Abstract
Honeybee colonies are vulnerable to parasites and pathogens ranging from viruses to vertebrates. An increasingly prevalent disease of managed honeybees is caused by the microsporidian Nosema ceranae. Microsporidia are basal fungi and obligate parasites with much-reduced genomic and cellular components. A recent genome-sequencing effort for N. ceranae indicated the presence of machinery for RNA silencing in this species, suggesting that RNA interference (RNAi) might be exploited to regulate Nosema gene expression within bee hosts. Here we used controlled laboratory experiments to show that double-stranded RNA homologous to specific N. ceranae ADP/ATP transporter genes can specifically and differentially silence transcripts encoding these proteins. This inhibition also affects Nosema levels and host physiology. Gene silencing could be mediated solely by Nosema or in concert with known systemic RNAi mechanisms in their bee hosts. These results are novel for the microsporidia and provide a possible avenue for controlling a disease agent implicated in severe honeybee colony losses. Moreover, since microsporidia are pathogenic in several known veterinary and human diseases, this advance may have broader applications in the future for disease control.
Microsporidia are fungal intracellular parasites of insects and other higher eukaryotes (18, 22, 23, 35). Two described species of microsporidia, Nosema ceranae and Nosema apis, parasitize adult honeybees (7). Of these, N. ceranae has shown a dramatic recent range expansion (11, 16, 20, 24), and this species has been shown to be highly pathogenic to honeybees (4, 14). Indeed, natural infection has been associated with a syndrome of gradual depopulation, copious colony death in autumn or winter, and poor honey production (13, 15). Transmission of Nosema in honeybee colonies is mainly via the fecal-oral route, in which pathogens are spread by transferring feces of diseased hosts to uninfected hosts via ingestion (2, 30). Millions of new spores can be found within the midgut of infected bees within a few weeks after initial infection, and the spores excreted with feces become new sources of infection within colonies (2). Antibiotic control strategies are losing efficacy due to development of resistance (38), and the presence of antibiotic residues may have adverse effects on human health (29). Therefore, finding novel methods to control Nosema disease is a top priority.
New genomic resources for N. ceranae (5) allow for a systematic gene-silencing approach, a strategy that has proven effective in reducing the abundance of another honeybee pathogen, Israeli acute paralysis virus (IAPV) (27). The process of posttranscriptional gene silencing (PTGS) is an evolutionarily conserved cellular defense mechanism found across kingdoms that is used to reduce the expression of foreign genes (31). For gene silencing to be effective as a preventive or curative strategy, amplification and systemic spread of the silencing signal is important. In some invertebrates, including honeybees, a systemic-interference-defective (SID) gene encodes a transmembrane protein that is an important participator in the systemic RNA interference (RNAi) pathway (19). Apparently, these SID1-like proteins channel double-stranded RNAs (dsRNAs) between cells, enabling systemic spread of the silencing signal (1). Although a canonical invertebrate RNA-dependent RNA polymerase (RdRP) homologue has not yet been described, there is evidence that such RdRP activity may occur via other enzymes, leading to amplification of the silencing signal in insects (26).
Microsporidia show reduced genomes and cell components, arguably a reflection of their current lifestyles as obligate parasites that rely on their hosts for energetic and metabolic needs. Reflecting this extreme reduction, Encephalitozoon cuniculi, a microsporidian found in humans, was shown to lack nearly all of the introns and intergenic spacers common to eukaryotic genomes (21). Microsporidia lost typical mitochondria during their evolution and, instead, possess an analogous “mitosome” that is extremely reduced in both size and biochemical complexity. To date, only 20 mitosomal proteins have been identified, in contrast to the mitochondrion of the yeast Saccharomyces cerevisiae, which contains about 1,000 proteins (17, 18).
Several bacterial intracellular parasites, such as Rickettsia spp., possess a certain type of nucleotide transporter which is used for import of ATP from their eukaryotic host cell. Genes for proteins homologous to those described from bacteria are apparent in the N. ceranae genome. The use of bacterium-like nucleotide transporters to acquire ATP from another eukaryotic cell is a unique strategy for a eukaryotic parasite. There are no homologues of these proteins in either vertebrate or invertebrate species sequenced to date. We hypothesized that the presence of these proteins in N. ceranae makes them good targets for a gene-silencing approach.
Here we demonstrate that host ingestion of dsRNA for Nosema-specific ADP/ATP transporters can impact Nosema parasites, leading to specific gene silencing, parasite load reduction, and changes in honeybee physiology consistent with reduced Nosema disease. The results suggest an avenue for controlling this honeybee disease agent. To the best of our knowledge, this is the first description of effective and specific gene silencing in a microsporidian parasite.
MATERIALS AND METHODS
Bioinformatic analysis.
Reciprocal comparisons between Nosema and other eukaryotes were carried out by translating BLAST algorithms (www.ncbi.nlm.nih.gov). For more-extensive database searches, the public BLAST and PSI-BLAST software, available from NCBI, was used. Protein domains were identified and analyzed by using the Pfam protein family database (10), InterProScan (39), SMART (25), and HHPred (32). Protein multiple sequence alignments were created using ClustalW (24a) and visualized using Jalview (37). Relationships among ATP/ADP transporters were inferred using MEGA4 by neighbor-joining analyses based on 475 aligned positions and a bootstrap consensus tree (500 replicates, alignments available on request).
Production of dsRNA and sequences.
Primers for the dsRNAs and the full ATP/ADP transporter-homologous dsRNA sequences are provided in the supplemental material (see Methods S1). Synthesis of dsRNA was done essentially as described by Maori et al. (27).
Targeting ADP/ATP transporter sequences by using dsRNA.
We carried out several independent tests of the ability of N. ceranae ATP/ADP transporter-homologous dsRNAs to silence their targets, Nc006 (GenBank accession number EEQ83030.1) and Nc123 (GenBank accession number EEQ82057.1). Using viable N. ceranae spores collected from the midguts of infected adult worker bees, we inoculated naïve, newly emerged workers at 24 to 48 h following emergence and then reared these bees in sterile plastic cups (9). Bees were fed a sucrose solution supplemented with a final concentration of 20 μg/ml double-stranded RNA for the two targets singly or together at an equimolar ratio (see Methods S1 in the supplemental material), and an additional set of bees was maintained on sucrose solution alone, without exogenous dsRNA. After 6, 10, or 14 days, workers were sacrificed and screened for total Nosema infection (as measured by transcripts for rRNA or beta-tubulin targets assayed by quantitative PCR) (Table 1) (8) and for the specific reduction of the targeted transcripts. Transcript abundances were calculated relative to the bee gene encoding actin (Table 1), and statistical analyses were carried out using these relative threshold cycle (CT) values.
TABLE 1.
Primers used for quantitative PCR
| Primer | Target | Orientation | Sequence |
|---|---|---|---|
| B90111 | Honeybee actin | Forward | AGGAATGGAAGCTTGCGGTA |
| B90121 | Honeybee actin | Reverse | AATTTTCATGGTGGATGGTGC |
| Ntub7025 | N. ceranae tubulin | Forward | AGAACCAGGAACGATGGAGA |
| Ntub7026 | N. ceranae tubulin | Reverse | TCCTTGCAAACAATCTGCAC |
| JCNcer.F | N. ceranae 16S rRNA | Forward | CAATATTTTATTATTTTGAGAGA |
| JCNcer.R | N. ceranae 16S rRNA | Reverse | TATATTTATTGTATTGCGCGTGCA |
| NA70011 | Nc006 (forward) | Forward | CACCTGAAAACAACTTACCTAC |
| NA70021 | Nc006 (reverse) | Reverse | GTATCTTGCCTTACCATCAC |
| NA70031a | Nc123 (forward) | Forward | GGAAAAGATGAGAATATGGAAGAAG |
| NA70041b | Nc123 (reverse) | Reverse | CCAGTTACCCTTGTTTGTGTAGG |
Nosema infection levels in caged bees.
Nurse bees were collected from a single hive that was previously determined to be free from N. ceranae. Bees were transferred into individual boxes, ∼30 per box. All four ADP/ATP transporter dsRNA sequences were fed daily to the bees at a dose of 20 μg/ml of each dsRNA in a total of 1.5 ml per day 66% (wt/vol) sucrose solution. At study day 5, bees were infected with >100,000 spores per bee via addition of the spores to the sucrose solution, while controls were maintained on sucrose solution alone. Dead bees were collected each day from the cages for another 14 days, and Nosema spores inside their midguts were counted according to the method described in reference 3.
Response threshold to sucrose.
At the end of each experiment, 12 bees from each treatment box from the remaining live bees were taken for hunger bioassays. Bees infected with N. ceranae are known to have a lower threshold level of responsiveness to sugar in an assay for the proboscis extension reflex (PER) (28), a sign of nutritional stress. At the end of the feeding regimen in the boxes, the bees were strapped essentially as described by Mayack and Naug (28). An antenna of a strapped bee was touched with a droplet of sucrose, and whether bees then had a proboscis extension reflex or not was recorded. Each bee was assayed for the PER with a concentration (wt/vol) series of 0.1%, 0.3% (low sucrose concentration), 1%, 3% (intermediate), 10%, and 30% (high). In between every two successive concentrations, the antennae were touched with water to control for possible sensitization from repeated stimulation.
Statistical analysis.
The levels of N. ceranae spores in dead bees and differences in response threshold to sucrose in live bees were compared by paired t tests for the control and treatment groups (JMP software; SAS).
Nucleotide sequence accession number.
The draft genome assembly and protein list for N. ceranae are present at the NCBI Genome Project Resources site, under accession number ACOL01000000.
RESULTS
DICER and Argonaute candidates in the N. ceranae genome.
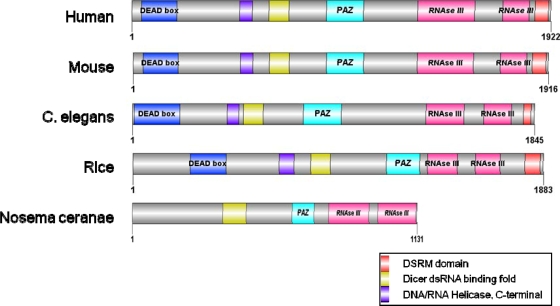
We identified an atypical N. ceranae DICER homologue (Fig. 1; see also Fig. S1 in the supplemental material), as well as a single Argonaute protein. This Argonaute homologue of Nosema contains several highly conserved domains and has high sequence similarity to the human Argonaute orthologue (see Fig. S2 in the supplemental material).
FIG. 1.
The Nosema ceranae putative DICER domain structure. Note the apparent absence of the common DEAD and helicase C domains. C. elegans, Caenorhabditis elegans.
Bacterium-like ADP/ATP transporters in the Nosema genome.
We identified four ADP/ATP transporters in the N. ceranae genome. Two of these transporters, Nc017 and Nc006, appear to be orthologous to specific proteins in E. cuniculi, while two additional N. ceranae transporters, Nc123 and Nc014, are paralogues whose split was considerably after the separation of lineages leading to Nosema and Encephalitozoon. The latter two appear to be most closely related to the mitosome-associated protein NTT3 (see Fig. S3 in the supplemental material) from Encephalitozoon (EcNTT3) (34). EcNTT3 seems to act by transporting ATP into the mitosome, in contrast to the actions of ADP/ATP transporters in most eukaryotes, which transport ATP out of the mitochondria into the cytoplasm (34).
Evidence that dsRNA fed to honeybee hosts targets specific N. ceranae transcripts.
When honeybees were fed dsRNA for genes encoding specific transporters, there was a reduction in transcript abundance for the predicted target in four separate trials. When feeding one of two ADP/ATP transporter dsRNAs (Nc006 or Nc123) and checking the transcript levels of the target and the alternate paralogue, the targeted gene showed consistently reduced abundance, differing in abundance by up to 15-fold from the alternate paralogue (Table 2).
TABLE 2.
Proportional transcript abundance of Nosema Nc006 relative to Nc123 for bees fed dsRNAs for Nc006, Nc123, and controlsa
| Bees fed dsRNA typeb | Nc006/Nc123 transcript ratio at dayb: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 6 |
10 |
13 |
14 |
|||||
| x̄ | SE | x̄ | SE | x̄ | SE | x̄ | SE | |
| Control (a) | 0.37 (a) | 0.11 | 0.44 (a) | 0.07 | 0.94 (a) | 0.15 | 1.26 (a) | 0.22 |
| Nc006 (ab) | 0.21 (a) | 0.07 | 0.42 (a) | 0.10 | 0.67 (a) | 0.13 | 1.08 (a) | 0.07 |
| Nc123 (b) | 0.641 (a) | 0.30 | 9.86 (b) | 3.02 | 3.38 (b) | 0.59 | 5.31 (b) | 1.68 |
Sampled at 6, 10, 13, and 14 days with samples of 7, 10, 9, and 9 bees, respectively.
Different parenthetical letters indicate significant differences.
Effects of RNAi-mediated knockdown of ADP/ATP transporters on host infection and behavior.
We compared N. ceranae levels in dead bees that succumbed to infection in all experiments from 7 to 15 days postinfection. The spore counts of infected bees indicated that bees treated with the dsRNA homologous to all four ATP/ADP transporters had a >3-fold lower spore count than untreated controls (for the dsRNA-treated bees, a mean of 4,205,532 spores/bee and standard error [SE] of 1,386,233 [n = 46], and for untreated bees, a mean of 13,240,035 spores/bee and SE of 1,735,100 [n = 29]; t test P < 0.0001). After controlled inoculation with 6,000 Nosema spores, bees given dsRNA for ADP/ATP transporters consistently showed lower infection levels, as measured by transcript abundance of N. ceranae 16S rRNA. Control bees showed an actin-normalized log2 abundance of −2.37 (n = 14; SE = 0.33), those fed Nc006 alone had levels of −4.11 (n = 8; SE = 0.80), those fed Nc123 had loads of −5.08 (n = 8; SE = 0.59), and those fed both Nc006 and Nc123 had a mean abundance of −4.97 (n = 8; SE = 0.39). Both the Nc123 and combined dsRNA treatments led to significantly lower Nosema loads than those for controls (t test P < 0.05).
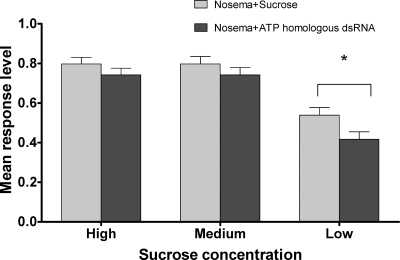
Our results also corroborate the study by Mayack and Naug (28) in showing that metabolic stress following N. ceranae infection can lead to an increased responsiveness of bees to very low concentrations of sucrose. Mayack and Naug found that PER to <1% sucrose was significantly increased in N. ceranae-infected bees. Here we show that the dsRNA treatment reduces PER at a <1% sucrose concentration (Fig. 2), indicating that silencing the ADP/ATP transport proteins has a direct effect on energy drain inflicted on the bees (total responses = 1,000). While not significantly different at high (10 to 30%) and moderate (1 to 3%) sucrose concentrations, the PER responsiveness of treated and untreated bees was significantly different (P < 0.03; responses = 330) at lower concentrations (0.1 to 0.3%). It is noteworthy that N. ceranae counts done under the microscope after the bees participated in the bioassay did not show a significant difference between counts at that point of the experiment (results not shown).
FIG. 2.
Effect of Nosema ceranae ADP/ATP transporter dsRNA on the responsiveness of Nosema-infected bees to various sucrose solutions. Response level refers to the proportion of bees extending their proboscis to different sucrose concentrations. Standard errors are shown. *, responses are significantly different at P < 0.03.
DISCUSSION
The recently published N. ceranae genome (5) revealed candidate silencing pathway genes, suggesting that this organism, like its fungal relatives, is capable of gene regulation via RNA interference. In order to target the main energy highway and minimize potential off-target effects, we chose the bacterial relic ADP/ATP transporter genes (see Fig. S3 in the supplemental material). The proteins encoded by these four genes play an important and probably crucial role in providing the necessary biochemical and metabolic needs of this obligatory parasite (34). We showed, by differentially silencing a specific gene, that highly specific N. ceranae endogenous gene silencing occurs via honeybee ingestion of the ADP/ATP transporter dsRNAs. Such specific endogenous silencing requires at least some involvement of the silencing pathway of N. ceranae.
We present strong evidence that host-ingested dsRNA can inhibit growth and development of an intracellular parasite. Why the various parameters (specific silencing and effect on N. ceranae viability and honeybee vitality) occasionally level off is currently not clear to us. Beyond the obvious aspects pertaining to the experimental setup and the level and stability of inoculants, the robust effect in the first few days, as well as the subsequent decline of the effect of the dsRNA, can result from a compensatory mechanism in N. ceranae. Genomic and proteomic studies of the N. ceranae-honeybee pathogenesis interaction should identify additional targets for silencing, which may provide a more robust overall reduction of N. ceranae pathogenesis. This study and a recent demonstration of RNAi-mediated knockdown of an RNA virus of bees (27) demonstrate that oral feeding of specific dsRNA can reduce target RNA and parasite loads in honeybees. Since infection with N. ceranae, arguably in concert with other parasites or stresses, has been shown to bring colonies to the state of collapse (6, 12, 36), a simultaneous gene-silencing approach of N. ceranae and other pathogens may have potential to address global colony declines. We further note the implications of this study to provide impetus for using RNAi to control microsporidian infections in other hosts, including humans.
Supplementary Material
Acknowledgments
We thank Scott Cornman and Alex Inberg for bioinformatics advice and Dawn Lopez for laboratory assistance.
This work was funded in part through a research grant from the Florida Department of Agriculture.
Footnotes
Published ahead of print on 9 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aronstein, K., T. Pankiw, and E. Saldivar. 2006. SID-I is implicated in systemic gene silencing in the honey bee. J. Apic. Res. 45:20-24. [Google Scholar]
- 2.Bailey, L., and B. V. Ball. 1991. Honey bee pathology, 2nd ed. Academic Press, London, United Kingdom.
- 3.Cantwell, G. E. 1970. Standard methods for counting Nosema spores. Am. Bee J. 110:222-223. [Google Scholar]
- 4.Chen, Y. P., J. D. Evans, C. Murphy, R. Gutell, M. Zuker, D. Gundersen-Rindal, and J. S. Pettis. 2009. Morphological, molecular, and phylogenetic characterization of Nosema ceranae, a microsporidian parasite isolated from the European honey bee, Apis mellifera. J. Eukaryot. Microbiol. 56:142-147. doi: 10.1111/j.1550-7408.2008.00374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornman, R. S., Y. P. Chen, M. C. Schatz, C. Street, Y. Zhao, B. Desany, M. Egholm, S. Hutchison, J. S. Pettis, W. I. Lipkin, and J. D. Evans. 2009. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox-Foster, D. L., S. Conlan, E. C. Holmes, G. Palacios, J. D. Evans, N. A. Moran, P. L. Quan, T. Briese, M. Hornig, D. M. Geiser, V. Martinson, D. vanEngelsdorp, A. L. Kalkstein, A. Drysdale, J. Hui, J. Zhai, L. Cui, S. K. Hutchison, J. F. Simons, M. Egholm, J. S. Pettis, and W. I. Lipkin. 2007. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 3:283-287. [DOI] [PubMed] [Google Scholar]
- 7.Ellis, J. D., and P. A. Munn. 2005. The worldwide health status of honey bees. Bee World 86:88-101. [Google Scholar]
- 8.Evans, J. D. 2006. Beepath: an ordered quantitative-PCR array for exploring honey bee immunity and disease. J. Invertebr. Pathol. 93:135-139. [DOI] [PubMed] [Google Scholar]
- 9.Evans, J. D., Y. P. Chen, G. Di Prisco, J. Pettis, and V. Williams. 2009. Bee cups: single-use cages for honey bee experiments. J. Apic. Res. 48:300-302. [Google Scholar]
- 10.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fries, I. 2010. Nosema ceranae in European honey bees (Apis mellifera). J. Invertebr. Pathol. 103:S73-S79. [DOI] [PubMed] [Google Scholar]
- 12.Fries, I., F. Feng, A. Da Silva, S. B. Slemenda, and N. J. Pieniazek. 1996. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 32:356-365. [Google Scholar]
- 13.Fries, I., R. Martin, A. Meana, P. Garcia-Palencia, and M. Higes. 2006. Natural infections of Nosema ceranae in European honey bees. J. Apic. Res. 45:230-233. [Google Scholar]
- 14.Higes, M., P. Garcia-Palencia, R. Martin-Hernandez, and A. Meana. 2007. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 94:211-217. [DOI] [PubMed] [Google Scholar]
- 15.Higes, M., R. Martin-Hernandez, C. Botias, E. G. Bailon, A. V. Gonzalez-Porto, L. Barrios, M. J. Del Nozal, J. L. Bernal, J. J. Jimenez, P. G. Palencia, and A. Meana. 2008. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 10:2659-2669. [DOI] [PubMed] [Google Scholar]
- 16.Higes, M., R. Martin, and A. Meana. 2006. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 92:81-83. [DOI] [PubMed] [Google Scholar]
- 17.Hirt, R. P., B. Healy, C. R. Vossbrinck, E. U. Canning, and T. M. Embley. 1997. A mitochondrial Hsp70 orthologue in Vairimorpha necatrix: molecular evidence that microsporidia once contained mitochondria. Curr. Biol. 7:995-998. [DOI] [PubMed] [Google Scholar]
- 18.Hirt, R. P., J. M. Logsdon, Jr., B. Healy, M. W. Dorey, W. F. Doolittle, and T. M. Embley. 1999. Microsporidia are related to fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl. Acad. Sci. U. S. A. 96:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honeybee Genome Sequencing Consortium. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443:931-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, W.-F., J.-H. Jiang, Y.-W. Chen, and C.-H. Wang. 2007. A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie 38:30-37. [Google Scholar]
- 21.Katinka, M. D., S. Duprat, E. Cornillott, G. Meteinler, F. Thomarat, G. Prensier, V. Barbe, E. Peyretaillade, P. Brottier, P. Wincker, F. Delbac, H. El Alaoul, P. Peyret, W. Saurin, M. Gouy, J. Weissenbach, and C. P. Vivares. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450-453. [DOI] [PubMed] [Google Scholar]
- 22.Keeling, P. J., M. A. Luker, and J. D. Palmer. 2000. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol. Biol. Evol. 17:23-31. [DOI] [PubMed] [Google Scholar]
- 23.Keeling, P. J., and G. I. McFadden. 1998. Origins of microsporidia. Trends Microbiol. 6:19-23. [DOI] [PubMed] [Google Scholar]
- 24.Klee, J., A. M. Besana, E. Genersch, S. Gisder, A. Nanetti, D. Q. Tam, T. X. Chinh, F. Puerta, J. M. Ruz, P. Kryger, D. Message, F. Hatjina, S. Korpela, I. Fries, and R. J. Paxton. 2007. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 96:1-10. [DOI] [PubMed] [Google Scholar]
- 24a.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, L. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 25.Letunic, I., T. Doerks, and P. Bork. 2009. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229-D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipardi, C., and B. M. Paterson. 2009. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc. Natl. Acad. Sci. U. S. A. 106:15645-15650. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Maori, E., N. Paldi, S. Shafir, H. Kalev, E. Tsur, E. Glick, and I. Sela. 2009. IAPV, a bee-affecting virus associated with colony collapse disorder can be silenced by dsRNA ingestion. Insect Mol. Biol. 18:55-60. [DOI] [PubMed] [Google Scholar]
- 28.Mayack, C., and D. Naug. 2009. Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. J. Invertebr. Pathol. 100:185-188. [DOI] [PubMed] [Google Scholar]
- 29.Molina, J.-M., M. Tourneur, C. Sarfati, S. Chevret, A. De Gouvello, J. G. Gobert, S. Balkan, and F. Derouin. 2002. Fumagillin treatment of intestinal microsporidiosis. N. Engl. J. Med. 346:1963-1969. [DOI] [PubMed] [Google Scholar]
- 30.Morse, R. A., and K. Flottum (ed.). 1997. Honey bee pests, predators and diseases, 3rd ed. A. I. Root Co., Medina, OH.
- 31.Siomi, H., and M. Siomi. 2009. On the road to reading the RNA-interference code. Nature 457:396-404. [DOI] [PubMed] [Google Scholar]
- 32.Söding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951-960. [DOI] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Tsaousis, A. D., E. R. S. Kunji, A. V. Goldberg, J. M. Lucocq, R. P. Hirt, and T. M. Embley. 2008. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 453:553-556. [DOI] [PubMed] [Google Scholar]
- 35.Van de Peer, Y., A. Ben Ali, and A. Meyer. 2000. Microsporidia: accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi. Gene 246:1-8. [DOI] [PubMed] [Google Scholar]
- 36.vanEngelsdorp, D., J. D. Evans, C. Saegerman, C. Mullin, E. Haubruge, B. K. Nguyen, M. A. Frazier, J. Frazier, D. Cox-Foster, Y. Chen, R. Underwood, D. R. Tarpy, and J. S. Pettis. 2009. Colony collapse disorder: a descriptive study. PLoS One 4(8):e6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse, A. M., J. B. Procter, D. M. Martin, M. Clamp, and G. J. Barton. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams, G. R., M. A. Sampson, D. Shutler, and R. E. L. Rogers. 2008. Does fumagillin control the recently detected invasive parasite Nosema ceranae in western honey bees (Apis mellifera)? J. Invertebr. Pathol. 99:342-344. [DOI] [PubMed] [Google Scholar]
- 39.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.