient procedure for extracting high-quality mRNA from soil. Transcript sizes of up to 4 kb allow for both massive parallel pyrosequencing using the 454 Titanium version and conventional Sanger sequencing if full-length cDNA analysis is intended.
The procedure used to enrich mRNA depends on the objectives of the study. Subtractive hybridization appears to be most suitable for obtaining a high yield of enriched mRNA. Contrary to the exonuclease treatment, subtractive hybridization preserves full mRNA diversity, allowing the analysis of processed and unprocessed mRNA. If the extracts contain considerable amounts of archaeal and/or eukaryotic rRNA, the use of specifically designed capture probes may be recommended (28).
Applied to environmental RNA, the exonuclease treatment is less efficient in the removal of rRNA than subtractive hybridization, presumably due to the slight amounts of residual humic acids in the total RNA. Therefore, the use of only exonuclease for enrichment of soil mRNA is not recommended. Moreover, the exonuclease treatment presumably results in a considerable removal of processed mRNAs that are 5′-monophosphorylated. Thus, the research may be directed toward the analysis of unprocessed mRNAs that are triphosphorylated. If the analysis of unprocessed mRNA is the specific aim, the sequential approach of subtractive hybridization and exonuclease treatment may be the method of choice.
Supplementary Material
Acknowledgments
Yongkyu Kim received a Ph.D. scholarship from the International Max Planck Research School for Environmental, Cellular and Molecular Microbiology. The study was supported by the European Science Foundation (METHECO, EuroDiversity 018) and the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 9 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Albert, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. D. Watson. 1994. Molecular biology of the cell, 3rd ed. Garland Publishing Inc., New York, NY.
- 2.Andersson, C. R., A. Isaksson, and M. G. Gustafsson. 2006. Bayesian detection of periodic mRNA time profiles without use of training examples. BMC Bioinformatics 7:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bürgmann, H., F. Widmer, W. V. Sigler, and J. Zeyer. 2003. mRNA extraction and reverse transcription-PCR protocol for detection of nifH gene expression by Azotobacter vinelandii in soil. Appl. Environ. Microbiol. 69:1928-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celesnik, H., A. Deana, and J. G. Belasco. 2007. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell 27:79-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., M. G. Dumont, A. Cébron, and J. C. Murrell. 2007. Identification of active methanotrophs in a landfill cover soil through detection of expression of 16S rRNA and functional genes. Environ. Microbiol. 9:2855-2869. [DOI] [PubMed] [Google Scholar]
- 6.Cullen, D. W., and P. R. Hirsch. 1998. Simple and rapid method for direct extraction of microbial DNA from soil for PCR. Soil Biol. Biochem. 30:983-993. [Google Scholar]
- 7.Dong, D., A. Yan, H. Liu, X. Zhang, and Y. Xu. 2006. Removal of humic substances from soil DNA using aluminium sulfate. J. Microbiol. Methods 66:217-222. [DOI] [PubMed] [Google Scholar]
- 8.Ebie, Y., N. Noda, H. Miura, M. Matsumura, S. Tsuneda, A. Hirata, and Y. Inamori. 2004. Comparative analysis of genetic diversity and expression of amoA in wastewater treatment processes. Appl. Microbiol. Biotechnol. 64:740-744. [DOI] [PubMed] [Google Scholar]
- 9.Frias-Lopez, J., Y. Shi, S. Yanmei, G. W. Tyson, M. L. Coleman, and S. C. Schuster. 2008. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. U. S. A. 105:3805-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert, J. A., D. Field, Y. Huang, R. Edwards, W. Li, P. Gilna, and I. Joint. 2008. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3:e3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2001. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA and rRNA-based microbial community composition. Appl. Microbiol. Biotechnol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henckel, T., U. Jäckel, S. Schnell, and R. Conrad. 2000. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 66:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurt, R. A., X. Qiu, L. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, X., and J. G. Belasco. 2004. Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc. Natl. Acad. Sci. U. S. A. 101:9211-9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kipton, H., J. Powell, and R. M. Town. 1992. Solubility and fractionation of humic acid; effect of pH and ionic medium. Anal. Chim. Acta 267:47-54. [Google Scholar]
- 16.Knauth, S., T. Hurek, D. Brar, and B. Reinhold-Hurek. 2005. Influence of different Oryza cultivars on expression of nifH gene pools in roots of rice. Environ. Microbiol. 7:1725-1733. [DOI] [PubMed] [Google Scholar]
- 17.Kolb, S., C. Knief, P. F. Dunfield, and R. Conrad. 2005. Abundance and activity of uncultured methanotrophic bacteria involved in consumption of atmospheric methane in two forest soils. Environ. Microbiol. 7:1150-1162. [DOI] [PubMed] [Google Scholar]
- 18.Kononova, M. M. 1966. Soil organic matter, 2nd ed., p. 400-404. Pergamon Press, Oxford, United Kingdom.
- 19.Lu, Y., D. Rosencrantz, W. Liesack, and R. Conrad. 2006. Structure and activity of bacterial community inhabiting rice roots and the rhizosphere. Environ. Microbiol. 8:1351-1360. [DOI] [PubMed] [Google Scholar]
- 20.Miller, D. N. 2001. Evaluation of gel filtration resins for the removal of PCR-inhibitory substances from soils and sediments. J. Microbiol. Methods 44:49-58. [DOI] [PubMed] [Google Scholar]
- 21.Noonberg, S. B., G. K. Scott, and C. C. Benz. 1995. Effect of pH on RNA degradation during guanidinium extraction. Biotechniques 19:731-733. [PubMed] [Google Scholar]
- 22.Popp, N. 2006. Ph.D. thesis. Technische Universität Bergakademie, Freiberg, Germany.
- 23.Poretsky, R. S., I. Hewson, S. Sun, A. E. Allen, J. P. Zehr, and M. A. Moran. 2009. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ. Microbiol. 11:1358-1375. [DOI] [PubMed] [Google Scholar]
- 24.Sharma, P. K., N. Capalash, and J. Kaur. 2007. An improved method for single step purification of metagenomic DNA. Mol. Biotechnol. 36:61-63. [DOI] [PubMed] [Google Scholar]
- 25.Sharma, S., M. K. Aneja, J. Mayer, J. C. Munch, and M. Schloter. 2005. Diversity of transcripts of nitrite reductase genes (nirK and nirS) in rhizospheres of grain legumes. Appl. Environ. Microbiol. 71:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shrestha, P. M., M. Kube, R. Reinhardt, and W. Liesack. 2009. Transcriptional activity of paddy soil bacterial communities. Environ. Microbiol. 11:960-970. [DOI] [PubMed] [Google Scholar]
- 27.Smalla, K., N. Cresswell, L. C. Mendoca-Hagler, A. Wolters, and J. D. van Elsas. 1993. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J. Appl. Bacteriol. 74:78-85. [Google Scholar]
- 28.Stewart, F. J., E. A. Ottesen, and E. F. DeLong. 2010. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 4:896-907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- 29.Swift, R. S. 1996. Organic matter characterization, p. 1018-1020. In D. L. Sparks, A. L. Page, P. A. Helmke, R. H. Loeppert, P. N. Soltanpour, M. A. Tabatabai, C. T. Johnston, and M. E. Sumner (ed.), Methods of soil analysis. Part 3. Chemical methods. Soil Science Society of America, Madison, WI.
- 30.Tsai, Y., and B. H. Olson. 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58:2292-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urich, T., A. Lanzén, J. Qi, D. H. Huson, C. Schleper, and S. C. Schuster. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Y., J. Shimodaira, T. Miyasaka, S. Morimoto, T. Oomori, N. Ogawa, M. Fukuda, and T. Fujii. 2008. Detection of bphAa gene expression of Rhodococcus sp. strain RHA1 in soil using a new method of RNA preparation from soil. Biosci. Biotechnol. Biochem. 72:694-701. [DOI] [PubMed] [Google Scholar]
- 33.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 34.Zuman, P., and E. B. Rupp. 2006. Humic acids—are they natural products? Croat. Chem. Acta 79:57-65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.