Abstract
In the fungus Penicillium chrysogenum, penicillin (PEN) production is compartmentalized in the cytosol and in peroxisomes. Here we show that intact peroxisomes that contain the two final enzymes of PEN biosynthesis, acyl coenzyme A (CoA):6-amino penicillanic acid acyltransferase (AT) as well as the side-chain precursor activation enzyme phenylacetyl CoA ligase (PCL), are crucial for efficient PEN synthesis. Moreover, increasing PEN titers are associated with increasing peroxisome numbers. However, not all conditions that result in enhanced peroxisome numbers simultaneously stimulate PEN production. We find that conditions that lead to peroxisome proliferation but simultaneously interfere with the normal physiology of the cell may be detrimental to antibiotic production. We furthermore show that peroxisomes develop in germinating conidiospores from reticule-like structures. During subsequent hyphal growth, peroxisome proliferation occurs at the tip of the growing hyphae, after which the organelles are distributed over newly formed subapical cells. We observed that the organelle proliferation machinery requires the dynamin-like protein Dnm1.
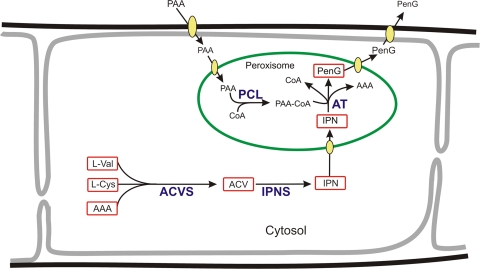
Penicillins (PENs) belong to the group of β-lactam antibiotics that are produced as secondary metabolites by specific actinomycetous bacteria and fungal species (26). For the industrial production of PEN, the filamentous fungus Penicillium chrysogenum is used. The biosynthesis of penicillin G (PenG) has been characterized in detail at the genetic and biochemical levels using P. chrysogenum and a related fungus, Aspergillus nidulans, as model organisms (7, 28). Starting from three amino acids, α-amino adipic acid, cysteine, and valine, PenG is formed in three unique enzymatic conversions (Fig. 1). These amino acids are first condensed to a tripeptide mediated by the function of a nonribosomal peptide synthetase, δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine (ACV) synthetase (ACVS). The resulting tripeptide, ACV, is cyclized by isopenicillin N synthase (IPNS) to form a β-lactam, isopenicillin N (IPN). As a final step, the enzyme acyl coenzyme A (CoA):6-amino penicillanic acid acyltransferase (AT) replaces the α-aminoadipyl side chain of IPN with a more hydrophobic one. In industrial fermentations, phenylacetic acid (PAA) or phenoxyacetic acid (POA) is applied to produce PenG or penicillin V (PenV), respectively.
FIG. 1.
Schematic overview of the penicillin biosynthetic pathway. ACVS, δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase; IPNS, isopenicillin N synthase; AT, acyl-CoA:6-amino penicillanic acid acyltransferase; PCL, phenylacetyl CoA ligase; PAA, phenylacetic acid.
In filamentous fungi, the PEN biosynthetic machinery is compartmentalized (Fig. 1). The first two enzymes, ACVS and IPNS, are both located in the cytosol (19, 32). As the pH of the cytosol in filamentous fungi is between 6.5 and 7.0 (9, 31), these enzymes are in their optimal physiological surroundings. The AT and phenylacetyl CoA ligase (PCL) enzymes have specific targeting sequences that sort these enzymes to the lumen of their target compartment, the peroxisome (18, 19). The pH of this organelle was shown to be 7.5, which is close to the pH optima of both AT and PCL (31). Apparently, the compartmentalization of these enzymes creates defined microenvironments and enables the generation of favorable substrate and cofactor concentrations for enzyme function.
Peroxisomes (belonging to the family of microbodies) are ubiquitously present in eukaryotic cells. They typically consist of a protein-rich matrix surrounded by a single membrane and are 0.1 to 1 μm in size. Although their function is often species and cell type specific, two widely distributed functions can be distinguished, namely, H2O2 metabolism and β-oxidation of fatty acids (for reviews, see references 25, 29, and 30). Muller et al. (18, 19) demonstrated the role of peroxisomes in PEN biosynthesis for the first time. Subsequently, it was speculated that a correlation may exist between the volume fraction of these organelles and PEN production rates (18, 27). This speculation was reinforced by Kiel and colleagues (13), who showed that the artificial proliferation of peroxisomes via the overexpression of the pex11 gene was associated with a 2- to 3-fold increase in PEN production rates. Here we further elaborate on these studies and show that peroxisomes de facto are required for efficient PEN biosynthesis in P. chrysogenum. In addition, we present details on the origin and subsequent partitioning of the organelles over newly formed subapical cells during hyphal development.
MATERIALS AND METHODS
Strains and growth conditions.
The P. chrysogenum strains used in this study are listed in Table 1. NRRL1951, Wis54-1255, and DS17690 derivatives were grown, as described previously (15), on PEN production (PP) medium supplemented with 0.25% (wt/vol) PAA or POA. In specific experiments, 0.1% oleic acid and 0.05% Tween 80 were also added to enhance peroxisome proliferation. Growth was done at 25°C at 200 rpm for a maximum of 7 days. Regeneration agar [6.0 ml 87% glycerol, 7.5 ml beet molasses, 5.0 g yeast extract, 18 g NaCl, 0.05 g MgSO4·7H2O, 0.06 g KH2PO4, 1.6 mg NH4Fe(SO4)2·12H2O, 0.1 mg CuSO4·5H2O, 0.25 g CaSO4·2H2O, and 20.0 g Oxoid agar per liter] was used to generate conidiospores from each strain. To identify and characterize non-oleate-utilizing mutants, minimal medium (MM) agar was used with oleic acid (1 or 10 g/liter), lauric acid or hexane (both 1 g/liter), sodium acetate (50 mM), or a mixture of ethanol and glycerol (both 10 g/liter) as the carbon source.
TABLE 1.
P. chrysogenum strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| NRRL1951 (ATCC 9480) | Wild-type P. chrysogenum | American Type Culture Collection |
| NRRL1951 GFP-SKL | NRRL1951 expressing GFP-SKL under the control of PgpdA | This study |
| Wis54-1255 (ATCC 28089) | P. chrysogenum with improved PEN production | American Type Culture Collection |
| Wis54-1255 GFP-SKL | Wis54-1255 expressing GFP-SKL under the control of PgpdA | This study |
| S2201 | Non-oleate-utilizing mutant of Wis54-1255 | This study |
| S2202 | Non-oleate-utilizing mutant of Wis54-1255 | This study |
| S2203 | Non-oleate-utilizing mutant of Wis54-1255 | This study |
| S2204 | Non-oleate-utilizing mutant of Wis54-1255 | This study |
| S2201 GFP-SKL | S2201 expressing GFP-SKL under the control of PgpdA | This study |
| S2202 GFP-SKL | S2202 expressing GFP-SKL under the control of PgpdA | This study |
| S2203 GFP-SKL | S2203 expressing GFP-SKL under the control of PgpdA | This study |
| S2204 GFP-SKL | S2204 expressing GFP-SKL under the control of PgpdA | This study |
| DS17690 | High-PEN-producing P. chrysogenum strain | 8 |
| DS17690 GFP-SKL | DS17690 expressing GFP-SKL under the control of PgpdA | This study |
| DS54465 | DS17690 ΔhdfA with increased homologous recombination | 23 |
| DS54465 GFP-SKL | DS54465 ΔniaD::PgpdA-GFP-SKL-TpenDE | This study |
| DS17690 Pex3-GFP DsRed-SKL | DS17690 expressing Pex3-GFP and DsRed-SKL both under the control of PgpdA | This study |
| Δdnm1 GFP-SKL | DS54465 GFP-SKL Δdnm1::amdS | This study |
| DsRed-SKL | DS17690 with DsRed-SKL under the control of PpcbC | 12 |
| Dnm1++++ | DsRed-SKL with integrated multiple copies of a PpcbC-dnm1-His8-TpenDE cassette | This study |
DNA procedures.
Plasmids and oligonucleotide primers used in this study are indicated in Tables S1 and S2, respectively, in the supplemental material. Standard recombinant DNA manipulations (22) were used throughout this study. PCRs were carried out by using high-fidelity polymerases (Roche). Cloned PCR fragments were sequenced to confirm the correct DNA sequence. For analyses of P. chrysogenum DNA and deduced amino acid sequences, the Clone Manager 5 program (Scientific and Educational Software, Durham, NC) was used.
The transformation of P. chrysogenum protoplasts was performed as described previously (1). Protoplasts were plated onto solid medium containing acetamide as the sole nitrogen source to identify amdS-positive transformants. Single colonies were selected, sporulated, streaked out to single spores, and subsequently analyzed for the expression of the transformed constructs by fluorescence microscopy and/or Western blotting. Nitrate reductase (niaD)-deficient strains were isolated by using chlorate plates using adenine as a nitrogen source (6).
Plasmid constructions.
For the visualization of peroxisomes by fluorescence microscopy, the peroxisomal targeting signal 1 (PTS1) Ser-Lys-Leu (SKL) was fused to the extreme C terminus of green fluorescent protein (GFP) and DsRed. For the construction of plasmid pGBRH2-GFP-SKL, a 0.74-kb SalI (blunted by Klenow treatment)-BamHI fragment containing the eGFP-SKL gene from plasmid pFEM36 was inserted between the BamHI and SmaI sites of P. chrysogenum expression vector pGBRH2. The related plasmid pBBK-001, carrying DsRed-SKL, was described previously (12). In pGBRH2-GFP-SKL and pBKK-001, the GFP-SKL and DsRed-SKL genes are under the control of the strong P. chrysogenum pcbC promoter. For lower-level, constitutive expression, we created derivatives in which the pcbC promoter was replaced by the A. nidulans gpdA promoter. Primers BB-JK009 and BB-JK010 were used to amplify a 0.9-kb fragment comprising PgpdA from pNiGANi, which was then digested with Asp718i and BamHI and cloned between the Asp718i and BamHI sites of both pBBK-001 and pGBRH2-GFP-SKL, yielding pBBK-007 and pWHM-001, respectively. For the random integration of the PpcbC-GFP-SKL-TpenDE and PgpdA-GFP-SKL-TpenDE cassettes into the various P. chrysogenum strains, the expression cassettes were isolated as NotI fragments from pGBRH2-GFP-SKL and pWHM-001 and cotransformed into P. chrysogenum protoplasts with a 6.2-kb NotI-SpeI fragment from plasmid pNiGANi comprising the A. nidulans amdS gene. Transformants were selected on acetamide plates, and green fluorescent colonies were selected for further use.
Multisite Gateway technology (Invitrogen) was used to construct a plasmid carrying a pex3-GFP fusion gene under the control of the A. nidulans gpdA promoter. First, the P. chrysogenum pex3 gene lacking a stop codon was isolated by PCR using primers attB1-f-pex3 and attB2-r-pex3-nostop using P. chrysogenum cDNA as a template (11). The resulting 1.8-kb fragment was then recombined into vector pDONR221, yielding pENTR221-pex3-nostop. Subsequently, this plasmid was recombined with plasmids pENTR41-PgpdA, pENTR23-GFP-TpenDE, and pDEST R4-R3 to yield pEXP-Pcpex3-GFP. Subsequently, a P. chrysogenum strain carrying both pex3-GFP and DsRed-SKL was obtained by cotransforming DS17690 protoplasts with a 6.0-kb SspI-NdeI fragment comprising the PgpdA-pex3-GFP-TpenDE cassette from pEXP-PcPex3-GFP, a 2.2-kb NotI fragment comprising the PgpdA-DsRed-SKL-TpenDE cassette from pBBK-007, and the 6.2-kb NotI-SpeI fragment from plasmid pNiGANi comprising A. nidulans amdS. Transformants were selected on acetamide plates. Subsequently, colonies displaying both green and red fluorescence were selected.
Construction of a P. chrysogenum dnm1 deletion strain.
To enable site-specific gene deletion in P. chrysogenum, strain DS54465, an ΔhdfA derivative of DS17690, was used (23). Since this strain cannot be provided with fluorescent markers by random integration, we constructed an niaD-[PgpdA-GFP-SKL-TpenDE]-niaD cassette that enables the specific integration of the fluorescent marker in the P. chrysogenum niaD locus. First, we amplified the PgpdA-GFP-SKL-TpenDE cassette with primers attB1-f-PgpdA and attB2-r-TpenDE using pWHM-001 as a template. The resulting 2.2-kb fragment was recombined into pDONR221, yielding pENTR221-PgpdA-GFP-SKL-TpenDE. Subsequently, this plasmid was recombined with pENTR41-5′niaD, pENTR23-3′niaD, and pDEST R4-R3 to yield pEXP 5′niaD-PgpdA-GFP-SKL-TpenDE-3′niaD. This plasmid was linearized with NdeI and transformed into DS54465 protoplasts. NiaD− transformants were selected on chlorate plates. A strain producing green fluorescent peroxisomes was designated DS54465 GFP-SKL and used for further study.
To delete the P. chrysogenum dnm1 gene, its promoter (1.0 kb) and terminator (1.2 kb) regions were PCR amplified by using primers BB-JK201 plus BB-JK202 and BB-JK203 plus BB-JK204, respectively, and recombined into Gateway vectors pDONR P4-P1R and pDONR P2R-P3, resulting in plasmids pENTR41-DNM1-Prom and pENTR23-DNM1-Term, respectively. Finally, plasmids pENTR41-DNM1-Prom, pENTR221-AMDS, and pENTR23-DNM1-Term were recombined with the destination vector pDEST R4-R3, resulting in plasmid pEXP43-5Dnm1-AMDS-3Dnm1.This plasmid was linearized with PscI and transformed into P. chrysogenum DS54465 GFP-SKL protoplasts. Transformants were selected for their ability to grow on plates containing acetamide as the sole nitrogen source. The correct integration of the deletion cassette was tested via PCR using primers BB-JK205 and PgpdA.rev, resulting in a fragment of 1.1 kb (data not shown).
Construction of a P. chrysogenum dnm1 overexpression strain.
In order to obtain dnm1 overexpression, the P. chrysogenum dnm1 gene was first amplified by PCR with primers BB-JK209 and BB-JK210 using P. chrysogenum cDNA (11) as a template, resulting in a 2.5-kb DNA fragment containing the entire dnm1 coding sequence lacking a stop codon. The PCR product was recombined into vector pDONR221, resulting in plasmid pENTR221-PcDNM1. Subsequently, plasmids pENTR41-PpcbC, pENTR221-PcDNM1, and pENTR23-His8-TpenDE were recombined into vector pDEST R4-R3/AMDS, resulting in the expression vector pEXP-PcDNM1-HIS8. This plasmid was linearized with SmaI and transformed into P. chrysogenum DsRed-SKL protoplasts. Transformants were selected for their ability to grown on acetamide as the sole nitrogen source. The presence of the overexpression cassette was further checked by PCR using primers BB-JK211 and BB-JK212, resulting in a 361-bp cDNA fragment and a 448-bp genomic fragment (data not shown). Strains with multiple copies of the dnm1 overexpression cassette were selected. Furthermore, the overproduction of the Dnm1 protein was demonstrated by Western blotting using specific antibodies against the Saccharomyces cerevisiae Dnm1 protein or the His8 tag (data not shown).
Isolation of non-oleate-utilizing mutants.
P. chrysogenum Wis54-1255 spores were mutagenized in potassium phosphate buffer (100 mM K2HPO4, 100 mM KH2PO4 [pH 6.8]) at 37°C using nitrochinoline oxide (1 mg/liter, final concentration). The incubation was terminated after 20 min by the addition of 2.5% (wt/vol) sodium thiosulfate. Spores were washed once with sterile water and resuspended in sterile water. Spores were diluted to one spore per droplet and spotted onto R-agar plates. After 7 days the colonies were transferred into MM with 1% (wt/vol) oleic acid, MM with 0.25% acetate, and MM with ethanol-glycerol to screen for non-oleate-utilizing strains.
Electron microscopy.
Hyphae of P. chrysogenum were fixed in either 1.5% KMnO4 or a mixture of glutaraldehyde (0.5%, vol/vol) and formaldehyde (2.5%, vol/vol) as described previously (33). Immunocytochemistry was performed on ultrathin sections of Unicryl-embedded samples by using antibodies raised against AT, GFP, or thiolase, as described previously (33).
Quantitative analysis of β-lactam levels.
Quantitative 1H nuclear magnetic resonance (NMR) experiments were performed at 600 MHz on a Bruker Avance 600 spectrometer. A known quantity of internal standard (maleic acid) dissolved in phosphate buffer was added to a known quantity of filtrate prior to lyophilization. The residue was dissolved in D2O and measured at 300 K. The delay between scans (30 s) was more than five times the T1 of all compounds, so the ratio between the integrals of the compounds of interest and the integral of the internal standard is an exact measure for the quantity of the PEN samples.
Extracellular titers of PenG were determined by high-performance liquid chromatography (HPLC) using an isocratic flow of 310 g/liter acetonitrile, 640 mg/liter KH2PO4, and 340 mg/liter H3PO4. Peaks were separated on a Shim-Pack XR-ODS 3.0-mm-internal-diameter by 75-mm column (Shimadzu) at a flow rate of 0.5 ml/min and detected at a wavelength of 254 nm. Intracellular β-lactam levels were determined by using mass spectrometry (5).
Fluorescence microscopy.
Wide-field fluorescence imaging was performed by using a Zeiss Axio Observer Z1 fluorescence microscope (Carl Zeiss, Oberkochen, Germany). Images were taken by using a Coolsnap HQ2 camera (Roper Scientific Inc.). The GFP signal was visualized with a 470/40-nm-band-pass excitation filter, a 495-nm dichromatic mirror, and a 525/50-nm-band-pass emission filter. DsRed fluorescence was visualized with a 545/25-nm-band-pass excitation filter, a 570-nm dichromatic mirror, and a 605/70-nm-band-pass emission filter. z-stack images were made at an interval of 0.5 μm. Image analysis was carried out by using ImageJ (http://rsb.info.nih.gov/nih-image/).
Confocal laser scanning microscopy (CLSM) was performed by using a Zeiss LSM510 microscope (Zeiss Netherlands BV, Sliedrecht, Netherlands) equipped with a Zeiss Plan Apochromatic 63× 1.4-numerical-aperture (NA) objective. GFP fluorescence was analyzed by the excitation of the cells with a 488-nm Ar/Kr laser with 1% output, and fluorescence was detected by use of a BP 500-530 photo multiplier tube (PMT). DsRed fluorescence was analyzed by the excitation of the cells with a 543-nm He/Ne laser at 20%, and fluorescence was detected by using a 560-nm-long-pass filter. z-stack images were acquired with special intervals of 1 μm. Grouped z-stack images were created by using ImageJ. The number of fluorescent spots was determined for grouped z-stack images of hyphal tip cells. Statistical analysis was performed by using a Student's t test.
For time-lapse recordings, the temperatures of the objective and objective slide were kept at 25°C. Nine z-axis planes, each at 1-μm intervals, were acquired for each sample. The laser power (Ar-ion laser at 30 mW; 488 nm) was set at a 50% maximum value; the acousto-optical tunable filter (AOTF) was tuned down to 0.5%. Kymograms were created as described previously (34). ImageJ was used to create kymograms from time-lapse images. Slices were rotated to obtain a side view; subsequently, a z projection was created, yielding a kymogram.
RESULTS
Peroxisomes are required for efficient PEN biosynthesis.
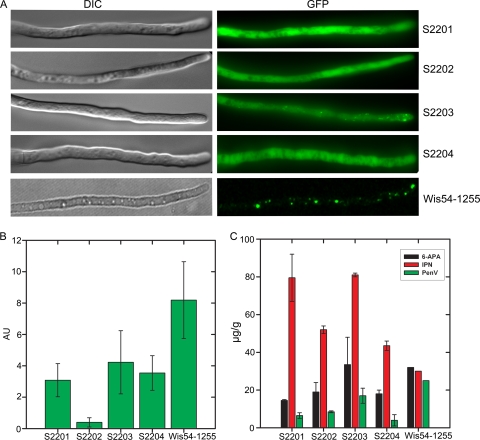
Earlier experiments suggested that peroxisomes are important for PEN production. To further elucidate the significance of peroxisomes in PEN biosynthesis, peroxisome-deficient mutants of P. chrysogenum strain Wis54-1255 were isolated essentially as described previously for baker's yeast (3), by selecting non-oleate-utilizing strains. The survival rate of Wis54-1255 after mutagenesis with nitrochinoline oxide was determined at several time points (data not shown). Incubations of 20 min gave the best results, with a survival rate of 3%. As a first screen for peroxisome deficiency, 5,614 surviving mutants were analyzed for their abilities to grow on oleic acid. False-positive mutants, due to a malfunctioning of the citric acid cycle or mitochondria, were weeded out on medium supplemented with acetate or ethanol-glycerol, respectively. Initially, six putative peroxisome-deficient P. chrysogenum strains were isolated. Electron microscopy analysis showed greatly reduced peroxisomal profiles for all six mutants (data not shown). Additionally, PEN production levels of the mutant strains appeared to be 3 to 10 times lower than that of the Wis54-1255 control. Upon purification and conidiospore preparation, only four mutants produced stable, viable spore stocks. These mutants were designated S2201 to S2204. The mutants were unable to grow on medium supplemented with oleic acid as the sole carbon source. Also, growth on the fatty acid lauric acid or on hexane was fully inhibited (data not shown). To investigate the presence of intact peroxisomes, the mutants and the Wis54-1255 control were provided with a PgpdA-GFP-SKL-TpenDE cassette, producing the peroxisomal marker protein GFP-SKL. After growth on PP medium, fluorescence microscopy analysis (Fig. 2A) revealed that the GFP fluorescence was cytosolic in mutant strains S2201, S2202, and S2204, while a partial mislocalization was observed for strain S2203. In contrast, all fluorescence was confined to spots representing peroxisomes in the Wis54-1255 control. This finding confirms that mutants S2201 to S2204 are indeed peroxisome-deficient strains. To quantify the effect of peroxisome deficiency on β-lactam production, the mutants were grown in PP medium supplemented with POA, and the production of PenV and its precursors both in spent medium and in cell extracts was monitored. NMR analysis of spent medium demonstrated that in the mutants, the PenV production capacity was severely diminished compared to that of the Wis54-1255 control (Fig. 2B). Intracellular β-lactam levels were determined by using mass spectrometry (5) (Fig. 2C). As expected, in the peroxisome-deficient mutants, the PenV precursor IPN accumulated significantly. These results convincingly demonstrate that peroxisomes are important for efficient PEN biosynthesis.
FIG. 2.
Analysis of peroxisome-deficient mutants. (A) Morphological phenotype of non-oleate-utilizing mutants. Shown is fluorescence microscopy analysis of cells of the non-oleate-utilizing mutants S2201 to S2204 and the parental strain Wis54-1255, all producing GFP-SKL, grown on PP medium. In the parental strain, the GFP fluorescence is present as distinct spots representing peroxisomes. In contrast, in cells of mutant strains S2201, S2202, and S2204, GFP-SKL is mislocalized to the cytosol, while S2203 cells show a partial mislocalization. (Left) Differential interference contrast (DIC) images. (Right) GFP fluorescence. (B and C) β-Lactam production by peroxisome-deficient mutants. The non-oleate-utilizing mutant strains S2201 to S2204 and the parental strain Wis54-1255 were cultivated on PP medium with POA, and PenV levels in spent media were determined by using NMR (B). Additionally, the intracellular amounts of the penicillin intermediates 6-APA (6-aminopenicillanic acid), IPN, and PenV were determined by mass spectrometry analysis (C). AU, arbitrary units. The error bars indicate the standard errors of the means (SEM).
A high-PEN-producing strain has elevated peroxisome numbers.
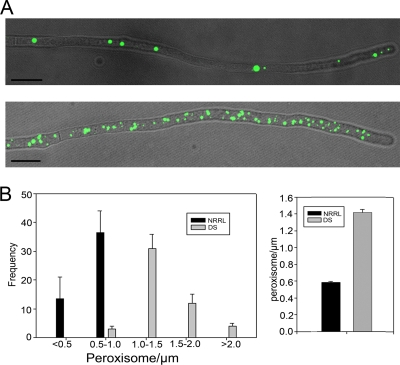
To study the relationship between peroxisome numbers and PEN production more closely, we compared peroxisome proliferations in two P. chrysogenum strains of increasing PEN production rates, namely, NRRL1951 and DS17690. NRRL1951 is the type strain from which all current production strains originate (21), including DS17690. The latter strain is a high-producing derivative obtained after various rounds of mutagenesis performed at the DSM. Transformants of the strains that expressed GFP-SKL under the control of the moderate A. nidulans gpdA promoter were used. Cells of each strain were grown in batch cultures on PP medium and analyzed by fluorescence microscopy. Figure 3A shows the peroxisome distribution in cells of the two strains. The data suggest that the number of organelles per cell increases significantly from NRRL1951 to the DS17960 strain. To seek further confirmation for this finding, quantitative measurements were performed by using grouped z stacks taken from subapical, GFP-SKL-producing cells of the two strains by confocal laser scanning microscopy (CLSM). The data, summarized in Fig. 3B, demonstrate the significant increase of fluorescent spots per unit length of hyphae from NRRL1951 (0.58 spots per μm hypha) to the DS17690 strain (1.40 spots per μm hypha). Thus, the increase in PEN production rates (at least 3 orders of magnitude [8a]) obtained after strain improvement is paralleled by a significant increase in peroxisome numbers. However, it must be noted that, conversely, an increase in peroxisome numbers does not always correlate with increased PEN levels (see below and Fig. 6C).
FIG. 3.
Strains with enhanced PEN production contain increased peroxisome numbers. (A) Fluorescence microscopy images showing the peroxisome abundance in a hypha of a low-PEN-producing strain (NRRL1951) (top) and high-PEN-producing strain (DS17960) (bottom) grown under PEN-inducing conditions. Peroxisomes are marked by GFP-SKL. The pictures show overlays of bright-field and fluorescence images. Scale bars represent 10 μm. (B) Quantitative determination of peroxisomes marked by GFP-SKL in NRRL1951 and DS17690. Fluorescent spots were counted from grouped z stacks taken from randomly selected subapical cells by confocal laser scanning microscopy. (Left) Distribution pattern of peroxisomes in both strains. (Right) Average number of organelles per μm hypha. For each sample, at least 50 hyphae were counted. The error bars represent the SEM.
FIG. 6.
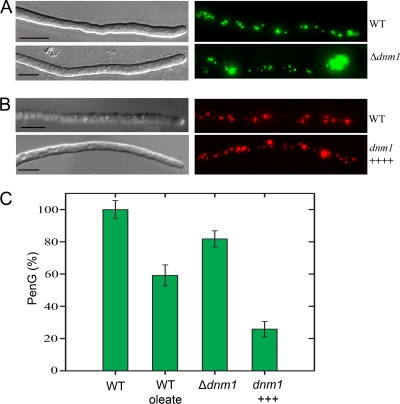
Modulation of Dnm1 levels affects peroxisome abundance and PEN production. (A) Deletion of dnm1 reduces overall organelle numbers and results in strongly expanded peroxisome clusters in the tip. (B) dnm1 overexpression (dnm1++++) leads to enhanced numbers of peroxisomes. Cells were grown in PEN production medium. (Left) DIC images. (Right) Fluorescence images. The bar represents 10 μm. (C) Overproduction of Dnm1 strongly affects PEN production. P. chrysogenum dnm1 deletion and overexpression strains and the WT host (DS17690) were grown on PEN production medium. Additionally, the WT strain was grown the same medium supplemented with oleate. Subsequently, PenG levels in spent media of the cultures were determined by using HPLC. The data show that a strong increase in Dnm1 levels results in a significant decrease in PenG levels. Moreover, the deletion of dnm1 or the addition of oleate to WT cultures also affects PenG production but to a much lesser extent. The error bars represent the SEM.
Development of peroxisomes in PEN-producing P. chrysogenum strains.
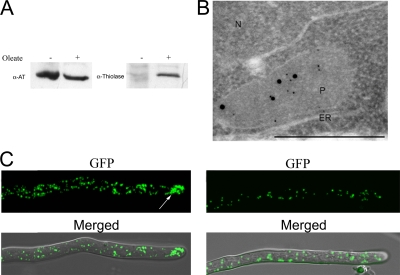
Previously, we demonstrated that P. chrysogenum contains true peroxisomes containing H2O2-producing oxidases and catalases (12). To investigate whether AT is targeted to these peroxisomes or to specialized organelles devoted solely to PEN biosynthesis (so-called “AT-somes” [18]), P. chrysogenum Wis54-1255 cells were grown on PP medium for 24 h to induce peroxisome proliferation, and oleic acid was subsequently added to the culture to simultaneously induce the synthesis of peroxisomal β-oxidation enzymes. During the adaptation of cells to oleic acid, the synthesis of one of the key enzymes of β-oxidation, thiolase, was indeed induced (Fig. 4A). Double-immunocytochemistry experiments using both anti-AT and anti-thiolase antibodies demonstrated that AT and thiolase were sorted to the same organelles (Fig. 4B). As observed for other fungi, the addition of oleic acid to PP medium resulted in a significantly enhanced number of peroxisomes per cell (Fig. 4C).
FIG. 4.
Peroxisomes in P. chrysogenum contain both thiolase and AT. (A) P. chrysogenum Wis54-1255 was grown on PEN production medium in the presence (+) and absence (−) of oleic acid. Western blots of crude cell extracts with equal amounts of protein loaded per lane were decorated with anti-AT (α-AT) and anti-thiolase antibodies. The data show that AT is produced under both conditions, whereas thiolase is present predominantly in cells grown in the presence of oleic acid. (B) Detail of a section of a P. chrysogenum Wis54-1255 cell grown on PEN production medium supplemented with oleic acid. After immunolabeling using anti-AT (5-nm gold particles) and anti-thiolase (15-nm gold particles), both particles were observed on the microbody profile. N, nucleus; P, microbody; ER, endoplasmic reticulum. The bar represents 0.2 μm. (C) Peroxisome abundance is induced by oleic acid. P. chrysogenum DS17690 cells producing GFP-SKL were grown on PEN production medium in the presence (left) and absence (right) of oleic acid. The addition of oleic acid leads to an increase in peroxisome numbers relative to those of cells grown on induction medium alone. The arrow in the left panel indicates a cluster of peroxisomes. The top panels show fluorescence images, and the bottom panels show overlays of bright-field and fluorescence images.
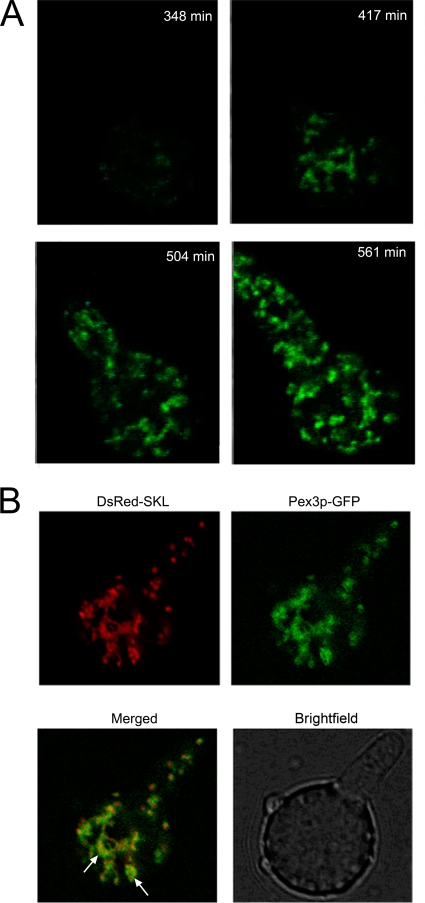
In the next series of experiments, we investigated the mode of peroxisome development in both germinating spores and developing hyphae. This was analyzed with the DS17690 GFP-SKL strain, as this strain contained the highest peroxisome numbers. Fresh conidiospores of this strain generally contained relatively few distinct GFP-SKL-containing spots, indicative of peroxisomes. About 10% of the spores lacked such structures. The germination of peroxisome-lacking conidiospores was monitored by time-lapse CLSM. Generally, germination started after 10 h of incubation in production medium. Germination was preceded by a swelling of the spores, a phenomenon that was accompanied by the induction of GFP-SKL synthesis that was organized in reticular networks. Upon germination, presented in Video S1 in the supplemental material, the intensity of these structures increased and extended into the germination tube in which, however, they were rapidly observed as separate organelles. Stills from this video are presented in Fig. 5A. Similar networks have been observed to form in conidiospores that contained few peroxisomes (data not shown). To investigate the reticular networks in more detail, the peroxisomal membrane protein Pex3 was fused to GFP and coproduced with DsRed-SKL. The data (Fig. 5B and Video S2) show that in germinating spores, Pex3-GFP colocalized with DsRed-SKL, thus confirming the peroxisomal nature of these reticular networks and the distinct organelles developing from them (Fig. 5B). We interpret these data to mean that organelles present in the germination tube arise from the peroxisome reticular network in the conidiospore and are administrated to the growing hyphal tip. We have analyzed the site of peroxisome proliferation further by electron microscopy and analyzed serial sections cut through the neck between the germinating spore and germination tube. Using Amira software, we reconstructed a three-dimensional (3-D) model in which the peroxisomes are marked in red (see Fig. S1 in the supplemental material). This model supports the presence of interconnected peroxisome structures.
FIG. 5.
Peroxisomes form from a network in germinating P. chrysogenum conidiospores. (A) Stills from Video S1 in the supplemental material at various time points during conidiospore germination. The first fluorescence is observed as a relatively small network structure at approximately 7 h of incubation of the spores in fresh PEN production medium. This structure rapidly increases in size at later stages of incubation and extends into the germination tube. (B) CLSM pictures of germinating conidiospores that simultaneously produce DsRed-SKL and Pex3-GFP. The merged image shows that the two fluorescent probes colocalize, indicating the peroxisomal nature of the complex network. Arrows point to the reticular networks that contain both DsRed-SKL and Pex3-GFP.
A remarkable feature of the growing hyphal tip is the continuous and virtually upward movement of organelles in the direction of the tip, including numerous GFP-SKL-containing peroxisomes (see Video S3 in the supplemental material). Kymogram analysis of the images demonstrated that the speed of tip growth exceeds the upward movement of the fluorescent spots (Video S3 and Fig. S2). This finding suggests that the main site of peroxisome development is in fact localized near the hyphal tips. Careful CLSM analysis indeed elucidated the presence of strongly fluorescent clusters of peroxisomes in these developing tips (Fig. 4C). The kymograms generated from CLSM images also demonstrated that in strain DS17960, cross wall formation in growing hyphae was not gradual during cell elongation but occurred simultaneously for several cross walls at the same time (Fig. S2). Frequently, several cross walls were formed at the same time, thereby partitioning the long hyphal tip in the corresponding compartments (Video S3 and Fig. S2). After septa had been formed, the upward movement of the fluorescent spots ceased (Fig. S2). The CLSM analysis also showed that the organelles incorporated into a subapical cell became strongly enhanced in fluorescence during prolonged cultivation as a result of the accumulation of the GFP-SKL protein (Fig. S2B). This finding suggests that these organelles are still competent to incorporate additional matrix components like newly synthesized PEN biosynthesis enzymes and, thus, further enhance PEN production rates in the corresponding cells in which they occur.
The peroxisome proliferation machinery in the hyphal tip requires the function of Dnm1.
The dynamin-like protein Dnm1 is known to be essential for peroxisome fission in yeast and mammals (reviewed in reference 20). To determine whether this protein has a comparable function in organelle proliferation in hyphal tip cells, we have constructed P. chrysogenum strains lacking or overproducing Dnm1.
P. chrysogenum GFP-SKL Δdnm1 cells and cells of the host GFP-SKL were grown for 48 h on PP medium and analyzed by CLSM. The data presented in Fig. 6A show that the peroxisomal structures in the tip of Δdnm1 cells have strongly expanded in conjunction with a reduction of the number of peroxisomes in the rest of the cells. The presence of these clusters in the apical cells was even more pronounced in mutant cells grown in the presence of oleic acid (not shown). These data suggest that Dnm1 indeed has an important function in the formation of peroxisomes at the hyphal tip. Consistent with this was the observation that the number of peroxisomes in a dnm1 overexpression strain of P. chrysogenum was enhanced relative to those of wild-type (WT) controls (Fig. 6B).
To determine whether the absence or the presence of overproduced Dnm1 also affected antibiotic production, we determined PenG levels in spent media of the mutant strains and the WT host cultured on PP medium (Fig. 6C). Remarkably, the PEN production levels were slightly reduced in the dnm1 strain, while a major reduction in PenG levels was observed upon dnm1 overexpression. Similarly, we observed that growth on oleate, another condition that results in peroxisome proliferation, also results in decreased PEN production levels in the WT host.
DISCUSSION
In this paper we describe the correlation that exists between peroxisome numbers and PEN production rates in the filamentous fungus P. chrysogenum. First, we show that PEN production by P. chrysogenum requires the function of intact peroxisomes, as the synthesis of this secondary metabolite is significantly reduced in peroxisome-deficient mutants of P. chrysogenum. Most likely, peroxisomes create a unique microenvironment that is ideally suited to perform the two final steps of PEN production, namely, the activation of the side chain by PCL (14) and the substitution of the α-aminoadipyl side chain for a hydrophobic one by AT (19, 35). For A. nidulans, a relationship between peroxisome function and PEN production was also observed previously (24). There, however, the correlation was less strict than that for P. chrysogenum, as peroxisome-deficient mutants of A. nidulans still showed high PEN titers (2). The present mutant studies and the significantly enhanced peroxisome numbers in high-producing strains like DS17960 demonstrate that intact organelles not only are crucial for efficient PEN production but also determine the ultimate production rates. However, it must be noted that not all our efforts to increase or decrease peroxisome numbers had the expected effect on β-lactam production. Although peroxisome numbers are highly upregulated by the addition of oleate to PP medium, we observed that PEN levels in spent medium were not increased but, rather, decreased (Fig. 6C). The reason for the decrease in PEN production levels may be the production of high levels of β-oxidation enzymes that compete with PEN production enzymes for peroxisomal CoA pools. Furthermore, the addition of oleate significantly affected the growth of P. chrysogenum cultures, which also results in reduced antibiotic production. A significant drop in PEN levels was also observed upon Dnm1 overproduction (Fig. 6C). We explain this surprising phenomenon by the effect that Dnm1 has on other organelles in this cell, i.e., its role in mitochondrial fission. Thus, our data imply that conditions that lead to peroxisome proliferation but simultaneously interfere with the normal physiology of the cell may be detrimental for antibiotic production. Previously, we demonstrated that the overproduction of Pex11, which exclusively affects peroxisomal profiles, led to an increase in PEN levels (13).
The second topic of our studies addressed the origin of peroxisomes in filamentous fungi. The analysis of germinating conidiospores demonstrated that the original peroxisome population following germination derived from a reticular network. As at the onset of germination, no distinct peroxisomes were detected in the spore, we cannot exclude that the initial organelle is formed via an alternative mode, e.g., from the endoplasmic reticulum (ER) (10, 17). However, upon spore swelling, the peroxisomal reticulum had already developed and most likely served as the source of the first organelles that migrated into the developing tip. During subsequent mycelial growth, peroxisome development occurred predominantly at the hyphal tip. Our data demonstrate that the fission of organelles from the reticular structure at the tip requires Dnm1. However, despite the absence of this dynamin-like protein (DLP), single peroxisomes could be observed in the apical and subapical cells. This finding suggests the formation of peroxisomes by alternative fission strategies (e.g., via the DLP Vps1, which is also conserved in filamentous fungi) or by the alternative formation of organelles from the ER. Recently, it was demonstrated that the fission of Woronin bodies, a special type of microbody required to close the septal pore upon hyphal damage, did not require the function of either Dnm1 or Vps1, although peroxisome numbers were apparently reduced in cells lacking dnm1 (16).
Remarkably, we found that septa in the P. chrysogenum strains under study are not formed successively, which is also the case for the NRRL1951 type strain, but can develop at the same time. Following cross wall formation, we observed an increase in peroxisome size after prolonged cultivation due to the uptake of matrix proteins that continued in subapical cells. We assume that this leads to a gradual increase in the PEN production capacity of each individual cell as the cell matures. As yet, the intriguing question of what the advantage is for the organism of creating several subhyphal compartments at the same time is still unresolved. Possibly, this allows the organism to carefully partition organelle numbers over the separate compartments in order to form optimally adapted cells for performing their various functions in, e.g., cell maintenance, energy generation, and metabolite production. Analyses that address these intriguing questions are ongoing.
Supplementary Material
Acknowledgments
W.H.M. was funded by the IBOS Programme (Integration of Biosynthesis and Organic Synthesis) of Advanced Chemical Technologies for Sustainability (ACTS), with financial contributions of the Dutch Ministry of Economic Affairs, the Netherlands Organization for Scientific Research (NWO), and DSM, Delft, Netherlands. L.G., S.F., and J.A.K.W.K. were financially supported by the Netherlands Ministry of Economic Affairs and the B-Basic partner organizations (www.b-basic.nl) through B-Basic, a public-private NWO-ACTS (Advanced Chemical Technologies for Sustainability) program. The project was carried out within the research program of the Kluyver Center for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
Published ahead of print on 2 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Cantoral, J. M., B. Diez, J. L. Barredo, E. Alvarez, and J. F. Martin. 1987. High-frequency transformation of Penicillium chrysogenum. Nat. Biotechnol. 5:494-497. [Google Scholar]
- 2.De Lucas, J. R., S. Valenciano, A. I. Dominguez, G. Turner, and F. Laborda. 1997. Characterization of oleate-nonutilizing mutants of Aspergillus nidulans isolated by the 3-amino-1,2,4-triazole positive selection method. Arch. Microbiol. 168:504-512. [DOI] [PubMed] [Google Scholar]
- 3.Erdmann, R., M. Veenhuis, D. Mertens, and W. H. Kunau. 1989. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 86:5419-5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reference deleted.
- 5.Gidijala, L., J. A. Kiel, R. D. Douma, R. M. Seifar, W. M. van Gulik, R. A. Bovenberg, M. Veenhuis, and I. J. van der Klei. 2009. An engineered yeast efficiently secreting penicillin. PLoS One 4:e8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gouka, R. J., W. van Hartingsveldt, R. A. Bovenberg, C. A. van den Hondel, and R. F. van Gorcom. 1991. Cloning of the nitrate-nitrite reductase gene cluster of Penicillium chrysogenum and use of the niaD gene as a homologous selection marker. J. Biotechnol. 20:189-199. [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez, S., F. Fierro, J. Casqueiro, and J. F. Martin. 1999. Gene organization and plasticity of the beta-lactam genes in different filamentous fungi. Antonie Van Leeuwenhoek 75:81-94. [DOI] [PubMed] [Google Scholar]
- 8.Harris, D. M., J. A. Diderich, Z. A. van der Krogt, M. A. Luttik, L. M. Raamsdonk, R. A. Bovenberg, W. M. van Gulik, J. P. van Dijken, and J. T. Pronk. 2006. Enzymic analysis of NADPH metabolism in beta-lactam-producing Penicillium chrysogenum: presence of a mitochondrial NADPH dehydrogenase. Metab. Eng. 8:91-101. [DOI] [PubMed] [Google Scholar]
- 8a.Hersbach, G. J. M., C. P. van der Beek, and P. W. M. van Dijck. 1984. The penicillins: properties, biosynthesis, and fermentation, p. 45-140. In E. Van Damme (ed.), Biotechnology of industrial antibiotics, vol. 3. Marcel Dekker, New York, NY.
- 9.Hesse, S. J., G. J. Ruijter, C. Dijkema, and J. Visser. 2002. Intracellular pH homeostasis in the filamentous fungus Aspergillus niger. Eur. J. Biochem. 269:3485-3494. [DOI] [PubMed] [Google Scholar]
- 10.Hoepfner, D., D. Schildknegt, I. Braakman, P. Philippsen, and H. F. Tabak. 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122:85-95. [DOI] [PubMed] [Google Scholar]
- 11.Kiel, J. A., R. E. Hilbrands, R. A. Bovenberg, and M. Veenhuis. 2000. Isolation of Penicillium chrysogenum PEX1 and PEX6 encoding AAA proteins involved in peroxisome biogenesis. Appl. Microbiol. Biotechnol. 54:238-242. [DOI] [PubMed] [Google Scholar]
- 12.Kiel, J. A., M. A. van den Berg, F. Fusetti, B. Poolman, R. A. Bovenberg, M. Veenhuis, and I. J. van der Klei. 2009. Matching the proteome to the genome: the microbody of penicillin-producing Penicillium chrysogenum cells. Funct. Integr. Genomics 9:167-184. [DOI] [PubMed] [Google Scholar]
- 13.Kiel, J. A., I. J. van der Klei, M. A. van den Berg, R. A. Bovenberg, and M. Veenhuis. 2005. Overproduction of a single protein, Pc-Pex11p, results in 2-fold enhanced penicillin production by Penicillium chrysogenum. Fungal Genet. Biol. 42:154-164. [DOI] [PubMed] [Google Scholar]
- 14.Lamas-Maceiras, M., I. Vaca, E. Rodriguez, J. Casqueiro, and J. F. Martin. 2006. Amplification and disruption of the phenylacetyl-CoA ligase gene of Penicillium chrysogenum encoding an aryl-capping enzyme that supplies phenylacetic acid to the isopenicillin N-acyltransferase. Biochem. J. 395:147-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lara, F., R. del Carmen Mateos, G. Vazquez, and S. Sanchez. 1982. Induction of penicillin biosynthesis by L-glutamate in Penicillium chrysogenum. Biochem. Biophys. Res. Commun. 105:172-178. [DOI] [PubMed] [Google Scholar]
- 16.Liu, F., S. K. Ng, Y. Lu, W. Low, J. Lai, and G. Jedd. 2008. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J. Cell Biol. 180:325-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullen, R. T., and R. N. Trelease. 2006. The ER-peroxisome connection in plants: development of the “ER semi-autonomous peroxisome maturation and replication” model for plant peroxisome biogenesis. Biochim. Biophys. Acta 1763:1655-1668. [DOI] [PubMed] [Google Scholar]
- 18.Muller, W. H., R. A. Bovenberg, M. H. Groothuis, F. Kattevilder, E. B. Smaal, L. H. Van der Voort, and A. J. Verkleij. 1992. Involvement of microbodies in penicillin biosynthesis. Biochim. Biophys. Acta 1116:210-213. [DOI] [PubMed] [Google Scholar]
- 19.Muller, W. H., T. P. van der Krift, A. J. Krouwer, H. A. Wosten, L. H. van der Voort, E. B. Smaal, and A. J. Verkleij. 1991. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 10:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagotu, S., M. Veenhuis, and I. J. van der Klei. 2010. Divide et impera: the dictum of peroxisomes. Traffic 11:175-184. [DOI] [PubMed] [Google Scholar]
- 21.Newbert, R. W., B. Barton, P. Greaves, J. Harper, and G. Turner. 1997. Analysis of a commercially improved Penicillium chrysogenum strain series: involvement of recombinogenic regions in amplification and deletion of the penicillin biosynthesis gene cluster. J. Ind. Microbiol. Biotechnol. 19:18-27. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Snoek, I. S., Z. A. van der Krogt, H. Touw, R. Kerkman, J. T. Pronk, R. A. Bovenberg, M. A. van den Berg, and J. M. Daran. 2009. Construction of an hdfA Penicillium chrysogenum strain impaired in non-homologous end-joining and analysis of its potential for functional analysis studies. Fungal Genet. Biol. 46:418-426. [DOI] [PubMed] [Google Scholar]
- 24.Sprote, P., A. A. Brakhage, and M. J. Hynes. 2009. Contribution of peroxisomes to penicillin biosynthesis in Aspergillus nidulans. Eukaryot. Cell 8:421-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramani, S. 1998. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol. Rev. 78:171-188. [DOI] [PubMed] [Google Scholar]
- 26.Thykaer, J., and J. Nielsen. 2003. Metabolic engineering of beta-lactam production. Metab. Eng. 5:56-69. [DOI] [PubMed] [Google Scholar]
- 27.Valenciano, S., J. R. Lucas, A. Pedregosa, I. F. Monistrol, and F. Laborda. 1996. Induction of beta-oxidation enzymes and microbody proliferation in Aspergillus nidulans. Arch. Microbiol. 166:336-341. [DOI] [PubMed] [Google Scholar]
- 28.van de Kamp, M., A. J. Driessen, and W. N. Konings. 1999. Compartmentalization and transport in beta-lactam antibiotic biosynthesis by filamentous fungi. Antonie Van Leeuwenhoek 75:41-78. [DOI] [PubMed] [Google Scholar]
- 29.van den Bosch, H., R. B. Schutgens, R. J. Wanders, and J. M. Tager. 1992. Biochemistry of peroxisomes. Annu. Rev. Biochem. 61:157-197. [DOI] [PubMed] [Google Scholar]
- 30.van der Klei, I. J., H. Yurimoto, Y. Sakai, and M. Veenhuis. 2006. The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim. Biophys. Acta 1763:1453-1462. [DOI] [PubMed] [Google Scholar]
- 31.van der Lende, T. R., P. Breeuwer, T. Abee, W. N. Konings, and A. J. Driessen. 2002. Assessment of the microbody luminal pH in the filamentous fungus Penicillium chrysogenum. Biochim. Biophys. Acta 1589:104-111. [DOI] [PubMed] [Google Scholar]
- 32.van der Lende, T. R., M. van de Kamp, M. Berg, K. Sjollema, R. A. Bovenberg, M. Veenhuis, W. N. Konings, and A. J. Driessen. 2002. Delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase, that mediates the first committed step in penicillin biosynthesis, is a cytosolic enzyme. Fungal Genet. Biol. 37:49-55. [DOI] [PubMed] [Google Scholar]
- 33.Waterham, H. R., V. I. Titorenko, P. Haima, J. M. Cregg, W. Harder, and M. Veenhuis. 1994. The Hansenula polymorpha PER1 gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J. Cell Biol. 127:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterman-Storer, C. 2002. Fluorescent speckle microscopy (FSM) of microtubules and actin in living cells. Curr. Protoc. Cell Biol. 4:4.10. [DOI] [PubMed] [Google Scholar]
- 35.Whiteman, P. A., E. P. Abraham, J. E. Baldwin, M. D. Fleming, C. J. Schofield, J. D. Sutherland, and A. C. Willis. 1990. Acyl coenzyme A:6-aminopenicillanic acid acyltransferase from Penicillium chrysogenum and Aspergillus nidulans. FEBS Lett. 262:342-344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.