Abstract
Human hepatitis E virus (HEV) is considered an emerging pathogen in industrialized countries. In Italy, the true burden of HEV infection is unknown. Molecular HEV screening of raw sewage samples from 11 wastewater treatment plants yielded 19 positives (16%; 18 genotype I, 1 genotype III) evenly distributed throughout Italy. Evidence that HEV could be establishing itself in our region is accumulating and may justify more active surveillance to monitor its spread.
Hepatitis E is a self-limited, enterically transmitted acute viral hepatitis that occurs most frequently in epidemic outbreaks and often spreads by way of fecally contaminated drinking water (5, 20). Hepatitis E virus (HEV) infections are caused by a positive-sense, nonenveloped RNA virus of the Hepevirus genus. The four major genotypes (GI to GIV), all belonging to a single serotype, are known to infect humans. While GI and GII are restricted to humans, GIII and GIV are zoonotic and may infect animals (swine, chickens, deer, mongooses, and rabbits), as well as humans, in both industrialized and nonindustrialized countries (18, 19). GI consists of epidemic strains circulating in Africa and Asia. GII is found in Mexico and Africa. GIII is widely distributed, mainly—but not exclusively—in the United States, Europe, and Japan. GIV is present in Asia (16). An HEV strain belonging to a fifth genotype has been identified in birds (12).
HEV is transmitted by the fecal-oral route. Large waterborne outbreaks with high attack rates among young adults have been described in regions characterized by poor sanitary conditions (22). Hepatitis E is responsible for over 50% of cases of acute viral hepatitis in countries where the disease is endemic (Central and Southeast Asia, North and West Africa, and Mexico), where seroprevalence rates range from 15% to 60% (8). North America and Europe have traditionally been considered areas where HEV is not endemic, with acute infection diagnosed rarely and largely confined to travelers returning from areas where the disease is endemic. The high rates of HEV IgG positivity reported in different studies, however, suggest that unrecognized or subclinical infection is common (8). In Europe, increasing numbers of HEV infections not associated with travel have been recently reported (15).
HEV infection may vary in severity from asymptomatic to fulminant. Case fatality rates range between 0.5% and 4% overall but may reach 25% among pregnant women (1). In industrialized countries, the case fatality rate seems to be higher than in areas where the disease is endemic, since infection occurs more frequently in elderly people with chronic liver disease, a subgroup of patients with a case fatality rate approaching 70% (26).
HEV, which is shed in the feces of infected individuals, has been detected in sewage samples, suggesting that HEV contamination of aquatic environments may also be present (2, 6, 7, 23). In Italy, the true burden of HEV infection is still unknown and there are no available studies on the presence of this virus in sewage. The prevalence of anti-HEV antibodies among healthy individuals has been found to be approximately 1% in the northern regions and up to 5% in the southern regions, including Sicily and Sardinia. Higher prevalence rates have been found among drug users (especially HIV-infected individuals), hemodialysis patients, and patients with chronic hepatitis C, suggesting that HEV may be transmitted not only by the fecal-oral route (the main mode of transmission) but also parenterally (27).
The objective of the present study was to investigate the occurrence of HEV through the molecular screening of raw sewage samples collected from urban wastewater treatment plants (WTPs) in different regions of Italy.
MATERIALS AND METHODS
Samples (118 inflow grab samples) were collected on a monthly basis from April 2008 to March 2009 at 11 WTPs located in the following regions throughout Italy: Campania, Umbria, Tuscany, Piedmont, Friuli-Venezia Giulia, Basilicata, Lombardy, Emilia Romagna, Veneto, Latium, and Sardinia (this region was enrolled later in the project) (Table 1). Due to incomplete compliance and to the fact that one of the regions, Sardinia, was enrolled only in December 2008, 118 samples were collected rather than the expected 132.
TABLE 1.
Environmental samples used in this studya
| Sample | City | WTP | Collection date |
|---|---|---|---|
| 1179 | Trieste | Servola | 7 May 2008 |
| 1183 | Bologna | Corticella | 21 May 2008 |
| 1185 | Venice | Fusina | 21 May 2008 |
| 1191 | Brescia | Verziano | 10 June 2008 |
| 1249 | Rome | Roma Sud | 30 September 2008 |
| 1307 | Venice | Fusina | 15 December 2008 |
| 1309 | Perugia | Pian della Genna | 16 December 2008 |
| 1315 | Turin | Castiglione T.se | 17 December 2008 |
| 1317 | Turin | Castiglione T.se | 13 January 2009 |
| 1321 | Brescia | Verziano | 19 January 2009 |
| 1325 | Bologna | Corticella | 21 January 2009 |
| 1327 | Venice | Fusina | 19 January 2009 |
| 1329 | Perugia | Pian della Genna | 20 January 2009 |
| 1341 | Brescia | Verziano | 9 February 2009 |
| 1345 | Naples | Cuma | 12 February 2009 |
| 1349 | Rome | Roma Sud | 25 February 2009 |
| 1357 | Venice | Fusina | 11 March 2009 |
| 1359 | Brescia | Verziano | 11 March 2009 |
| 1363 | Potenza | Tiera di Vaglio | 11 March 2009 |
GenBank database accession no. FN796464 to FN796481.
RNA was extracted from 10 ml of sewage using the NucliSens miniMAG (bioMérieux Italia S.p.A., Rome, Italy) nucleic acid isolation kit. RNAs were then eluted in 100 μl elution buffer and stored in aliquots at −80°C until use, as previously described (13). A feline calicivirus (FCV; strain CVF9) was used as an internal control for some of the samples. A known amount of CVF9 (106 50% cell culture infective doses) was added to the samples prior to processing. Average recovery, used as a measure of extraction efficiency, was calculated as the mean ratio of genome copies (GCs) detected after and before concentration (GCs after/GCs before) by using a specific real-time PCR for CVF9 (PCR 652). Both the primers and the TaqMan probe were designed using Primer Express Software v3.0 (Applied Biosystems) (Table 2). The calibration curve used to calculate the number of FCV GCs was based on a linearized plasmid prepared from a recombinant pCR4-TOPO vector containing 630 bp of the polymerase region of strain CVF9 (PCR 650).
TABLE 2.
PCRs and primers used in this study
| PCR no. and primer | Sequence (5′ → 3′) | Product length (bp) | Primer position (5′ → 3′)b | Virus |
|---|---|---|---|---|
| 653 | ||||
| 1661 | TTAYGGKGATGCCTTTGATGACACC | 302 | 4329-4353 | HEV, GI |
| 1662 | TRATAACGGCCATRTTCCAGACAGTATTCC | 4630-4601 | ||
| 654 | ||||
| 1663 | TGTTTGAGAATGACTTTTCTGAGTTTGAYT | 175 | 4394-4423 | HEV, GI |
| 1664 | TTCCAAAACCCTCGCAGAGAC | 4568-4548 | ||
| 660 | ||||
| 1669 | GGYGACGCYTATGAGGAGT | 299 | 4362-4380 | HEV, GIII |
| 1670 | GCTATRATYGCCATRTTCCA | 4660-4641 | ||
| 661 | ||||
| 1671 | TGGTRTTTGARAATGAYTT | 163 | 4420-4438 | HEV, GIII |
| 1672 | AGAGACTCCTTCGGCGCCTG | 4582-4563 | ||
| 650 | ||||
| 1654 | CCCGTGGAGAAGGTTAGTGA | 630 | 4256-4272 | FCV |
| 1655 | CTTGTCAACCCGAGTTGGTT | 4885-4866 | ||
| 651 | ||||
| 1656 | CAAGGTATTTGCGGTCGATT | 340 | 4453-4472 | FCV |
| 1657 | ACCATCATCCCCGTAAGTCA | 4792-4773 | ||
| 652 | ||||
| 1658 | GGCCTATACAGGTTGGTATCAACA | 100 | 4380-4403 | FCV |
| 1659 | TTGGAATAATCGACCGCAAAT | 4479-4459 | ||
| 1660a | ATAGCCCCAGCGTCGAAGCGC | 4407-4427 |
Primer 5′ labeled with 6-carboxyfluorescein and 3′ labeled with 6-carboxytetramethylrhodamine.
The above PCR, using the reagents and enzymes employed for HEV detection, was also used on HEV-negative samples in order to exclude the presence of inhibitors.
Samples were analyzed by reverse transcriptase PCR (RT-PCR), followed by nested PCR, using both published and newly designed primers targeting portions of the RdRp gene (Table 2). Among the primers used to detect GI (RT-PCR 653, primers 1661 and 1662; nested PCR 654, primers 1663 and 1664), two (1662 and 1663) were originally described by McCaustland et al. (17). In the present study, however, several degeneracies, based on information drawn from public sequence databases, were introduced into the above primers to obtain a broader range of detection. Primers for GIII (RT-PCR 660, primers 1669 and 1670; nested PCR 661, primers 1671 and 1672) were designed by our group to target the same fragment as that targeted by the GI assay. This was done through multiple alignment of GenBank HEV GIII sequences.
Two microliters of the extracted RNA and 22 pmol of each primer were used in a final mixture of 25 μl using GoTaq Green Master Mix 2x, RNase inhibitor, and Moloney murine leukemia virus RT by Promega as previously described (14). GIII amplifications were carried out in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems) under the following conditions: reverse transcription at 42°C for 45 min, 1 cycle of template denaturation at 94°C for 5 min, and 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s, followed by 1 cycle of elongation for 10 min at 72°C. Following the reaction, 1 μl of this mixture was subjected to a second round of PCR involving 35 cycles of amplification under the same conditions. For GI, the annealing temperature was raised to 54°C. The second round is able to amplify a 175-bp fragment of HEV GI (PCR 654) and a 163-bp fragment of HEV GIII (PCR 661) (Table 2). Standard precautions were taken to prevent PCR contamination.
PCR amplicons were directly sequenced with a capillary automatic sequencer (ABI PRISM 310 Genetic Analyzer; Applied Biosystems). Some products were also cloned into the pCR4-TOPO vector (Invitrogen), following the manufacturer's protocol, and sequenced using vector-specific primers.
Bioinformatic analysis included all of the nucleotide sequences obtained here, as well as 27 prototype sequences obtained from GenBank (GI to GIV sequences and an avian HEV used as an outgroup). The analysis was performed as follows. The raw forward and reverse ABI files were aligned and assembled into a single consensus sequence using GAP4 of the Staden package. All sequences were submitted to BLAST analysis for genotyping at http://blast.ncbi.nlm.nih.gov/Blast.cgi. The nucleotide alignment was tested using Modeltest v3.1 (25), coupled with PAUP* v4.0 software to find the best-fit model of nucleotide substitution. The model selected (general time reversible) was used for the Bayesian analysis as implemented in the BEAST (Bayesian evolutionary analysis sampling trees) program available at http://beast.bio.ed.ac.uk.
Nucleotide sequence accession numbers.
All of the sequences determined in this study were deposited in the GenBank database under accession no. FN796464 to FN796481.
RESULTS
Extraction efficiency was good, with an average FCV recovery exceeding 50%. Of 118 sewage samples, 19 (16%) were positive for HEV RNA. PCR inhibitors were not detected in the negative samples.
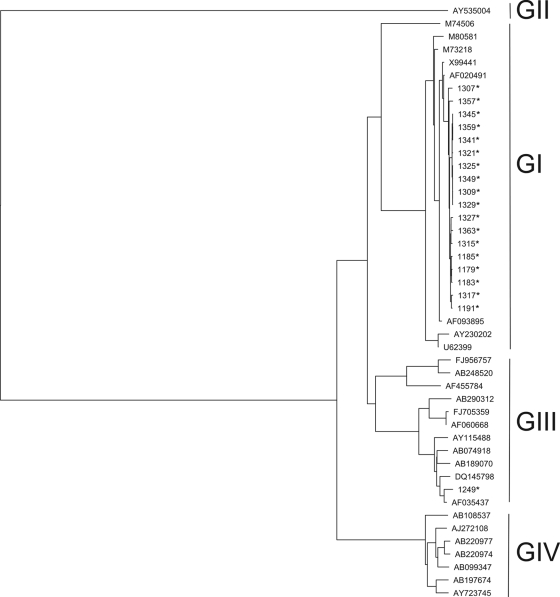
Figure 1 shows the genetic relationships between the HEV sequences obtained in this study and prototype sequences obtained from GenBank (multialigment of 127-bp fragments). The 19 HEV isolates formed two clusters, one comprising GI strains (18 isolates) and the other comprising a single GIII strain. The mean pairwise distance between the GI sequences detected in this study (18 taxa) was 0.7%. The HEV GIII strain showed sequence similarity to a strain isolated in the United States from a non-travel-related case (AF035437) and with strain CAM-3F13 (DQ145798), detected in swine feces in Cambodia in 2006.
FIG. 1.
Phylogenetic analysis of 19 HEV strains isolated in the present study and 27 prototype HEV strains obtained from GenBank. *, HEV-positive sewage sample.
The vast majority of positive samples were collected in winter or spring (no positive samples were found in July, August, October, or November; only one positive in September, peak of five positives in January).
Geographically, HEV sequences were detected in 9 out of the 11 regions analyzed, distributed throughout the country.
DISCUSSION
The purpose of the present study was to provide preliminary information regarding the occurrence of HEV in the population of Italy. This was done through molecular analysis of urban sewage samples. Similar studies have been conducted in other European countries (Spain and France), as well as in the United States. In these studies, HEV strains belonging to GIII and also, sporadically, to GI were found in urban sewage and biosolids (2, 6, 7, 23). While the detection of virions by molecular methods does not necessarily entail the presence of infectious particles, infectious HEV particles have been reported to occur in sewage (24), indicating the existence of a possible public health risk from the contamination of surface water with HEV.
Despite the scarcity of information on the occurrence of HEV in Italy, different authors have demonstrated a sustained circulation of the virus in this country. The virus is generally diagnosed in travelers returning from areas where the disease is endemic, but non-travel-related cases have also been documented, indicating that a reservoir of HEV may exist (3, 11, 30).
Our results revealed a high proportion of HEV-positive sewage samples in Italy (16%), evenly distributed across the entire country. GI was found in all but one of our positive samples. This high proportion of GI in Italy has yet to be explained and its public health implications studied, seeing that this genotype has been associated with significant epidemics in nonindustrialized countries and has thus far been identified in Western Europe relatively rarely. Increased migration, tourism, and international trade, particularly of food products, may all contribute to the spread of HEV into new regions. GIII, found in only one sample in our study, comprises both human and animal (mainly swine and wild boar) strains and includes strains responsible for sporadic cases of hepatitis in industrialized countries. The GIII strain detected in this work (sample 1249) showed high sequence identity (95%) with a strain isolated from a pig in 2006. In Italy, HEV strains have often been detected in swine populations (4, 9, 10). These data are in line with recent reports from Spain and Netherlands (28, 29) suggesting that HEV infection is widespread in European pig farms.
Different studies have demonstrated that human and swine HEV sequences, especially when obtained in the same geographical area, are highly homologous or even identical (16, 21), suggesting not only the possibility of zoonotic transmission but also that of food-borne transmission.
Results from this study indicate that HEV strains could be more widespread than previously thought. More studies are necessary to gather information on the occurrence and diversity of the strains circulating in humans and animals in order to gain a better understanding of the epidemiology of HEV and plan adequate preventive measures.
Acknowledgments
This study was funded by the project Rapid Diagnosis of Viruses in Sewage (CCM no. 4393, ISS prot. 8M09) of the Italian Center for Disease Control and Prevention (CCM—Centro Nazionale per la Prevenzione e il Controllo delle Malattie) and by the Italian Ministry of Labor, Health, and Social Policy.
We thank Emilia Luca and Marta Fratini for technical assistance and Simona Sermoneta for insightful linguistic and critical revision of the manuscript. We are also grateful to Vito Martella, University of Bari, Italy, for providing the calicivirus strain used as a control.
Footnotes
Published ahead of print on 2 July 2010.
REFERENCES
- 1.Aggarwal, R., and S. Naik. 2009. Epidemiology of hepatitis E: current status. J. Gastroenterol. Hepatol. 24:1484-1493. [DOI] [PubMed] [Google Scholar]
- 2.Bofill-Mas, S., N. Albinana-Gimenez, P. Clemente-Casares, A. Hundesa, J. Rodriguez-Manzano, A. Allard, M. Calvo, and R. Girones. 2006. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 72:7894-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cacopardo, B., R. Russo, W. Preiser, F. Benanti, G. Brancati, and A. Nunnari. 1997. Acute hepatitis E in Catania (eastern Sicily) 1980-1994. The role of hepatitis E virus. Infection 25:313-316. [DOI] [PubMed] [Google Scholar]
- 4.Caprioli, A., F. Martelli, F. Ostanello, I. Di Bartolo, F. M. Ruggeri, L. Del Chiaro, and F. Tolari. 2007. Detection of hepatitis E virus in Italian pig herds. Vet. Rec. 161:422-423. [DOI] [PubMed] [Google Scholar]
- 5.Chandra, V., S. Taneja, M. Kalia, and S. Jameel. 2008. Molecular biology and pathogenesis of hepatitis E virus. J. Biosci. 33:451-464. [DOI] [PubMed] [Google Scholar]
- 6.Clemente-Casares, P., S. Pina, M. Buti, R. Jardi, M. Martin, S. Bofill-Mas, and R. Girones. 2003. Hepatitis E virus epidemiology in industrialized countries. Emerg. Infect. Dis. 9:448-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemente-Casares, P., J. Rodriguez-Manzano, and R. Girones. 2009. Hepatitis E virus genotype 3 and sporadically also genotype 1 circulate in the population of Catalonia, Spain. J. Water Health 7:664-673. [DOI] [PubMed] [Google Scholar]
- 8.Dalton, H. R., R. Bendall, S. Ijaz, and M. Banks. 2008. Hepatitis E: an emerging infection in developed countries. Lancet Infect. Dis. 8:698-709. [DOI] [PubMed] [Google Scholar]
- 9.Di Bartolo, I., F. Martelli, N. Inglese, M. Pourshaban, A. Caprioli, F. Ostanello, and F. M. Ruggeri. 2008. Widespread diffusion of genotype 3 hepatitis E virus among farming swine in Northern Italy. Vet. Microbiol. 132:47-55. [DOI] [PubMed] [Google Scholar]
- 10.Di Martino, B., F. Di Profio, V. Martella, E. Di Felice, C. E. Di Francesco, C. Ceci, and F. Marsilio. 2010. Detection of hepatitis E virus in slaughtered pigs in Italy. Arch. Virol. 155:103-106. [DOI] [PubMed] [Google Scholar]
- 11.Grieco, A., L. Miele, G. Gasbarrini, and R. Grillo. 2001. Sporadic HEV hepatitis in Italy. Gut 48:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, F. F., G. Haqshenas, H. L. Shivaprasad, D. K. Guenette, P. R. Woolcock, C. T. Larsen, F. W. Pierson, F. Elvinger, T. E. Toth, and X. J. Meng. 2002. Heterogeneity and seroprevalence of a newly identified avian hepatitis E virus from chickens in the United States. J. Clin. Microbiol. 40:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Rosa, G., M. Iaconelli, M. Pourshaban, M. Fratini, and M. Muscillo. 2010. Molecular detection and genetic diversity of norovirus genogroup IV—a year-long monitoring of sewage throughout Italy. Arch. Virol. 155:589-593. [DOI] [PubMed] [Google Scholar]
- 14.La Rosa, G., M. Iaconelli, M. Pourshaban, and M. Muscillo. 2010. Detection and molecular characterization of noroviruses from five sewage treatment plants in central Italy. Water Res. 44:1777-1784. [DOI] [PubMed] [Google Scholar]
- 15.Lewis, H. C., O. Wichmann, and E. Duizer. 2010. Transmission routes and risk factors for autochthonous hepatitis E virus infection in Europe: a systematic review. Epidemiol. Infect. 138:145-166. [DOI] [PubMed] [Google Scholar]
- 16.Lu, L., C. Li, and C. H. Hagedorn. 2006. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 16:5-36. [DOI] [PubMed] [Google Scholar]
- 17.McCaustland, K. A., S. Bi, M. A. Purdy, and D. W. Bradley. 1991. Application of two RNA extraction methods prior to amplification of hepatitis E virus nucleic acid by the polymerase chain reaction. J. Virol. Methods 35:331-342. [DOI] [PubMed] [Google Scholar]
- 18.Meng, X. J. 2010. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 140:256-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng, X. J. 2010. Recent advances in hepatitis E virus. J. Viral Hepat. 17:153-161. [DOI] [PubMed] [Google Scholar]
- 20.Mushahwar, I. K. 2008. Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J. Med. Virol. 80:646-658. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto, H. 2007. Genetic variability and evolution of hepatitis E virus. Virus Res. 127:216-228. [DOI] [PubMed] [Google Scholar]
- 22.Panda, S. K., D. Thakral, and S. Rehman. 2007. Hepatitis E virus. Rev. Med. Virol. 17:151-180. [DOI] [PubMed] [Google Scholar]
- 23.Pina, S., M. Buti, M. Cotrina, J. Piella, and R. Girones. 2000. HEV identified in serum from humans with acute hepatitis and in sewage of animal origin in Spain. J. Hepatol. 33:826-833. [DOI] [PubMed] [Google Scholar]
- 24.Pina, S., J. Jofre, S. U. Emerson, R. H. Purcell, and R. Girones. 1998. Characterization of a strain of infectious hepatitis E virus isolated from sewage in an area where hepatitis E is not endemic. Appl. Environ. Microbiol. 64:4485-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posada, D. 2003. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr. Protoc. Bioinformatics Chapter 6:Unit 6.5. [DOI] [PubMed]
- 26.Renou, C., E. Nicand, A. Pariente, J. F. Cadranel, and N. Pavio. 2009. How to detect and diagnose an autochthonous hepatitis E? Gastroenterol. Clin. Biol. 33:F27-F35. [DOI] [PubMed] [Google Scholar]
- 27.Romanò, L., S. Paladini, and A. R. Zanetti. 2009. Epatite E in Italia: solo patologia da importazione? IX Workshop SEIEVA Sistema Epidemiologico Integrato dell'Epatite Virale Acuta Porretta Terme (BO), 10-12 December 2009.
- 28.Rutjes, S. A., W. J. Lodder, M. Bouwknegt, and A. M. de Roda Husman. 2007. Increased hepatitis E virus prevalence on Dutch pig farms from 33 to 55% by using appropriate internal quality controls for RT-PCR. J. Virol. Methods 143:112-116. [DOI] [PubMed] [Google Scholar]
- 29.Seminati, C., E. Mateu, B. Peralta, N. de Deus, and M. Martin. 2008. Distribution of hepatitis E virus infection and its prevalence in pigs on commercial farms in Spain. Vet. J. 175:130-132. [DOI] [PubMed] [Google Scholar]
- 30.Zanetti, A. R., G. G. Schlauder, L. Romano, E. Tanzi, P. Fabris, G. J. Dawson, and I. K. Mushahwar. 1999. Identification of a novel variant of hepatitis E virus in Italy. J. Med. Virol. 57:356-360. [DOI] [PubMed] [Google Scholar]