Abstract
Salmonella represents an important zoonotic pathogen worldwide, but the transmission dynamics between humans and animals as well as within animal populations are incompletely understood. We characterized Salmonella isolates from cattle and humans in two geographic regions of the United States, the Pacific Northwest and the Northeast, using three common subtyping methods (pulsed-field gel electrophoresis [PFGE], multilocus variable number of tandem repeat analysis [MLVA], and multilocus sequence typing [MLST]). In addition, we analyzed the distribution of antimicrobial resistance among human and cattle Salmonella isolates from the two study areas and characterized Salmonella persistence on individual dairy farms. For both Salmonella enterica subsp. enterica serotypes Newport and Typhimurium, we found multidrug resistance to be significantly associated with bovine origin of isolates, with the odds of multidrug resistance for Newport isolates from cattle approximately 18 times higher than for Newport isolates from humans. Isolates from the Northwest were significantly more likely to be multidrug resistant than those from the Northeast, and susceptible and resistant isolates appeared to represent distinct Salmonella subtypes. We detected evidence for strain diversification during Salmonella persistence on farms, which included changes in antimicrobial resistance as well as genetic changes manifested in PFGE and MLVA pattern shifts. While discriminatory power was serotype dependent, the combination of PFGE data with either MLVA or resistance typing data consistently allowed for improved subtype discrimination. Our results are consistent with the idea that cattle are an important reservoir of multidrug-resistant Salmonella infections in humans. In addition, the study provides evidence for the value of including antimicrobial resistance data in epidemiological investigations and highlights the benefits and potential problems of combining subtyping methods.
Salmonella is an important human and animal pathogen worldwide. In the United States, Salmonella causes an estimated 1.4 million human cases, 15,000 hospitalizations, and more than 400 deaths each year (44, 75). Human infections can be acquired through contact with animals or humans shedding Salmonella or through contaminated environments, but the majority of human infections are food-borne, and a large number of human outbreaks have been linked to foods of animal origin (20). Beef represents one well-recognized source of human infection (71). In addition, a number of human cases have been linked to dairy products or cattle contact, for instance at state fairs or on dairy farms (for example, see references 25, 35, and 61).
Salmonella enterica subsp. enterica serotypes Typhimurium and Newport are commonly isolated from human cases, including those linked to cattle (20, 61). In 2006, Salmonella serotypes Typhimurium and Newport were isolated from 17 and 8% of reported human salmonellosis cases in the United States, respectively, making them the first and third most common human disease-associated serotypes in the United States (15). S. enterica serotype 4,5,12:i:− is both genetically and antigenically closely related to Salmonella serotype Typhimurium, of which it represents a monophasic variant (62). Salmonella enterica serotype 4,5,12:i:− is characterized by a deletion of flagellar genes fliA and fliB, which prevents expression of the phase 2 flagellar antigen (60). In the United States, the prevalence of Salmonella serotype 4,5,12:i:− has increased considerably over the past 10 years, and in 2006, Salmonella serotype 4,5,12:i:− represented the sixth most commonly isolated serotype from humans in the United States (15, 60).
Salmonella serotype Newport represents two distinct clonal groups or lineages—one predominantly associated with isolates from cattle (i.e., Newport lineage A) and one associated with isolates from birds (i.e., Newport lineage B) (1, 33). Members of both lineages cause human infections (1, 33). The two Newport lineages can be clearly distinguished by multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE), and some correlation between genetic lineage and antimicrobial resistance profile seems to exist (1, 33). In general, Newport lineage B isolates are pansusceptible or resistant to only a few antimicrobial drugs. In contrast, lineage A is strongly associated with multidrug resistance and includes a Newport subtype commonly referred to as Newport MDR-AmpC (1, 33).
The prevalence of antimicrobial resistance among Salmonella serotype Newport and Typhimurium isolates has increased worldwide during the last 2 decades, predominantly as a result of emerging multidrug-resistant (MDR) strains (14, 52, 65). During the 1990s, Salmonella serotype Typhimurium phage type DT104 with pentaresistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (ACSSuT) increased considerably in prevalence around the world, and some isolates acquired resistance to additional antimicrobial agents, including trimethoprim or ciprofloxacin (52). MDR Salmonella serotype Typhimurium DT104 has been isolated from a wide variety of host species and caused numerous large human outbreaks around the world (65). Salmonella serotype Newport MDR-AmpC, characterized by resistance to ACSSuT and carrying a plasmid encoding resistance to amoxicillin-clavulanic acid, cefoxitin, ceftiofur, and cephalothin emerged in the United States during the late 1990s, where it quickly became widespread among humans and cattle, leading to several large human outbreaks (14).
Whether antimicrobial drug use in animals facilitates the emergence of MDR human pathogens is still subject to debate. Some studies report a temporal association between the introduction of new antimicrobial agents in veterinary medicine and the emergence of antimicrobial resistance (for instance, see references 22 and 58), but questions regarding the underlying evolutionary mechanisms, the origin and distribution of naturally occurring resistance genes, and the role of antimicrobial usage among humans remain (for example, see references 2 and 66 for reviews on this topic). Moreover, some studies report a higher prevalence of antimicrobial resistance among Salmonella isolates from farm animals than humans. Gebreyes et al. (26), for instance, found a higher prevalence of antimicrobial resistance among Salmonella isolates from pigs than humans, but potential effects attributable to differences in serotype distribution are difficult to assess in this study. In recent years, risk factors for MDR have received considerable attention. Infections with MDR Salmonella strains can lead to treatment failures, may be of longer duration, and may result in more severe clinical disease. Hence, such infections lead more often to hospitalization or death than infections with susceptible Salmonella strains, but serotype or subtype differences between resistant and susceptible Salmonella strains complicate the interpretation of clinical data (34, 41, 68).
Subtyping methods allow characterization of Salmonella isolates and include phenotypic methods (e.g., serotyping or phage typing) as well as molecular subtyping methods, such as pulsed-field gel electrophoresis (PFGE), ribotyping, multilocus variable number of tandem repeat analysis (MLVA), and multilocus sequence typing (MLST) (5). PFGE is widely used and robust, and rigorous standardization allows comparison between laboratories (5). However, the method is time-intensive and laborious, requires careful standardization and analysis, does not allow phylogenetic inference, and can in rare cases be affected by endogenous nucleases or DNA methylation (for a review of this topic, see reference 5). MLVA and MLST are rapid, allow for easy data exchange between laboratories, and provide some phylogenetic information (5). MLVA is highly discriminatory but subject to rapid diversification and therefore most appropriate for the analysis of closely related isolates. While MLST lacks discriminatory power within Salmonella serotypes, it is highly reproducible and allows for phylogenetic analysis of more distantly related isolates (1, 5, 33). PFGE and MLST can be performed regardless of serotype, but MLVA protocols are serotype specific and have so far only been validated for a limited number of Salmonella serotypes. Moreover, MLVA can be complicated by inaccurate sizing of DNA fragments, and the degree of reliability can be considerably influenced by nucleotide composition and fragment length (5). Overall, these subtyping methods differ considerably in discriminatory power and sometimes yield conflicting results, and the most appropriate subtyping method or combination thereof strongly depends on serotype and chosen application (19, 56, 72, 76). Other genetic or phenotypic characteristics, such as antimicrobial resistance patterns or the presence of specific plasmids, have also been used successfully for subtyping in outbreak investigations and other epidemiological studies and can provide valuable additional information (7, 8, 40, 63, 64).
Here we describe the distribution and subtype diversity of Salmonella serotypes Newport, 4,5,12:i:−, and Typhimurium among cattle and humans in two geographic regions of the United States, and we assess common risk factors for multidrug resistance. In addition, we utilize three Salmonella subtyping methods (PFGE, MLVA, and MLST), analyze their usefulness for characterizing isolates representing three common human-associated Salmonella serotypes, and compare the combined discriminatory power of PFGE and MLVA to that of PFGE and antimicrobial resistance patterns.
MATERIALS AND METHODS
Isolate selection.
Nontyphoidal Salmonella enterica subsp. enterica isolates from bovine (n = 222) and human (n = 203) sources representing S. enterica serotypes Typhimurium (n = 190), 4,5,12:i:− (n = 40), and Newport (n = 195) were included in this study (see Table S1 in the supplemental material). These isolates were collected between January 2004 and May 2005 in two geographic regions of the United States, the Pacific Northwest and the Northeast, and human isolates were obtained from the Wadsworth Center, New York State Department of Health (n = 73) or from the Washington State Department of Health Public Health Laboratories (n = 130). Human-source Salmonella isolates specifically originated from reported cases among residents of New York State (69 isolates, including 2 isolates with unknown county of origin, 1 isolate from New York City, and 66 isolates from New York State) and New Jersey (n = 4); isolates from New York City and New Jersey residents represent exceptional cases where isolates were submitted to the New York State Department of Health. For the Northwest, human isolates were obtained from Washington State (n = 130) residents. The main source populations for human isolates thus are New York State (except New York City) and Washington State with approximate human populations of 11 and 6.6 million residents, respectively (70).
Northeastern bovine-source Salmonella isolates (n = 154) were collected from dairy cattle on 57 farms in New York (n = 113) and Vermont (n = 41) as part of a previously reported study to determine Salmonella incidence in cattle herds in the northeastern United States (18) and obtained via the New York State Animal Health Diagnostic Center. Pacific Northwest bovine isolates (n = 68) were collected in Washington (n = 58), Idaho (n = 8), and Oregon (n = 2) from cattle on 39 farms, primarily via the Washington Animal Diagnostic Laboratory. The majority of isolates from cattle were isolated from clinical cases, except for 17 cattle isolates from the Pacific Northwest, which were obtained from cattle without clinical signs of salmonellosis, and originated from previous field studies (see Table S1 in the supplemental material). Of the 227 isolates collected in the Northeast (Table S1), 152 have been described and characterized previously using PFGE and MLST based on a three-gene MLST scheme (based on the manB, fimA, and mdh genes) (1, 60). Isolate designation of these previously characterized isolates is identical to their original designation to allow comparison between the studies.
Serotyping of all bovine and human isolates included in this study was performed according to the Kaufman-White scheme using the standard procedure described by Ewing et al. (24). Where discrimination between Salmonella serotypes 4,5,12:i:− and Typhimurium appeared questionable, serotype designation was confirmed by PCR amplification of the phase 2 flagellin gene fljB as described previously (21, 60).
PFGE analysis.
PFGE analysis with restriction enzyme XbaI (Invitrogen, Carlsbad, CA) was performed according to the CDC PulseNet protocol (55) using Salmonella serovar Braenderup isolate H9812 (Centers for Disease Control and Prevention) as the reference strain as described previously (37). PFGE patterns were analyzed using BioNumerics software package version 2.5 (Applied Maths, Austin, TX). Similarity analyses were based on Dice coefficients with a maximum space tolerance of 1.5%, and cluster analysis was performed using the unweighted pair group method with arithmetic mean (UPGMA) algorithm in BioNumerics. Clusters were inspected visually to ascertain correct classification.
MLVA typing analysis.
Isolates were stored at −80°C, and single colonies were subcultured on LB agar plates overnight prior to MLVA analysis. MLVA analysis was performed as described previously, with analysis of Salmonella serotype Typhimurium and 4,5,12:i:− isolates based on previously described variable number of tandem repeat (VNTR) loci STTR5, STTR6, STTR9, and STTR10pl, while Salmonella serotype Newport isolates were characterized using VNTR loci STTR5, STTR6, NEWPORTA, NEWPORTB, NEWPORTL, and NEWPORTM, also as described previously (19, 42). Briefly, boiled-lysate suspensions of single bacterial colonies were added to 25-μl PCR mixtures and subjected to denaturation (15 min at 94°C), followed by 25 cycles, with 1 cycle consisting of 30 s at 94°C, 1 min at 55°C, and 90 s at 72°C. Following a final extension step of 10 min at 72°C, size standard (LIZ600; Applied Biosystems, Foster City, CA) and Hi Di formamide were added to the PCR mixtures, and the mixtures were loaded into a 3730 DNA analyzer with Pop-7 polymer (Applied Biosystems) for capillary electrophoresis. Electropherograms were subsequently analyzed using GeneMarker (Softgenetics LCC, State College, PA), and MLVA patterns were assigned on the basis of the composite data for the respective VNTR loci (see Table S3 in the supplemental material). Fragment sizes for each VNTR locus were analyzed as categorical data in BioNumerics. Minimum spanning trees (MSTs) based on the MLVA data were constructed for Salmonella serotype Newport as well as Salmonella serotype Typhimurium and 4,5,12:i:− isolates using categorical coefficients and allowing for hypothetical nodes.
Data from MLVA (categorical coefficients) and PFGE (Dice similarity coefficients) were combined using the “average from experiments” function in BioNumerics and the UPGMA algorithm. Clusters were identified in the resulting dendrograms by choosing an arbitrary level of similarity at 75% to define a cluster, and cluster numbers were assigned to clusters containing two or more isolates. Correspondence between the clustering based on MLVA and PFGE analysis was assessed using Kendall's tau rank correlation coefficient and Pearson's correlation coefficient, again using BioNumerics.
Antimicrobial resistance patterns.
Susceptibility to a panel of antimicrobial agents was determined for all isolates using a disc diffusion method as described previously (4). The antimicrobial agents tested belonged to the following drug classes: beta-lactams (ampicillin [10 μg], ceftazidime [30 μg], and amoxicillin-clavulanic acid [20 and 10 μg, respectively]), quinolones (nalidixic acid [30 μg]), phenicols (chloramphenicol [30 μg]), aminoglycosides (kanamycin [30 μg], gentamicin [10 μg], and streptomycin [10 μg]), sulfonamides and dihydrofolate reductase inhibitors (triple-sulfa [sulfadiazine, sulfamethazine, and sulfamerazine] [300 μg] and trimethoprim-sulfamethoxazole [1.25 and 23.75 μg, respectively]), and tetracyclines (tetracycline [30 μg]). Resistance breakpoints were as reported by the Clinical and Laboratory Standards Institute (46, 47). To facilitate data analysis, isolates classified as intermediate were analyzed as susceptible. Multidrug resistance (MDR) was defined as resistance to more than two antimicrobial drug classes.
MLST analysis.
Isolates with highly prevalent PFGE patterns were further analyzed by sequence typing using a 7-gene MLST scheme (targeting aroC, dnaN, hemD, hisD, purE, sucA, and thrA) as described previously (38). A single representative isolate was selected at random (using the random generator available at www.random.org) for each PFGE pattern for characterization by MLST analysis. Bacterial genomic DNA was prepared using the Qiagen genomic DNA purification kit (Qiagen, Valencia, CA) and amplified by PCR, followed by Sanger sequencing of PCR products. Consensus sequences were assembled using the DNAStar software package (Lasergene, Madison, WI) and submitted to the MLST database at the Environmental Research Institute (ERI), University College Cork (http://mlst.ucc.ie/) for assignment of MLST allele numbers. Consensus sequences were subsequently trimmed to span the allelic region of interest specified previously (38), and concatenated sequences spanning all 7 genes were constructed using SEAL (http://tree.bio.ed.ac.uk/software/seal/). Sequences were aligned using SEAL, and phylogenetic trees based on individual genes or the concatenated sequences were inferred as described below. In addition, representative sequences for each inferred allelic type were aligned with Newport and Typhimurium sequences available in the MLST database (see Table S3 in the supplemental material), choosing one representative sequence for each allelic type present in the MLST database.
Unrooted maximum likelihood (ML) phylogenetic trees were estimated using PAUP*, with the best-fitting model of nucleotide substitution determined using Modeltest (53) (all parameter values available from the authors upon request). Support for individual nodes on the trees was assessed through bootstrap values based on 1,000 neighbor-joining trees using PAUP*.
Statistical analysis.
To avoid overrepresentation of Salmonella strains persisting on individual farms, a single representative isolate was chosen at random from groups of isolates that originated from the same farm, had indistinguishable PFGE and MLVA patterns, and showed identical antimicrobial resistance patterns (see Table S1 in the supplemental material). This reduced data set (n = 366) was used to describe the distribution of resistance against individual antimicrobial drugs and to compare the discriminatory power of the different subtyping methods. Simpson's index of diversity (D) was calculated as described previously to assess the serotype-specific discriminatory power of the different analytical methods (36, 57). D can vary between 0 and 1, with 1 indicating infinite diversity and 0 indicating no diversity. To assess the statistical significance of differences between individual D values, 95% confidence intervals (95% CIs) based on normal approximation were calculated as described previously (30).
Ninety-five percent confidence intervals for the proportion of isolates resistant to individual antimicrobial agents were calculated using the normal approximation to the binominal distribution. Due to the low number of Salmonella serotype 4,5,12:i:− isolates included in this study, these isolates were excluded from subsequent analysis of multidrug resistance, resulting in a second reduced data set (n = 338). The frequency distributions of Salmonella serotype Newport and Typhimurium isolates were analyzed using Fisher's exact test, with P values of <0.05 deemed statistically significant. To estimate the odds ratios for risk factors associated with multidrug resistance, logistic regression analysis was performed, with model selection performed using a backward stepwise selection algorithm. Overall model fit was assessed on the basis of deviance estimates and inspection of studentized residuals, and 95% Wald confidence intervals for odds ratios were estimated. All statistical analyses were performed using SAS version 9.2 (SAS, Cary, NJ).
To further analyze the distribution of Salmonella subtypes associated with PFGE patterns that were shared between humans and cattle (or between the two geographic regions in the United States), patterns with a frequency of <2 were excluded, since the probability of these patterns being shared equaled zero, and the remaining isolates were classified by the presence or absence of a shared PFGE pattern, resulting in a data set of 234 isolates comprising 116 Salmonella serotype Newport isolates and 118 Salmonella serotype Typhimurium isolates.
Nucleotide sequence accession number.
The newly determined Salmonella enterica serotype Typhimurium hemD sequence was submitted to GenBank and assigned accession no. GU784796.
RESULTS
PFGE and MLVA subtype diversity.
The initial data set of 425 isolates included 190 Salmonella enterica serotype Typhimurium isolates, which we classified into 90 PFGE patterns and 135 MLVA patterns, and 195 Salmonella enterica serotype Newport isolates, which we classified into 54 PFGE patterns and 59 MLVA patterns (see Table S1 in the supplemental material). Among the 40 Salmonella serotype 4,5,12:i:− isolates, we detected 13 PFGE and 15 MLVA patterns. Two PFGE patterns (L1 and L6) were detected among Salmonella serotype 4,5,12:i:− and Typhimurium isolates. To avoid overrepresentation of persisting Salmonella subtypes on farms, we selected a single representative for each combination of PFGE, MLVA, and antimicrobial resistance pattern on a given farm, resulting in a reduced data set of 182 Salmonella serotype Typhimurium isolates, 156 Salmonella serotype Newport isolates, and 28 Salmonella serotype 4,5,12:i:− isolates (see Table S1 in the supplemental material).
Distribution of antimicrobial resistance.
Among the 203 human Salmonella isolates and 163 independent bovine Salmonella isolates, 74 isolates were susceptible to all antimicrobial drugs tested (i.e., pansusceptible), representing 39 Salmonella serotype Typhimurium isolates, 19 Salmonella serotype Newport isolates, and 16 Salmonella serotype 4,5,12:i:− isolates (Table 1). These isolates included 5 of the 91 (6%) bovine Salmonella serotype Newport isolates, 14 of the 65 (22%) human Salmonella serotype Newport isolates, 11 of the 62 (18%) bovine Salmonella serotype Typhimurium isolates, and 28 of the 120 (23%) human Salmonella serotype Typhimurium isolates. In addition, 4 of the 10 (40%) bovine Salmonella serotype 4,5,12:i:− isolates and 12 of the 18 (67%) human Salmonella serotype 4,5,12:i:− isolates were pansusceptible. For most antimicrobial agents, resistance prevalence was significantly higher among cattle isolates than human isolates, while significantly more human isolates than cattle isolates were pansusceptible (Table 1). Resistance to gentamicin and nalidixic acid was detected only among human isolates and at a low frequency. In general, resistance prevalence was higher among Salmonella serotype Newport isolates than Salmonella serotype Typhimurium isolates, but resistance to kanamycin was significantly more prevalent among Salmonella serotype Typhimurium isolates than Salmonella serotype Newport isolates (Table 1).
TABLE 1.
Distribution of resistance to individual antimicrobial agents among sources and Salmonella enterica serotypes
| Antimicrobial agent or drug resistance category | Distribution of resistance to antimicrobial agent among: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All isolates |
Bovine isolates |
Human isolates |
Serotype Newport isolatesa |
Serotype Typhimurium isolatesa |
||||||
| No. of isolatesb | % of isolates (95% CI)c | No. of isolates | % of isolates (95% CI) | No. of isolates | % of isolates (95% CI) | No. of isolates | % of isolates (95% CI) | No. of isolates | % of isolates (95% CI) | |
| Antimicrobial agents | ||||||||||
| Triple-sulfad | 288 | 79 (74-83) | 143 | 88 (83-93) | 145 | 71 (65-78) | 137 | 88 (83-93) | 139 | 76 (70-83) |
| Tetracycline | 238 | 65 (60-70) | 139 | 85 (80-91) | 99 | 49 (42-56) | 122 | 78 (72-85) | 111 | 61 (54-68) |
| Ampicillin | 233 | 64 (59-69) | 142 | 87 (82-92) | 91 | 45 (38-52) | 120 | 77 (70-84) | 106 | 58 (51-65) |
| Streptomycin | 229 | 63 (58-68) | 128 | 79 (72-85) | 101 | 50 (43-57) | 121 | 78 (71-84) | 100 | 55 (48-62) |
| Chloramphenicol | 187 | 51 (46-56) | 110 | 67 (60-75) | 77 | 38 (31-45) | 98 | 63 (55-70) | 83 | 46 (38-53) |
| Ceftazidime | 153 | 42 (37-47) | 104 | 64 (56-71) | 49 | 24 (18-30) | 106 | 68 (61-75) | 46 | 25 (19-32) |
| Amoxicillin-clavulanic acid | 119 | 33 (28-37) | 86 | 53 (45-60) | 33 | 16 (11-21) | 79 | 51 (43-58) | 34 | 19 (13-24) |
| Kanamycin | 82 | 22 (18-27) | 53 | 33 (25-40) | 29 | 14 (9-19) | 19 | 12 (7-17) | 62 | 34 (27-41) |
| Sulfoxazole-trimethoprim | 43 | 12 (8-15) | 21 | 13 (8-18) | 22 | 11 (7-15) | 13 | 8 (4-13) | 30 | 16 (11-22) |
| Gentamicin | 8 | 0 | 8 | 1 | 5 | |||||
| Nalidixic acid | 4 | 0 | 4 | 2 | 2 | |||||
| Drug resistance category | ||||||||||
| Pansusceptible | 74 | 20 (16-25) | 20 | 12 (8-17) | 54 | 27 (21-33) | 19 | 12 (7-17) | 39 | 21 (16-28) |
| Intermediate resistance | 54 | 15 (11-18) | 4 | 3 (0-5) | 50 | 25 (19-31) | 15 | 10 (5-14) | 35 | 19 (14-25) |
| Multidrug resistant | 238 | 65 (60-70) | 139 | 85 (80-91) | 99 | 49 (42-56) | 122 | 78 (72-85) | 108 | 59 (52-67) |
Data for Salmonella serotype 4,5,12:i:− isolates not shown.
Number of isolates showing resistance to the specific antimicrobial drug out of 366 isolates included in the reduced data set.
Percentage of isolates resistant to the specific antimicrobial drug. The 95% binominal confidence interval (95% CI) based on normal approximation is shown in parentheses.
Triple-sulfa is sulfadiazine, sulfamethazine, and sulfamerazine.
One hundred twenty-two of 156 (78%) Salmonella serotype Newport isolates, 9 of 28 (32%) Salmonella serotype 4,5,12,:i:− isolates, and 108 of 182 (59%) Salmonella serotype Typhimurium isolates were multidrug resistant, defined as resistant to more than 2 classes of antimicrobial agents (Table 1). For both Salmonella serotype Newport and Typhimurium, multidrug resistance was significantly (P < 0.05) associated with origin from cattle (data not shown). To estimate odds ratios, a logistic regression model was fit to the data. Using a backward stepwise selection approach, we constructed a final model consisting of the predictors “source,” “serotype,” “geographic origin,” and the interaction term “source × serotype.” The year of isolation was not significant and therefore not included in the final model. In our data set, the odds of multidrug resistance were approximately 18 times higher for bovine Salmonella serotype Newport isolates than for human Newport isolates and more than 3 times higher for bovine Salmonella serotype Typhimurium isolates than for human Typhimurium isolates (Table 2). Among both human and bovine isolates, the odds of multidrug resistance were higher for Salmonella serotype Newport isolates than for Salmonella serotype Typhimurium isolates, even though the differences were only statistically significant among isolates from cattle (Table 2). Moreover, in the United States, the odds of multidrug resistance were approximately twice as high for isolates that originated from the Northwest compared to isolates from the Northeast.
TABLE 2.
Odds ratio estimates for odds of multidrug resistance based on logistic regression modela
| Comparison of isolates | Odds ratio | 95% CIb |
|---|---|---|
| Bovine vs. human serotype Newport | 18.2 | 6.3-52.6 |
| Bovine vs. human serotype Typhimurium | 3.5 | 1.7-7.1 |
| Bovine serotype Newport vs. bovine Typhimurium | 6.1 | 2.1-17.9 |
| Human serotype Newport vs. human serotype Typhimurium | 1.2 | 0.6-2.2 |
| Northwest vs. Northeast | 2.0 | 1.2-3.4 |
The odds of multidrug resistance in S. enterica isolates of different serotypes, from different hosts, and from different geographic areas of the United States were calculated. The overall model fit was satisfactory as determined on the basis of deviance estimates (P = 0.41) and visual inspection of studentized residuals.
95% Wald confidence interval for odds ratio.
Salmonella diversity on farms.
The median number of Salmonella isolates collected per farm equaled 1 and ranged from 1 to 32. Both Salmonella serotype Newport and Typhimurium were isolated from 2 farms in the Northeast and 5 farms in the Northwest, while both serotypes Newport and 4,5,12:i:− were isolated from 2 farms in the Northeast and none in the Northwest (see Table S2 in the supplemental material). On average, 1.3 PFGE patterns, 1.5 MLVA patterns, and 1.3 antimicrobial resistance patterns were detected among Salmonella isolates from the same farm in the Northeast (ranges, 1 to 3, 1 to 7, and 1 to 4, respectively; median, 1), and 1.2 PFGE patterns, 1.4 MLVA patterns, and 1.5 antimicrobial resistance patterns were detected among isolates from the same farm in the Northwest (ranges, 1 to 5, 1 to 5, and 1 to 7, respectively; median, 1).
On 27 of the 96 farms, representing 17 of 57 (30%) farms in the Northeast and 10 of 39 (26%) farms in the Northwest, multiple isolates with indistinguishable PFGE patterns were isolated (see Table S2 in the supplemental material). On the farms where the same PFGE pattern was detected more than once, the predominant PFGE pattern was shared by between 40% and 100% of isolates (median, 100%; average, 83%). The average number of isolates that were indistinguishable by PFGE, MLVA, and antimicrobial resistance profiles equaled 1.7 isolates per farm (range, 1 to 19 isolates; median, 1), and on average, 3.3 different combinations of PFGE, MLVA, and antimicrobial resistance patterns were isolated from these farms (range, 1 to 12; median, 1). Salmonella isolates that were indistinguishable by MLVA, PFGE, and antimicrobial resistance patterns were isolated from the same farm over time periods ranging from less than 1 month to more than 7 months; however, isolation dates were missing for some isolates, and longer durations are therefore possible.
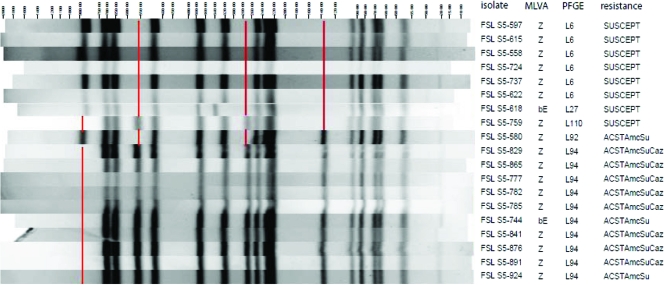
On 7 of the 27 farms, all isolates shared a single PFGE and MLVA pattern, while on another 8 farms, the isolates shared a single PFGE pattern but had MLVA patterns that differed from each other at one locus (see Table S2 in the supplemental material). On 10 of the 15 farms with isolates with a single PFGE pattern, all isolates shared the same antimicrobial resistance pattern, while on each of the remaining 5 farms, two closely related antimicrobial resistance profiles were detected. On farm NE152, all 4 isolates shared the same MLVA pattern but differed in PFGE pattern. On several of the remaining farms, some of the isolates shared PFGE and others shared MLVA patterns. For instance, among the 19 Salmonella serotype 4,5,12:i:− isolates from farm NE261, 5 PFGE patterns and 2 MLVA patterns were detected. These isolates could be classified into 2 clusters based on PFGE, with clusters differing by 4 bands (Fig. 1). One cluster represented susceptible isolates, and the other represented multidrug-resistant isolates, yet isolates from both clusters shared identical MLVA patterns, likely indicating close evolutionary relatedness. Most of the isolates shared MLVA pattern Z, but two isolates, collected in August 2004, shared MLVA pattern bE and yet belonged to two different PFGE clusters (Fig. 1). PFGE patterns for one multidrug-resistant isolate and one susceptible isolate (isolates S5-580 and S5-759) shared most bands with one cluster but some with the other, and were isolated in July and September 2004, respectively. All other susceptible isolates were collected between June and August of that year, while the remaining multidrug-resistant isolates were obtained between 31 August and December 2004.
FIG. 1.
Characterization of Salmonella enterica serotype 4,5,12:i:− isolates from farm NE299 by PFGE, MLVA, and antimicrobial resistance pattern. The isolate designations, MLVA pattern, PFGE pattern, and antimicrobial resistance pattern are shown to the right of the figure. The antimicrobial resistance pattern is shown as follows: SUSCEPT, susceptible; ACSTAmcSu, resistance to ampicillin, chloramphenicol, streptomycin, tetracycline, amoxicillin-clavulanic acid, and sulfonamides; ACSTAmcSuCaz, resistance to ampicillin, chloramphenicol, streptomycin, tetracycline, amoxicillin-clavulanic acid, sulfonamides, and ceftazidime. Absent PFGE bands distinguishing the two clusters of isolates are indicated by red lines.
On 9 farms, some isolates with identical MLVA and PFGE patterns but different antimicrobial resistance profiles were isolated (see Table S2 in the supplemental material). On 5 of these 9 farms, information on sample collection dates allowed inference as to whether resistance was likely acquired or lost. On 3 of the farms, isolates appeared to have gained additional resistance to ceftazidime, while on 1 farm, isolates seemed to have lost resistance to sulfa-trimethoprim (Table S2). On farm NE510, some isolates seem to have gained resistance to ceftazidime, while other isolates later appeared to have lost resistance to kanamycin or triple-sulfa. Conclusions on resistance loss or gain are difficult to draw though, as isolates with specific resistance patterns may have coexisted in a given farm or may have been present at a given time period without being detected. Further studies are therefore needed to determine the stability of resistance patterns on farms.
Discriminatory power of Salmonella subtyping methods.
Comparisons of Salmonella subtyping methods were based on the reduced data set comprising a total of 366 Salmonella isolates. Overall, a combination of MLVA and PFGE showed the highest discriminatory power (Table 3), as judged by Simpson's index of diversity (D). For Salmonella serotype Typhimurium, the discriminatory power of MLVA was significantly higher than that of PFGE, while no significant differences were detected for Salmonella serotype Newport or Salmonella serotype 4,5,12:i:− (Table 3). For Salmonella serotype Newport, the combination of PFGE with antimicrobial resistance patterns yielded a significantly higher discriminatory power than either PFGE or MLVA alone. For Salmonella serotype Typhimurium, this combination yielded a higher discriminatory power than PFGE, which was comparable to that of MLVA.
TABLE 3.
Simpson's index of diversity scores among the three Salmonella serotypes by different Salmonella subtyping methods
| Isolate (n)a | Simpson's index of diversityb for the following subtyping method(s): |
|||
|---|---|---|---|---|
| PFGEc | MLVAd | MLVA and PFGE (combined) | PFGE and antimicrobial resistance (combined)e | |
| Serotype 4,5,12:i:− (n = 28) | 0.833 (0.711-0.955) | 0.910 (0.843-0.977) | 0.971 (0.940-1.00) | 0.844 (0.717-0.971) |
| Serotype Typhimurium (n = 182) | 0.967 (0.953-0.981) | 0.993 (0.989-0.996) | 0.997 (0.996-0.999) | 0.988 (0.980-0.994) |
| Serotype Newport (n = 156) | 0.930 (0.911-0.948) | 0.913 (0.886-0.941) | 0.980 (0.970-0.990) | 0.972 (0.961-0.982) |
| Bovine isolates (n = 163) | 0.947 (0.933-0.960) | 0.950 (0.932-0.968) | 0.984 (0.976-0.992) | 0.979 (0.971-0.987) |
| Human isolates (n = 203) | 0.986 (0.980-0.992) | 0.994 (0.991-0.997) | 0.999 (0.997-0.999) | 0.992 (0.987-0.997) |
| Bovine serotype Typhimurium (n = 62) | 0.938 (0.910-0.966) | 0.976 (0.959-0.993) | 0.992 (0.984-0.999) | 0.980 (0.967-0.994) |
| Human serotype Typhimurium (n = 120) | 0.975 (0.960-0.990) | 0.994 (0.990-0.999) | 0.997 (0.994-1.00) | 0.990 (0.979-0.997) |
| Bovine serotype Newport (n = 91) | 0.859 (0.830-0.889) | 0.856 (0.811-0.900) | 0.954 (0.931-0.977) | 0.943 (0.922-0.964) |
| Human serotype Newport (n = 65) | 0.976 (0.959-0.994) | 0.968 (0.947-0.988) | 0.995 (0.990-1.00) | 0.985 (0.971-0.999) |
The number of isolates included in the final data set (n) (i.e., reduced data set of 366 isolates) is shown.
The mean estimates are shown in boldface type, and the 95% confidence intervals are shown in parentheses.
Salmonella serotype 4,5,12:i:−, Typhimurium, and Newport isolates exhibited 13, 90, and 54 PFGE patterns, respectively.
Salmonella serotype 4,5,12:i:−, Typhimurium, and Newport isolates exhibited 15, 135, and 59 MLVA patterns, respectively.
Salmonella serotype 4,5,12:i:−, Typhimurium, and Newport isolates exhibited 7, 41, and 22 antimicrobial resistance patterns, respectively.
Analysis of serotype and source diversity.
Simpson's index of diversity estimates for Salmonella isolates from humans were significantly higher than those for isolates obtained from cattle, regardless of the subtyping method (Table 3). This trend was consistent among both Salmonella serotype Typhimurium and Newport isolates, but the differences were significant only for Salmonella serotype Newport isolates. Among the isolates from cattle, Salmonella serotype Typhimurium isolates were significantly more diverse than Salmonella serotype Newport isolates, while the differences between the two serotypes were considerably less pronounced among the human isolates (Table 3).
Analysis of correspondence in clustering based on MLVA and PFGE.
To estimate the correlation between similarity coefficients based on MLVA and those based on PFGE, we use BioNumerics to calculate Kendall's tau rank correlation coefficient and Pearson's correlation coefficient. For Salmonella serotype Typhimurium, Kendall's tau rank correlation coefficient equaled 0.11 (95% CI, 0.105 to 0.119) while Pearson's correlation coefficient equaled 0.19. For Salmonella serotype Newport, the estimates were 0.46 (95% CI, 0.456 to 0.470) and 0.61, respectively, suggesting a better correspondence between the subtyping methods for Salmonella serotype Newport, possibly due to the more genetically homogeneous population structure of Salmonella serotype Newport.
Genetic relationship between isolates.
To further describe the phylogenetic relationship between the isolates, we analyzed 22 Salmonella isolates (2 Salmonella serotype 4,5,12:i:− isolates, 12 Salmonella serotype Typhimurium isolates, and 8 Salmonella serotype Newport isolates), representing the most common PFGE types, using a 7-gene MLST scheme (Table 4). All analyzed Newport isolates represented the same sequence type (ST) and clearly clustered separately from the Salmonella serotype Typhimurium and 4,5,12:i:− isolates (see Fig. S1 in the supplemental material). The Salmonella serotype Typhimurium isolates represented two STs; the more frequent ST was identical to that of the two Salmonella serotype 4,5,12:i:− isolates analyzed. The other ST was shared by two Salmonella serotype Typhimurium isolates (S5-511 and S5-831) and differed from the first ST in a single nucleotide in hemD. Isolate S5-511 was isolated from a human case, and isolate S5-831 was isolated from cattle; both isolates were collected in New York State and were pansusceptible. However, the isolates differed in two MLVA loci (STTR5 and STTR6) and by more than three bands in their PFGE patterns.
TABLE 4.
MLST alleles of representative Salmonella isolates with PFGE patterns detected at high frequencya
| Isolate | Salmonella serotype | PFGE pattern | No. of isolates with the same PFGE patternb | Antimicrobial resistance patternc | Host species | Allele no. for the following gene by MLST schemed |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hisD | hemD | sucA | thrA | purE | dnaN | aroC | ||||||
| S5-597e | 4,5,12:i:− | L6 | 13 | Suscept | Cattle | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| S5-777 | 4,5,12:i:− | L94 | 4 | ACSTAmcSuCaz | Cattle | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| R6-027 | Typhimurium | L154 | 6 | AKSTSu | Cattle | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| R6-104 | Typhimurium | L42 | 4 | AKSTSuCaz | Human | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| R6-107 | Typhimurium | L2 | 12 | ACKSTSuCaz | Human | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| R6-108 | Typhimurium | L40 | 4 | ACSxtSTSuCaz | Human | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| R6-114 | Typhimurium | L46 | 9 | S | Human | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| R8-3079 | Typhimurium | L21 | 6 | SSu | Human | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| R8-3130e | Typhimurium | L1 | 5 | Suscept | Human | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| S5-509 | Typhimurium | L25 | 26 | ACSTAmcSu | Human | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| S5-511 | Typhimurium | L96 | 4 | Suscept | Human | 9 | New1f | 9 | 2 | 5 | 7 | 10 |
| S5-640 | Typhimurium | L93 | 4 | Suscept | Human | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| S5-679 | Typhimurium | L104 | 9 | AKSTSu | Human | 9 | 12 | 9 | 2 | 5 | 7 | 10 |
| S5-831 | Typhimurium | L97 | 3 | Suscept | Cattle | 9 | New1f | 9 | 2 | 5 | 7 | 10 |
| R6-051 | Newport | L75 | 18 | ACKSTSuCaz | Cattle | 14 | 21 | 12 | 12 | 15 | 7 | 10 |
| R6-054 | Newport | L73 | 9 | ASTSuCaz | Cattle | 14 | 21 | 12 | 12 | 15 | 7 | 10 |
| R6-112 | Newport | L77 | 4 | ACSTSuCaz | Human | 14 | 21 | 12 | 12 | 15 | 7 | 10 |
| R8-3128 | Newport | L71 | 20 | ACSTAmcSuCaz | Human | 14 | 21 | 12 | 12 | 15 | 7 | 10 |
| S5-446 | Newport | L107 | 5 | Suscept | Human | 14 | 21 | 12 | 12 | 15 | 7 | 10 |
| S5-518 | Newport | L105 | 4 | ACSTAmcSuCaz | Human | 14 | 21 | 12 | 12 | 15 | 7 | 10 |
| S5-545 | Newport | L108 | 20 | ACSTAmcSuCaz | Cattle | 14 | 21 | 12 | 12 | 15 | 7 | 10 |
| S5-562 | Newport | L70 | 23 | ACSTAmcSu | Cattle | 14 | 21 | 12 | 12 | 15 | 7 | 10 |
hemD sequencing of an additional 18 pansusceptible Salmonella serotype Newport isolates, representing PFGE pattern L107 (4 isolates) as well as 15 other PFGE patterns that were each detected once in this study (see Table S1 in the supplemental material) identified allelic type (AT) hemD21 among the 5 isolates from cattle and ATs hemD21 and hemD45 among 5 and 8 human isolates, respectively.
Based on the final, reduced data set.
The antimicrobial resistance pattern is shown as follows: Suscept, susceptible; ACSTAmcSuCaz, resistance to ampicillin, chloramphenicol, streptomycin, tetracycline, amoxicillin-clavulanic acid, sulfonamides, and ceftazidime; AKSTSu, resistance to ampicillin, kanamycin, streptomycin, tetracycline, and sulfonamides; and Sxt, resistance to sulfoxazole-trimethoprim.
Allele numbers were assigned using the MLST database (http://mlst.ucc.ie/).
PFGE pattern detected among both Salmonella serotype Typhimurium and 4,5,12:i:− isolates.
Newly described AT.
We subsequently compared the concatenated nucleotide sequences for each sequence type to previously reported MLST types for Salmonella serotype Newport and Typhimurium isolates (see Table S3 and Fig. S1 in the supplemental material). The Salmonella serotype Newport isolates analyzed in this study were identical to 145 of the 389 published Newport sequences, thereby representing the most frequent Newport ST in the MLST database (http://mlst.ucc.ie/). Similarly, the predominant Salmonella serotype Typhimurium ST in our study exactly matched the ST for 251 of the 412 published serotype Typhimurium sequences, representing the most common Typhimurium ST in the MLST database. The less frequent Salmonella serotype Typhimurium ST in our study had no exact matches in the database, but it is very closely related to the other serotype Typhimurium ST detected in this study and clearly clustered with these isolates in phylogenetic analyses (see Fig. S1 in the supplemental material).
Relationship between the genetic similarity of isolates and the resistance type.
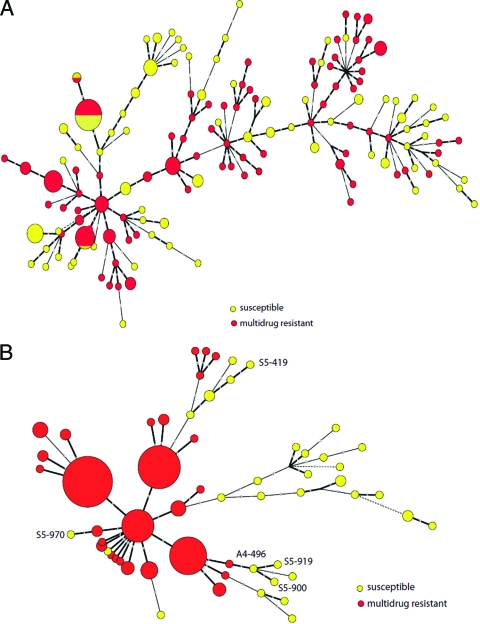
In the minimum spanning tree (MST) based on MLVA data for Salmonella serotype Typhimurium and 4,5,12:i:− isolates, multidrug-resistant and susceptible isolates occasionally clustered together, and resistant and susceptible isolates sometimes shared identical MLVA patterns (Fig. 2A). Among the Salmonella serotype Newport isolates, multidrug-resistant and susceptible isolates never shared identical MLVA patterns but sometimes showed similar MLVA patterns that clustered together. For example, three pansusceptible bovine Salmonella serotype Newport isolates (A4-496, S5-919, and S5-900), which were isolated from the same farm (NE299) between December 2004 and February 2005 and clustered together (Fig. 2B), also clustered with a susceptible human isolate (R8-3109) and with a bovine isolate (R6-028) resistant to six antimicrobials (ampicillin, chloramphenicol, streptomycin, tetracycline, sulfonamides, and ceftazidime [ACSTSuCaz]). Another pansusceptible bovine Salmonella serotype Newport isolate from farm NE299, S5-970, differed in MLVA locus STTR6 and clustered separately from the other 3 pansusceptible isolates from the same farm, even though the 4 isolates were indistinguishable by PFGE.
FIG. 2.
Minimum spanning tree (MST) based on MLVA data for Salmonella enterica serotype Typhimurium isolates (A) and Salmonella enterica serotype 4,5,12:i:− or Newport isolates (B) included in this study. The size of the circle is proportional to the number of isolates represented by each node, and the length of the branch is based on categorical coefficients and proportional to the number of differences between nodes. Hypothetical nodes are indicated by broken lines. Nodes encompassing multidrug-resistant isolates are shown in red, and those encompassing susceptible isolates are shown in yellow. Pansusceptible Salmonella serotype Newport isolates from farm NE299, which are characterized in the text, are indicated by their respective accession number in the tree. Interpretation of central nodes as ancestral is dependent on linear relationship between phylogenetic relationship and difference in the number of repeats, an assumption that might not be valid in all cases, particularly for isolates from Salmonella serotype Newport lineages A and B.
To further characterize the genetic relationship between the pansusceptible Salmonella serotype Newport isolates, we sequenced hemD in all pansusceptible serotype Newport isolates (Table 4); analysis of this gene has been shown to clearly classify Newport isolates into the two recognized lineages (33). Analysis of hemD sequences indeed grouped our isolates into the two separate lineages. One lineage contained pansusceptible Salmonella serotype Newport isolates from cattle and humans; these isolates had the same hemD allelic type (AT) (hemD21) as the multidrug-resistant bovine serotype Newport isolates characterized by MLST analysis in our study. The isolates belonging to the other lineage, all obtained from humans, shared AT hemD45; on the basis of the MLST database, this AT represented a perfect match with a large number of serotype Newport isolates that have been collected from reptiles, humans, birds, and a small number of domestic animals (see Table S4 in the supplemental material). Six of 14 (43%) pansusceptible human Salmonella serotype Newport isolates analyzed here belong to AT hemD21, contributing 6 of 11 (55%) pansusceptible serotype Newport isolates with AT hemD21; this AT was also associated with the multidrug-resistant bovine serotype Newport isolates analyzed by 7-gene MLST in our study, indicating it is associated with the cattle-associated Newport lineage. Isolates that grouped together based on hemD AT also were clearly distinct by PFGE analysis, corroborating previous findings that the two Salmonella serotype Newport lineages represent distinct subgroups (1, 33) (data not shown).
Distribution of isolates between sources.
To better characterize the distribution of related isolates, we performed a cluster analysis based on the combination of PFGE and MLVA results. On the basis of a clustering cutoff at 75% similarity, among the Salmonella serotype Newport isolates, we identified 10 combined PFGE and MLVA clusters that contained multiple isolates as well as 7 unique isolates (see Fig. S2 in the supplemental material). The largest cluster included 89 of the 93 bovine isolates and 31 of the 65 human isolates; 2 of the remaining clusters also contained bovine and human serotype Newport isolates (see Fig. S2A in the supplemental material). Among the Salmonella serotype Typhimurium isolates, we detected 28 unique isolates as well as 29 combined PFGE and MLVA clusters; 17 of these clusters contained human and cattle isolates (Fig. S2B).
To further characterize the subtype distribution, we analyzed the frequency distribution of isolates with PFGE patterns that were detected among both bovine and human isolates. We identified 17 PFGE patterns, 7 among Salmonella serotype Newport isolates and 10 among Salmonella serotype Typhimurium isolates, that were detected among both human and bovine isolates. Among the multidrug-resistant isolates, 13 PFGE patterns were shared between cattle and humans, 6 PFGE patterns among Salmonella serotype Newport isolates, and 7 patterns among Salmonella serotype Typhimurium isolates, representing a total of 139 isolates. Among the susceptible isolates, 8 PFGE patterns, representing 17 isolates, were shared between humans and cattle.
Distribution of isolates between the Pacific Northwest and the northeastern United States.
To better understand the subtype distribution between the two geographic regions in the United States, we analyzed the distribution of isolates with PFGE patterns that were detected in both geographic regions. Two of the 10 Salmonella serotype Newport combined PFGE and MLVA clusters and 18 of the 29 Salmonella serotype Typhimurium combined PFGE and MLVA clusters contained isolates from both geographic regions (see Fig. S2 in the supplemental material). Five PFGE patterns were identified among isolates from both geographic regions, including 4 patterns associated with Salmonella serotype Newport isolates and 1 pattern associated with Salmonella serotype Typhimurium isolates (data not shown). All PFGE patterns that were detected in both geographic regions were also associated with both isolates from cattle and humans, indicating that these Salmonella subtypes are probably widely distributed among cattle and humans across the United States.
Among the Salmonella serotype Typhimurium isolates, 5 PFGE patterns were shared between resistant and susceptible isolates (i.e., L15, L2, L21, L25, and L36) including one PFGE pattern (L25) found among susceptible and multidrug-resistant isolates in both geographic regions. Three PFGE patterns associated with multidrug-resistant Salmonella serotype Newport isolates (L70, L71, and L90) and one PFGE pattern associated with a susceptible Salmonella serotype Newport isolate (L90) were detected in both geographic regions.
DISCUSSION
A total of 425 Salmonella isolates from two geographic regions in the United States, the Pacific Northwest and the Northeast, were characterized by PFGE and MLVA analysis to obtain a better understanding of genetic diversity, epidemiology, and antimicrobial resistance among Salmonella serotype Newport, Typhimurium, and 4,5,12:i:− isolates from humans and cattle. Overall, our data showed the following. (i) Multidrug resistance is significantly more prevalent among isolates associated with cattle than isolates associated with humans but differs considerably by serotype and geographic region of the United States. (ii) Salmonella subtypes persist on farms, although diversification of molecular subtypes and resistance types may potentially occur over short time periods. (iii) Genetic diversity differs considerably among Salmonella serotypes and between Salmonella isolates from human and bovine host populations. (iv) The combination of PFGE with MLVA or antimicrobial resistance patterns can increase subtype discrimination for selected Salmonella serotypes.
Multidrug resistance is significantly more prevalent among cattle-associated isolates than human-associated isolates, but it differs considerably by serotype and geographic region of the United States.
The high frequency of antimicrobial resistance among Salmonella isolates from predominantly clinically sick cattle observed here is consistent with previous reports (18). These trends are also comparable to those reported by the National Antimicrobial Resistance Monitoring System (NARMS) in the United States, but the frequency of antimicrobial resistance observed in our study was considerably higher than that reported by NARMS, especially for human isolates (45). NARMS reported that approximately 62% of human Salmonella serotype isolates and 32% of cattle Salmonella serotype Typhimurium isolates, as well as 83% of human and 17% of cattle Newport isolates collected in 2006 were susceptible to all tested antimicrobial drugs (45). By comparison, we found much lower frequencies of pansusceptible isolates, with 23 and 18% of human and cattle Salmonella serotype Typhimurium isolates and 22 and 6% of human and cattle Salmonella serotype Newport isolates being pansusceptible, respectively. Most likely, these differences represent differences in the geographic origin of isolates between the NARMS report and our study. Geographic differences in the prevalence of antimicrobial resistance among Salmonella isolates have been described previously, and our study characterized isolates from only two states, while NARMS data are based on isolates from all 50 U.S. states. This is important, as Greene et al. (29) reported a positive correlation between cattle density per acre and the prevalence of multidrug resistance among human Salmonella serotype Newport isolates, and New York and Washington specifically represent two states with high cattle density and comparatively high prevalence of MDR as reported by Greene et al. (29). Furthermore, Salmonella isolates originating from New York City residents are generally submitted to the New York City Department of Health and were therefore only very rarely included in this study, potentially resulting in sampling biased toward rural human populations. In addition to different sampling schemes, differences in the antimicrobial drugs tested and the testing method used may also have contributed to the high frequency of drug resistance reported here. Importantly, a considerable number of human isolates in our study were resistant only to triple-sulfa, a combination of sulfonamides not tested for by NARMS. These isolates would hence likely be reported as pansusceptible by NARMS. While our study and NARMS used different resistance testing methods (i.e., broth microdilution versus disc diffusion), a previous study has reported good correspondence between resistance typing results based on these two methods, at least for Salmonella serotype Heidelberg, despite minor differences, suggesting a limited effect of these methodological differences (48).
Resistance to ACSSuT as well as to amoxicillin-clavulanic acid and ceftiofur was particularly prevalent in our study, but resistance to other antimicrobial agents, such as kanamycin or the combination of sulfisoxazole-trimethoprim, was also relatively common, indicating that the MDR subtypes acquired additional antimicrobial resistance genes, a phenomenon that has been described previously (27). The high prevalence of resistance to ceftazidime, an expanded-spectrum cephalosporin, reported in previous studies and confirmed here, is of particular concern, as extended-spectrum cephalosporins are the drugs of choice for treatment of nontyphoidal salmonellosis in humans if fluoroquinolones are contraindicated (31, 54).
In our study, MDR and susceptible isolates were generally characterized by distinct MLVA and PFGE subtypes, particularly for Salmonella serotype Newport. Multidrug resistance was significantly more common among bovine than human isolates, especially for Salmonella serotype Newport, with the odds of multidrug resistance approximately 18 times higher for bovine than human Newport isolates. While differences in sampling and surveillance between human and cattle isolates have to be considered, these factors are unlikely to have distorted the study significantly, as both human and cattle isolates originated almost exclusively from cases with clinical signs typical of salmonellosis. Rather, our findings suggest a significantly higher frequency of multidrug resistance among cattle than human Salmonella isolates, supporting the hypothesis that cattle may represent one possible reservoir for multidrug-resistant Salmonella and that cattle may serve as a source for emerging MDR strains (see references 2 and 66 for a review of this topic). This conclusion is corroborated by findings of other studies which indicate that consumption of beef or dairy products, as well as dairy farm contact, represent important risk factors for human Salmonella serotype Newport MDR-AmpC infection (74). Moreover, a number of human Salmonella serotype Newport MDR-AmpC outbreaks have been linked to beef or dairy product consumption, and a role of cattle as reservoir for multidrug-resistant Salmonella serotype Newport has been proposed previously (14). Bovine Salmonella serotype Typhimurium isolates, however, were also significantly more likely to be multidrug resistant than human isolates. Similar observations have also been reported by Busani et al. (9), who reported a significantly lower prevalence of resistance to ACSSuT among Salmonella serotype Typhimurium isolates from humans than from nonhuman sources and specifically detected a comparably high prevalence of the ACSSuT resistance type among serotype Typhimurium isolates from cattle. Differences in antimicrobial resistance prevalence between isolates from humans and animals were not formally tested in that study though, and resistance prevalence was highest among environmental samples. Somewhat higher resistance prevalence among animal than human Salmonella serotype Typhimurium isolates for some antimicrobial drugs has also been reported in other studies, and a role of cattle and other domestic animals as reservoir for human infection with multidrug-resistant Salmonella serotype Typhimurium strains has been proposed repeatedly (12, 13, 51, 52).
We also detected a significant effect of geographic region, with the odds of multidrug resistance approximately twice as high for isolates from the Pacific Northwest as for isolates from the Northeast. Among the cattle isolates, sampling differences between the two study sites may have contributed to this observation, with cattle isolates from the Northeast originating from herds enrolled in an active surveillance and sampling project, thereby potentially increasing the likelihood of detecting milder clinical cases or including animals exhibiting signs caused by other diseases (18). However, our findings are consistent with a previous study which reported an association between prevalence of multidrug resistance among human Salmonella serotype Newport isolates and cattle density per acres of farmland, even though in this previous study (29), the highest resistance prevalence was observed in the northeastern United States, particularly in Maine and Connecticut. Salmonella prevalence in dairy herds is known to vary considerably by geographic region and two 2002 studies found a considerably lower Salmonella prevalence in dairy herds in the northeastern United States compared to the western United States, potentially due to smaller average herd sizes in the Northeast (3, 11). While our data thus suggest a possible correlation between Salmonella prevalence and antimicrobial resistance prevalence, further studies will be needed to test this hypothesis and to better define risk factors for high prevalence of multidrug-resistant Salmonella.
Salmonella subtypes persist on farms, although diversification of molecular subtypes and resistance types may potentially occur over short time periods.
In our study, specific Salmonella strains appear to have persisted on farms for considerable amounts of time, with isolates characterized by indistinguishable MLVA, PFGE, and resistance profiles isolated from the same farm over periods of more than 7 months. Salmonella persistence on farms has been a consistent finding, but the observed duration of persistence varies (17). In our study, all but 17 isolates were collected from cattle with clinical signs of salmonellosis. As differences between herds with and without clinical signs of salmonellosis (for instance, differences in serotype distribution or Salmonella prevalence) have been reported, differences in duration of persistence also appear likely (18). The generalizability of our results may therefore be restricted to herds with clinical cases of salmonellosis.
On some farms, we collected isolates with different molecular patterns, typically closely related PFGE, MLVA, or antimicrobial resistance patterns. For example, Salmonella isolates from farm NE261 could be grouped into two clusters based on PFGE and antimicrobial resistance patterns, while MLVA results indicated that isolates from both clusters were probably closely related. The molecular methods applied in this study do not allow for definitive inferences concerning the underlying genetic events. However, the fact that prior to August 2004, the predominant persistent Salmonella serotype 4,5,12:i:− strain on farm NE261 was multidrug resistant, while a pansusceptible 4,5,12:i:− strain subsequently appeared to become most prevalent on this farm, paired with the observation that the new strain was characterized by a PFGE pattern that differed by the absence of one band and the presence of three new bands compared to the initial strain could be explained by the loss of a mobile element encoding multidrug resistance determinants. Importantly, one isolate from the same farm, collected in early September, was also pansusceptible but contained two bands unique to the cluster of resistant isolates while lacking one band unique to the pansusceptible cluster and one band unique to the multidrug-resistant cluster, consistent with a second molecular process, potentially the insertion or excision of a bacteriophage. The possibility that this and other similar observations represent a reintroduction of closely related Salmonella subtypes onto a given premise or that multiple closely related subtypes coexisted on the same farm without being detected initially cannot be excluded. However, this example illustrates the possibility of considerable diversification of Salmonella strains during persistence on farms or in production environments, which can complicate molecular epidemiological investigations (for instance, see references 43 and 69).
Genetic diversity differs considerably among Salmonella serotypes and between Salmonella isolates from human and bovine host populations.
In this study, Salmonella serotype Typhimurium isolates seemed more genetically diverse than Salmonella serotype Newport isolates, and differences were particularly pronounced among isolates from cattle. These observations agree with previous studies, which report a clonal nature of Salmonella serotype Newport subtype MDR-AmpC (7, 31, 33). Salmonella serotype Newport MDR-AmpC appears predominantly associated with cattle but has also been isolated from humans and a variety of other host species (see reference 10 for a review of this topic). In general, we found cattle isolates to be less genetically diverse than human isolates regardless of the serotype analyzed. These observations may be due to sampling differences, especially since cattle isolates originated from a limited number of farms, but the results appear to agree with previous study results, which show somewhat lower genetic diversity among animal Salmonella isolates than human Salmonella isolates, likely reflecting the fact that human infections can occur from a variety of sources, which are expected to harbor a considerable variety of Salmonella subtypes (28, 51, 59). However, future studies are needed to reliably capture the genetic diversity in Salmonella populations circulating among humans and cattle.
Combination of PFGE with MLVA or antimicrobial resistance patterns can increase subtype discrimination for selected Salmonella serotypes.
In our study, PFGE showed a high discriminatory power for all serotypes tested, supporting the value of this subtyping method (6, 16, 33). However, for Salmonella serotype Typhimurium, the discriminatory power of MLVA was significantly higher than that of PFGE, while for Salmonella serotype Newport, the discriminatory power of both methods was comparable, consistent with previous reports (19, 42, 67).
We detected moderate correspondence between isolate clustering based on PFGE and MLVA, also consistent with previous studies (19, 76). PFGE patterns can be impacted by a variety of genetic events, including plasmid gain or loss, transposition, integration, or loss of mobile genetic elements, such as bacteriophages, and point mutation (16). MLVA, in contrast, detects copy number differences in tandem repeat DNA motifs, which result from slipped-strand mispairing during DNA replication (73). VNTR mutation rates seem to depend on a variety of factors, including the number of tandem repeats and nucleotide composition, and variability appears to differ between VNTR loci (39, 49). MLVA and PFGE patterns are therefore affected by different evolutionary mechanisms, and results have to be interpreted accordingly. Clustering confirmed by both subtyping methods provides strong evidence for true genetic relatedness, while discordant results can be difficult to interpret. Similar to our study which suggested diversification of both PFGE and MLVA patterns on individual farms, the diversification of PFGE and MLVA patterns has been reported during human salmonellosis outbreaks, even though diversification rates are difficult to estimate (23, 50, 64, 67). In such instances, the use of additional information, e.g., epidemiologic data, antimicrobial resistance patterns, or prior knowledge of subtype diversity, can be very useful for inferring relationships between isolates. In fact, while in our study both antimicrobial resistance and MLVA patterns seemed to diversify during Salmonella persistence on farms, resistance patterns appeared to be less variable, and among isolates from the same farm and of the same PFGE type, resistance patterns varied in at most one antimicrobial drug. Moreover, in our study, PFGE and antimicrobial resistance-based clustering was occasionally more consistent with epidemiologic data than MLVA-based clustering, as seen for farm NE229. Combining PFGE with antimicrobial resistance profiles also yielded a considerable gain in discriminatory power over that of PFGE alone. Antimicrobial resistance data have contributed useful information in several epidemiologic investigations, but to our knowledge, our study is the first to formally quantify the gain in discriminatory power for three common Salmonella serotypes (7, 8, 64). In our analysis as well as in previous studies, however, the combination of PFGE and MLVA provided the highest discriminatory power (19, 42, 67). Overall, these observations further reinforce the need to choose the Salmonella subtyping methods most appropriate for a given study goal, while additional factors, such as subtype diversity or the availability of serotype-specific protocols, have to be taken into consideration (8, 67).
Even though MLST does not generally provide high discriminatory power within serotypes, we found sequence typing of a single gene sufficient to distinguish between the two Salmonella serotype Newport lineages (5). Importantly, MLST-based lineage attribution corresponded well with PFGE-based lineage attribution, corroborating previous findings (1, 33). Since Salmonella serotype Newport lineages A and B are associated with different animal hosts, lineage determination can provide useful insights for serotype Newport source attribution, thereby potentially allowing source attribution for the important fraction of human Salmonella infections for which the source currently remains unknown (32, 71). The ability to predict Salmonella serotype Newport lineage based on single-gene sequencing or PFGE as shown here can thus potentially contribute to more cost-effective and rapid Salmonella serotype Newport source attribution.
Conclusions.
Our study provides support for the hypothesis that cattle may represent a reservoir for multidrug-resistant Salmonella and may serve as a source for emerging MDR strains. However, the importance of that role may depend on a variety of factors, including serotype and geographic origin. Our data show that Salmonella can persist on individual dairy farms for prolonged periods of time, potentially with diversification of some subtype patterns over time. In some instances, new antimicrobial resistance may be acquired or lost during persistence, and interpretation of epidemiologic links based exclusively on subtyping results can be complicated. While PFGE and MLVA are highly reliable and generally extremely useful, they can be affected by a variety of genetic events, and discordant results must be interpreted carefully. Some Salmonella subtypes appear to be widespread among cattle and humans across the United States, but most appear to be associated with a specific geographic area and, to a lesser extent, host species.
Supplementary Material
Acknowledgments
Support for this project was provided by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract numbers N01-AI-30054-ZC-006-07 and N01-A1-30055. K.H. was supported by Morris Animal Foundation fellowship training grant D08FE-403.
Footnotes
Published ahead of print on 16 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alcaine, S. D., Y. Soyer, L. D. Warnick, W. L. Su, S. Sukhnanand, J. Richards, E. D. Fortes, P. McDonough, T. P. Root, N. B. Dumas, Y. Grohn, and M. Wiedmann. 2006. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl. Environ. Microbiol. 72:7575-7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcaine, S. D., L. D. Warnick, and M. Wiedmann. 2007. Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 70:780-790. [DOI] [PubMed] [Google Scholar]
- 3.Animal and Plant Inspection Service. 2005. Salmonella on U.S. dairy operations: prevalence and antimicrobial drug susceptibility. APHIS info sheet. Document N435.1005 Animal and Plant Inspection Service, Veterinary Services, Centers for Epidemiology and Animal Health, U.S. Department of Agriculture, Fort Collins, CO.
- 4.Bauer, A. W., W. M. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 5.Belkum, A. V., P. T. Tassios, L. Dijkshoorn, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, and M. Struelens, and the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM). 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3):1-46. [DOI] [PubMed] [Google Scholar]
- 6.Bender, J. B., C. W. Hedberg, D. J. Boxrud, J. M. Besser, J. H. Wicklund, K. E. Smith, and M. T. Osterholm. 2001. Use of molecular subtyping in surveillance for Salmonella enterica serotype Typhimurium. N. Engl. J. Med. 344:189-195. [DOI] [PubMed] [Google Scholar]
- 7.Berge, A. C. B., J. M. Adaska, and W. M. Sischo. 2004. Use of antibiotic susceptibility patterns and pulsed-field gel electrophoresis to compare historic and contemporary isolates of multidrug-resistant Salmonella enterica subsp. enterica serovar Newport. Appl. Environ. Microbiol. 70:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Best, E. L., M. D. Hampton, S. Ethelberg, E. Liebana, F. A. Clifton-Hadley, and E. J. Threlfall. 2009. Drug-resistant Salmonella Typhimurium DT 120: use of PFGE and MLVA in a putative international outbreak investigation. Microb. Drug Resist. 15:133-138. [DOI] [PubMed] [Google Scholar]
- 9.Busani, L., C. Graziani, A. Battisti, A. Franco, A. Ricci, D. Vio, E. Digiannatale, F. Paterlini, M. D'Incau, S. Owczarek, A. Caprioli, and I. Luzzi. 2004. Antibiotic resistance in Salmonella enterica serotypes Typhimurium, Enteritidis and Infantis from human infections, foodstuffs and farm animals in Italy. Epidemiol. Infect. 132:245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butaye, P., G. B. Michael, S. Schwarz, T. J. Barrett, A. Brisabois, and D. G. White. 2006. The clonal spread of multidrug-resistant non-typhi Salmonella serotypes. Microbes Infect. 8:1891-1897. [DOI] [PubMed] [Google Scholar]
- 11.Callaway, T. R., J. E. Keen, T. S. Edrington, L. H. Baumgard, L. Spicer, E. S. Fonda, K. E. Griswold, T. R. Overton, M. E. Van Amburgh, R. C. Anderson, K. J. Genovese, T. L. Poole, R. B. Harvey, and D. J. Nisbet. 2005. Fecal prevalence and diversity of Salmonella species in lactating dairy cattle in four states. J. Dairy Sci. 88:3603-3608. [DOI] [PubMed] [Google Scholar]
- 12.Casin, I., J. Breuil, A. Brisabois, F. D. R. Moury, F. Grimont, and E. Collatz. 1999. Multidrug resistant human and animal Salmonella Typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded beta-lactamase PSE-1. J. Infect. Dis. 179:1173-1182. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 1997. Multidrug-resistant Salmonella serotype Typhimurium-United States, 1996. MMWR Morb. Mortal. Wkly. Rep. 46:308-310. [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. 2002. Outbreak of multidrug-resistant Salmonella Newport-United States, January-April 2002. MMWR Morb. Mortal. Wkly. Rep. 51:545-548. [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2008. Salmonella surveillance: annual summary, 2006. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, GA.
- 16.Cooke, F. J., D. J. Brown, M. Fookes, D. Pickard, A. Ivens, J. Wain, M. Roberts, R. A. Kingsley, N. R. Thomson, and G. Dougan. 2008. Characterization of the genomes of a diverse collection of Salmonella enterica serovar Typhimurium definitive phage type 104. J. Bacteriol. 190:8155-8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cummings, K. J., L. D. Warnick, K. A. Alexander, C. J. Cripps, Y. T. Grohn, K. L. James, P. L. McDonough, and K. E. Reed. 2009. The duration of fecal Salmonella shedding following clinical disease among dairy cattle in the northeastern USA. Prev. Vet. Med. 92:134-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummings, K. J., L. D. Warnick, K. A. Alexander, C. J. Cripps, Y. T. Grohn, P. L. McDonough, D. V. Nydam, and K. E. Reed. 2009. The incidence of salmonellosis among dairy herds in the northeastern United States. J. Dairy Sci. 92:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis, M. A., K. N. Baker, D. R. Call, L. D. Warnick, Y. Soyer, M. Wiedmann, Y. Grohn, P. L. McDonough, D. D. Hancock, and T. E. Besser. 2009. Multilocus variable-number tandem-repeat method for typing Salmonella enterica serovar Newport. J. Clin. Microbiol. 47:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyle, M. E., C. Kaspar, J. Archer, and R. Klos. 2009. White paper on human illness caused by Salmonella from all food and non-food vectors. FRI Briefings. Food Research Institute, University of Wisconsin—Madison, Madison, WI. http://fri.wisc.edu/briefs/FRI_Brief_Salmonella_Human_Illness_6_09.pdf.
- 21.Echeita, M. A., S. Herrera, and M. A. Usera. 2001. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:− appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 39:2981-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endtz, H. P., G. J. Ruijs, B. van Klingeren, W. H. Jansen, T. van der Reyden, and R. P. Mouton. 1991. Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J. Antimicrob. Chemother. 27:199-208. [DOI] [PubMed] [Google Scholar]
- 23.Ethelberg, S., G. Sorensen, B. Kristensen, K. Christensen, L. Krusell, A. Hempel-Jorgensen, A. Perge, and E. M. Nielsen. 2007. Outbreak with multi-resistant Salmonella Typhimurium DT104 linked to carpaccio, Denmark, 2005. Epidemiol. Infect. 135:900-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewing, W. H. 1972. The nomenclature of Salmonella, its usage, and definitions for the three species. Can. J. Microbiol. 18:1629-1637. [DOI] [PubMed] [Google Scholar]
- 25.Fish, N. A., M. C. Finlayson, and R. P. Carere. 1967. Salmonellosis: report of a human case following direct contact with infected cattle. Can. Med. Assoc. J. 96:1163-1165. [PMC free article] [PubMed] [Google Scholar]
- 26.Gebreyes, W. A., S. Thakur, P. Dorr, D. A. Tadesse, K. Post, and L. Wolf. 2009. Occurrence of spvA virulence gene and clinical significance for multidrug-resistant Salmonella strains. J. Clin. Microbiol. 47:777-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glynn, M. K., C. Bopp, W. Dewitt, P. Dabney, M. Mokhtar, and F. J. Angulo. 1998. Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333-1338. [DOI] [PubMed] [Google Scholar]
- 28.Gorman, R., and C. C. Adley. 2004. Characterization of Salmonella enterica serotype Typhimurium isolates from human, food, and animal sources in the Republic of Ireland. J. Clin. Microbiol. 42:2314-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greene, S. K., A. M. Stuart, F. M. Medalla, J. M. Whichard, R. M. Hoekstra, and T. M. Chiller. 2008. Distribution of multidrug-resistant human isolates of MDR-ACSSuT Salmonella Typhimurium and MDR-AmpC Salmonella Newport in the United States, 2003-2005. Foodborne Pathog. Dis. 5:669-680. [DOI] [PubMed] [Google Scholar]
- 30.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta, A., J. Fontana, C. Crowe, B. Bolstorff, A. Stout, S. Van Duyne, M. P. Hoekstra, J. M. Whichard, T. J. Barrett, and F. J. Angulo. 2003. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 188:1707-1716. [DOI] [PubMed] [Google Scholar]
- 32.Hald, T., D. M. Lo Fo Wong, and F. M. Aarestrup. 2007. The attribution of human infections with antimicrobial resistant Salmonella bacteria in Denmark to sources of animal origin. Foodborne Pathog. Dis. 4:313-326. [DOI] [PubMed] [Google Scholar]
- 33.Harbottle, H., D. G. White, P. F. McDermott, R. D. Walker, and S. Zhao. 2006. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J. Clin. Microbiol. 44:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helms, M., P. Vastrup, P. Gerner-Smidt, and K. Molbak. 2002. Excess mortality associated with antimicrobial drug-resistant Salmonella Typhimurium. Emerg. Infect. Dis. 8:490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmberg, S. D., J. G. Wells, and M. L. Cohen. 1984. Animal-to-man transmission of antimicrobial-resistant Salmonella: investigations of U.S. outbreaks, 1971-1983. Science 225:833-835. [DOI] [PubMed] [Google Scholar]
- 36.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella Typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 39.Lai, Y., and F. Sun. 2003. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 20:2123-2131. [DOI] [PubMed] [Google Scholar]
- 40.Lawson, A. J., M. Desai, S. J. O'Brien, R. H. Davies, L. R. Ward, and E. J. Threlfall. 2004. Molecular characterisation of an outbreak strain of multiresistant Salmonella enterica serovar Typhimurium DT104 in the UK. Clin. Microbiol. Infect. 10:143-147. [DOI] [PubMed] [Google Scholar]
- 41.Lee, L. A., N. D. Puhr, E. K. Maloney, N. H. Bean, and R. V. Tauxe. 1994. Increase in antimicrobial-resistant Salmonella infections in the United States, 1989-1990. J. Infect. Dis. 170:128-134. [DOI] [PubMed] [Google Scholar]
- 42.Lindstedt, B. A., E. Heir, E. Gjernes, and G. Kapperud. 2003. DNA fingerprinting of Salmonella enterica subsp. enterica serovar Typhimurium with emphasis on phage type DT104 based on variable number of tandem repeat loci. J. Clin. Microbiol. 41:1469-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Urtaza, J., and E. Liebana. 2005. Use of pulsed-field gel electrophoresis to characterize the genetic diversity and clonal persistence of Salmonella Senftenberg in mussel processing facilities. Int. J. Food Microbiol. 105:153-163. [DOI] [PubMed] [Google Scholar]
- 44.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Antimicrobial Resistance Monitoring System. 2009. National Antimicrobial Resistance Monitoring System—enteric bacteria (NARMS): 2006 Executive Report. Food and Drug Administration, U.S. Department of Health and Human Services, Rockville, MD.
- 46.National Committee for Clinical Laboratory Standards. 2003. Approved standard M2-A8. Performance standards for antimicrobial disk susceptibility tests, 8th ed. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 47.National Committee for Clinical Laboratory Standards. 2003. M100-S13(M2). Disk diffusion supplemental tables. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 48.Nayak, R., V. Call, P. Kaldhone, C. Tyler, G. Anderson, S. Phillips, K. Kerdahi, and S. L. Foley. 2007. Comparison of Salmonella enterica serovar Heidelberg susceptibility testing results. Clin. Med. Res. 5:98-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noller, A. C., M. C. McEllistrem, K. A. Shutt, and L. H. Harrison. 2006. Locus-specific mutational events in a multilocus variable-number tandem repeat analysis of Escherichia coli O157:H7. J. Clin. Microbiol. 44:374-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nygard, K., B. A. Lindstedt, W. Wahl, L. Jensvoll, C. Kjelso, K. Molbak, M. Torpdahl, and G. Kapperud. 2007. Outbreak of Salmonella Typhimurium infection traced to imported cured sausage using MLVA-subtyping. Euro Surveill. 12:E070315.5. [DOI] [PubMed] [Google Scholar]
- 51.Oloya, J., D. Doetkott, and M. L. Khaitsa. 2009. Antimicrobial drug resistance and molecular characterization of Salmonella isolated from domestic animals, humans, and meat products. Foodborne Pathog. Dis. 6:273-284. [DOI] [PubMed] [Google Scholar]
- 52.Poppe, C., N. Smart, R. Khakhria, W. Johnson, J. Spika, and J. Prescott. 1998. Salmonella Typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39:559-565. [PMC free article] [PubMed] [Google Scholar]
- 53.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 54.Rankin, S. C., H. Aceto, J. Cassidy, J. Holt, S. Young, B. Love, D. Tewari, D. S. Munro, and C. E. Benson. 2002. Molecular characterization of cephalosporin-resistant Salmonella enterica serotype Newport isolates from animals in Pennsylvania. J. Clin. Microbiol. 40:4679-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 56.Ross, I. L., and M. W. Heuzenroeder. 2009. A comparison of two PCR-based typing methods with pulsed-field gel electrophoresis in Salmonella enterica serovar Enteritidis. Int. J. Med. Microbiol. 299:410-420. [DOI] [PubMed] [Google Scholar]
- 57.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 58.Smith, K. E., J. M. Besser, C. W. Hedberg, F. T. Leano, J. B. Bender, J. H. Wicklund, B. P. Johnson, K. A. Moore, and M. T. Osterholm for the Investigation Team. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N. Engl. J. Med. 340:1525-1532. [DOI] [PubMed] [Google Scholar]
- 59.Soyer, Y., S. D. Aclaine, D. Schoonmaker-Bopp, L. D. Warnick, P. McDonough, N. B. Dumas, Y. T. Grohn, and M. Widemann. 2010. Pulse field gel electrophoresis diversity of human and bovine clinical Salmonella 2010 pulse field gel electrophoresis diversity of human and bovine clinical Salmonella isolates. Foodborne Pathog. Dis. 7:707-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soyer, Y., A. Moreno Switt, M. A. Davis, J. Maurer, P. L. McDonough, D. J. Schoonmaker-Bopp, N. B. Dumas, T. Root, L. D. Warnick, Y. T. Grohn, and M. Wiedmann. 2009. Salmonella 4,5,12:i:−: an emerging Salmonella serotype that represents multiple distinct clones. J. Clin. Microbiol. 47:3546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steinmuller, N., L. Demma, J. B. Bender, M. Eidson, and F. J. Angulo. 2006. Outbreaks of enteric disease associated with animal contact: not just a foodborne problem anymore. Clin. Infect. Dis. 43:1596-1602. [DOI] [PubMed] [Google Scholar]
- 62.Switt, A. I., Y. Soyer, L. D. Warnick, and M. Wiedmann. 2009. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i. Foodborne Pathog. Dis. 6:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taguchi, M., K. Seto, M. Kanki, T. Tsukamoto, H. Izumiya, and H. Watanabe. 2005. Outbreak of food poisoning caused by lunch boxes prepared by a company contaminated with multidrug resistant Salmonella Typhimurium DT104. Jpn. J. Infect. Dis. 58:55-56. [PubMed] [Google Scholar]
- 64.Tassios, P. T., C. Chadjichristodoulou, M. Lambiri, A. Kansouzidou-Kanakoudi, Z. Sarandopoulou, J. Kourea-Kremastinou, L. S. Tzouvelekis, and N. J. Legakis. 2000. Molecular typing of multidrug-resistant Salmonella Blockley outbreak isolates from Greece. Emerg. Infect. Dis. 6:60-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Threlfall, E. J. 2000. Epidemic Salmonella Typhimurium DT 104-a truly international multiresistant clone. J. Antimicrob. Chemother. 46:7-10. [DOI] [PubMed] [Google Scholar]
- 66.Tollefson, L., and B. E. Karp. 2004. Human health impact from antimicrobial use in food animals. Med. Mal. Infect. 34:514-521. [PubMed] [Google Scholar]
- 67.Torpdahl, M., G. Sorensen, B. A. Lindstedt, and E. M. Nielsen. 2007. Tandem repeat analysis for surveillance of human Salmonella Typhimurium infections. Emerg. Infect. Dis. 13:388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Travers, K., and M. Barza. 2002. Morbidity of infections caused by antimicrobial-resistant bacteria. Clin. Infect. Dis. 34(Suppl. 3):S131-S134. [DOI] [PubMed] [Google Scholar]
- 69.Uesugi, A. R., M. D. Danyluk, R. E. Mandrell, and L. J. Harris. 2007. Isolation of Salmonella Enteritidis phage type 30 from a single almond orchard over a 5-year period. J. Food Prot. 70:1784-1789. [DOI] [PubMed] [Google Scholar]
- 70.U.S. Census Bureau. June 2010, accession date. Annual estimates of the resident population for the United States, regions, states, and Puerto Rico: April 1, 2000 to July 1, 2009. NST-EST2009-01. Population Division, U.S. Census Bureau, Washington, DC. http://www.census.gov/popest/states/NST-ann-est.html.
- 71.U.S. Department of Agriculture. 2007. A statistical model for attributing human salmonellosis to meat, poultry, and eggs. Food Safety Summit, Arlington, Virginia, April 2007. Food Safety Inspection Services, U.S. Department of Agriculture, Washington, DC. http://www.fsis.usda.gov/PPT/RBI_guo.ppt#261,2,Attributing Human Salmonellosis to Food Sources.
- 72.Valdezate, S., M. Arroyo, R. Gonzalez-Sanz, R. Ramiro, S. Herrera-Leon, M. A. Usera, M. De la Fuente, and A. Echeita. 2007. Antimicrobial resistance and phage and molecular typing of Salmonella strains isolated from food for human consumption in Spain. J. Food Prot. 70:2741-2748. [DOI] [PubMed] [Google Scholar]
- 73.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varma, J. K., R. Marcus, S. A. Stenzel, S. S. Hanna, S. Gettner, B. J. Anderson, T. Hayes, B. Shiferaw, T. L. Crume, K. Joyce, K. E. Fullerton, A. C. Voetsch, and F. J. Angulo. 2006. Highly resistant Salmonella Newport-MDRAmpC transmitted through the domestic US food supply: a FoodNet case-control study of sporadic Salmonella Newport infections, 2002-2003. J. Infect. Dis. 194:222-230. [DOI] [PubMed] [Google Scholar]
- 75.Voetsch, A. C., T. J. Van Gilder, F. J. Angulo, M. M. Farley, S. Shallow, R. Marcus, P. R. Cieslak, V. C. Deneen, and R. V. Tauxe. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl. 3):S127-S134. [DOI] [PubMed] [Google Scholar]
- 76.Witonski, D., R. Stefanova, A. Ranganathan, G. E. Schutze, K. D. Eisenach, and M. D. Cave. 2006. Variable-number tandem repeats that are useful in genotyping isolates of Salmonella enterica subsp. enterica serovars Typhimurium and Newport. J. Clin. Microbiol. 44:3849-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.