Abstract
Marine bacteria are a rich, yet underexplored, resource of compounds with inhibitory bioactivity against a range of eukaryotic target organisms. Identification of those inhibitors, however, requires a culturable or genetically tractable producer strain, a prerequisite that is not often fulfilled. This study describes a novel functional genomic screen that is based on expression of inhibitors in a heterogeneous recombinant host (i.e., Escherichia coli). Functional libraries were screened by selective grazing by the nematode Caenorhabditis elegans, in a simple, rapid, high-throughput manner. We applied our approach to discover inhibitors of C. elegans produced by the marine bacterium Pseudoalteromonas tunicata D2, a model organism for exploring a range of antagonistic activities between bacteria and eukaryotes and a known producer of several toxic compounds. Expression of P. tunicata DNA in E. coli and grazing selection by the nematode Caenorhabditis elegans identified two clones, with slow- and fast-killing modes of action. Genomic analysis of the slow-killing clone revealed that the activity was due to a small molecule, tambjamine, while the fast-killing activity involved a gene encoding for a novel protein. Microscopic analysis showed substantial colonization of the intestinal lumen, or rapid death of the nematode without colonization, for the two activities, respectively. The novel functional genomic screen presented here therefore detects new eukaryotic inhibitors with different chemical structures, kinetics, and predicted modes of actions.
Marine environments harbor highly diverse microbial communities, with an estimated more than one million different species (60). The vast majority of these are still functionally undescribed and unexplored, and only a fraction of the total number of species can currently be investigated by culture-dependent methods (47). Surface-associated marine microorganisms thrive in challenging habitats, often characterized by space and nutrient limitation, competition with other microorganisms, and colonizing higher organisms, as well as the targeted predation pressure by protozoa, nematodes, and other grazers. In response to this highly competitive environment, microorganisms have evolved strategies such as the production of toxins, attachment structures, biofilm formation, and host resilience in order to prevent the settlement and growth of competitive colonizers and for protection against bacterivorous predators. In fact, some of these adaptive traits are now recognized as virulence factors against a range of eukaryotic organisms, including plants, animals, and humans (24, 25, 44, 46). Despite this realization, there is limited information available on the presence and function of virulence factors in marine microbial organisms, nor is the full potential to mine such organisms for novel compounds with bioactivity realized.
The marine bacterial genus Pseudoalteromonas contains numerous species, which synthesize biologically active molecules. Many of these species have been demonstrated to produce an array of low- and high-molecular-weight compounds with antimicrobial, algicidal, neurotoxic, and other pharmaceutically relevant activities (7). P. tunicata strain D2 is the most comprehensively studied species within the genus (7). This species colonizes sessile eukaryotes such as algae and tunicates and is a producer of several compounds with inhibitory activities against a range of organisms. Although the identity of several of these compounds remains to be elucidated, they target a range of bacteria, fungi, invertebrate larvae, diatoms, algal spores, and protozoa (15, 28, 29). Furthermore, a recent analysis of the P. tunicata D2 genome revealed properties characteristic of pathogens such as curli, several proteases, and homologs to virulence regulators (59). Hence, P. tunicata D2 is a powerful model system in which to investigate bioactive compounds and their mode of action, including those that serve as virulence factors.
In order to detect and identify bacterial bioactive compounds that target multicellular eukaryotes, the nematode Caenorhabditis elegans can be utilized as a model system. This free-living worm provides several practical experimental advantages, including its ability to feed solely on bacteria, a short life cycle, and easy cultivation in large numbers. Comprehensive studies have reported the nematode as a versatile model metazoan in which to assess the virulence of many human, animal, plant, and insect pathogens (53). Some of the characteristics of the C. elegans immune system are conserved in higher eukaryotic organisms; moreover, diverse bacterial virulence factors necessary for killing of the nematode are used as virulence strategies regardless of the host (53). Despite the progress made using this model, current methods that help elucidate microbial genes involved in toxin-mediated killing or virulence are time-consuming or require expensive automation. Furthermore, a large fraction of potentially pathogenic bacteria elude investigations because they are not cultivable by using conventional laboratory techniques (47) or because of incompatible culture conditions for the pathogen and C. elegans (e.g., C. elegans is cultured at 25°C, while the Yersinia pestis virulence factors are upregulated only at 37°C [55]). Therefore, new high-resolution and simple methods are required to study genes and effector molecules mediating the inhibitory or toxic activity displayed by both cultured and uncultured bacteria.
In the present study we investigate the presence and activity of toxins in P. tunicata D2 with a rapid, culture-independent, eukaryotic screening assay. Our novel approach is based on the ability of C. elegans, using a sophisticated chemosensory system, to perceive and behaviorally respond to a range of chemical cues, including deterrence from noxious substances and attraction to nutrients or signals (2, 4, 26, 27, 45, 51, 52, 61, 62). The high-throughput screen successfully detected antinematode bioactive compounds and rapidly identified the responsible P. tunicata D2 genes and gene products in a recombinant Escherichia coli clone library. To our knowledge, this is the first time that a functional genomic library screening has been used to identify antinematode bioactive compounds.
MATERIALS AND METHODS
Strains and culture conditions.
All of the bacterial strains and vectors used in the present study are listed in Table 1 . The large insert library was constructed in E. coli EPI300-T1R using a fosmid CopyControl vector (Epicentre, Wisconsin) as described by Burke et al. (11).
TABLE 1.
Bacterial strains and vectors used in this study
| Strain or vector | Relevant characteristic or genotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| EPI300-T1R | F−mcrA Δ(mrr hsdRMS mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG trfA tonA dhfr | Epicentre |
| OP50 | Uracil auxotroph | 9 |
| P. tunicata D2 | Marine isolate | 29 |
| P. aeruginosa PA01 | Gene cluster hcnABC, involved in the synthesis of the toxic compound hydrogen cyanide | 18 |
| Vectors | ||
| pCC1FOSa | Fosmid backbone for P. tunicata D2 library; Cmr | Epicentre |
| p519ngfp | High-copy-number plasmid with constitutive GFP expression; Kmr | 54 |
| pBAD-TOPO ZP_01132244a | D2 wild-type gene ZP_01132244 cloned downstream the PBAD promoterb; Ampr | This study |
Copy number inducible by arabinose.
According to the manufacturer's protocol (pBAD TOPO TA expression kit; Invitrogen).
All strains were grown on Luria broth (LB10), nematode growth medium (NGM) (56), marine broth (Difco Laboratories, Maryland), or complex marine medium (VNSS) (37) as indicated and stored in 30% (vol/vol) glycerol at −80°C. Solid medium was prepared by the addition of 19 g of agar (Oxoid, Australia)/liter. All strains were grown at 37°C with the exception of P. tunicata D2, which was grown at 25°C. Where required, ampicillin (100 μg/ml), chloramphenicol (12.5 μg/ml), kanamycin (100 μg/ml), tetracycline (10 μg/ml), and l-arabinose (0.02% [wt/vol]) were added to the media.
C. elegans strain Bristol N2 (9) was maintained at 25°C on NGM agar plates spread with E. coli OP50 as a food source (9).
Fosmid library screening.
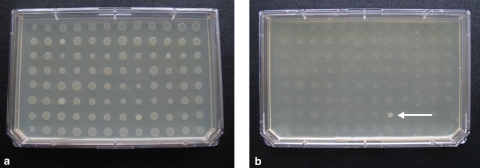
Nine hundred clones of a P. tunicata D2 genome library in E. coli (average insert size, 35 kb) were spotted from frozen stock cultures on LB10 agar plates (Nunc, Denmark) by using a 96-pin replicator tool and allowed to grow for 5 days at 25°C (see Fig. 2a). Ninety-six colonies of PA01 strain and OP50 strain were spotted in the same manner on LB10 agar plates, followed by incubation at 37 and 25°C, respectively. Chloramphenicol and l-arabinose were added to the media where required. At day 5, 5 μl of M9 medium (40) with four to five L4-stage nematodes were added next to each colony with a multichannel pipette. The plates were stored at room temperature and checked every 24 h. A total of three replicates were included. A clone was considered positive if the colony was not grazed after 5 days of incubation with C. elegans. Putative positive clones were confirmed by testing their ability to resist the grazing of the nematode on separate 3.5-cm petri dishes with LB10 agar medium plus chloramphenicol and l-arabinose.
FIG. 2.
Nematode selective grazing. E. coli fosmid libraries with a large DNA insert of P. tunicata D2 on agar plate before the incubation with nematodes (a) and after 5 days of incubation (b) are shown. The clone HG8 (arrow, panel b) is still present and visible after the incubation with nematodes, whereas all of the other library clones were grazed within that time.
Nematode killing assay.
Grazing-resistant clones were grown overnight at 37°C in LB10, and 10-μl portions of the culture were spread onto 3.5-cm LB10 agar plates supplemented with selective antibiotic and l-arabinose as needed, followed by incubation at 25°C for 4 days. A random library clone was used as a negative control under the same conditions. P. tunicata D2 was grown overnight at 25°C in VNSS, and the culture was spread onto a 3.5-cm VNSS agar plate, followed by incubation at 25°C for 4 days. For an additional negative control, the E. coli OP50 strain was grown overnight at 37°C in LB10, and 10 μl of the culture was spread onto a 3.5-cm VNSS agar plate, followed by incubation at 25°C for 4 days. L4-stage nematodes (30 to 40 per plate) were added to the plate, and each assay was carried out in triplicate. All of the strains were incubated at 25°C and scored for live and dead worms every 24 h for 5 days. A worm was considered dead when it failed to respond to touch.
In order to confirm that the killing activity was due to the insert DNA, fosmid DNA was extracted from positive clones by using a QIAprep miniprep kit (Qiagen, Netherlands) according to the manufacturer's protocol and transformed in EPI300-T1R using electroporation (GenePulser X Cell; Bio-Rad, California), and these clones were tested for nematode killing. In addition, clones were tested in the absence of arabinose, which is an inducer to increase the fosmid copy number about 50-fold (Epicentre).
Fosmid analysis and transposon mutant library screening.
The fosmid insert ends from active clones were sequenced by using EpiFOS forward and reverse primers (Epicentre) on an ABI 3730 DNA sequencer at the University of New South Wales Automated DNA Analysis Facility. Sequences were subjected to BLAST analysis against the P. tunicata D2 genome using the Integrated Microbial Genome database (38).
The fosmid clones HG8 and AA11 were mutagenized by using an in vitro transposon mutagenesis kit (EZ-Tn5 insertion kit; Epicentre). For each fosmid a library of 96 E. coli transposon mutants was generated, replicated on LB10 Omnitray plates (Nunc, Denmark), and screened as described for the fosmid library. Clones that were partially or totally grazed by the worms were chosen for further characterization in the nematode killing assay. The genes disrupted were identified by outward sequencing from the transposon using the TET-1 forward and reverse primers (Epicentre). Prediction of the signal peptide in the ZP_01132244 protein was carried out by using the SignalP 3.0 Server (3).
Construction of complementation plasmid.
Primers were designed to amplify gene ZP_01132244 for expression in the pBAD-TOPO TA expression system in accordance with the manufacturer's instructions (Invitrogen, California). Primers ZP_01132244F (GAGGAATAATAAATGAAATTTATCTATAGAAATACG-3′) and ZP_01132244R (5′-TTAATCAAGCAAGGTTAACTTAGTG-3′) were used to amplify ZP_01132244 gene under the following conditions: 20 ng of template fosmid DNA, 10 pmol of F primer and 10 pmol of R primer, 1× RedTaq buffer, 10 nmol of deoxynucleoside triphosphate mix, and 2 U of RedTaq in a 20-μl reaction, cycled at 94°C for 1 min, followed by 25 cycles of 94°C for 30 s, 53°C for 30 s, and 2°C for 2 min, followed in turn by a final extension at 72°C for 5 min. The PCR products were quantified via gel electrophoresis and then ligated to the pBAD-TOPO cloning vector (Table 1) according to the manufacturer's instructions (Invitrogen). Ligation products were transformed into chemically competent HG8 Tn::ZP_01132244 cells and selected on LB10 agar with ampicillin, tetracycline, and chloramphenicol. Colony PCR was used to confirm that the genes had been cloned in the correct orientation. A 10-pmol portion of the ZP_01132244R primer described above and 10 pmol of the forward primer from the pBAD-TOPO vector (Invitrogen) were used for PCRs as described above with a small number of cells from the clone as the DNA template.
Microscopy of C. elegans.
A green fluorescent protein (GFP)-expressing plasmid (Table 1) was transformed in E. coli EPI300-T1R clones using electroporation (GenePulser X Cell). Nematodes were exposed to GFP-labeled bacteria, collected after death (except for the negative control which exposure lasted 5 days), and placed on a microscope slide in 5% formaldehyde. LB10 agar plates with the addition of chloramphenicol, kanamycin, and l-arabinose were used as required. Worms were examined with an Olympus DP70 digital camera system with differential interference microscope optics and epifluorescence microscopy.
RESULTS
Functional genomics screen for the identification of toxicity against C. elegans.
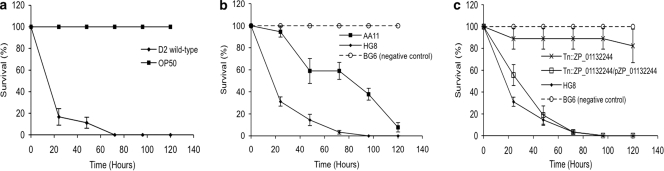
We first assessed whether the marine bacterium Pseudoalteromonas tunicata D2, a known producer of several toxic compounds (15, 28, 29), was able to kill C. elegans. When nematodes were placed on a bacterial lawn of P. tunicata D2, there were no live animals present after 5 days, and more than 70% were killed in the first 24 h (see Fig. 3a); the bacterial lawn remained intact, and no progeny was generated at that time. In contrast, nematodes grown on E. coli OP50, the laboratory food source of C. elegans (9), were able to feed and reproduce and at day 5 the bacterial lawn was removed.
FIG. 3.
Kinetics of C. elegans killing. The ability to kill the nematode was tested for P. tunicata D2 wild-type (a), the positive clones AA11 and HG8 (b), and a clone HG8/ZP_01132244 mutant complemented with the wild-type gene ZP_01132244 (c). The numbers of survival worms are expressed as a percentage. Each data point represents means ± the standard error of three replicates.
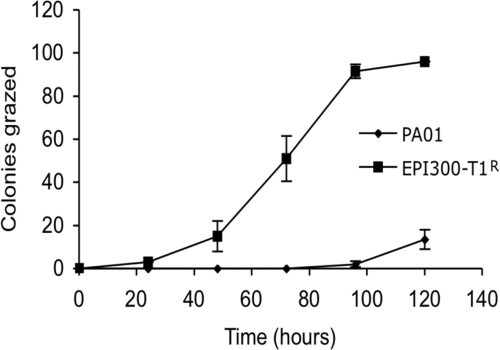
To develop a functional genomic library screen for the rapid identification of bioactive compounds and to unravel the means by which P. tunicata D2 kills C. elegans, we first established that worms in our system graze nontoxic colonies of the E. coli expression host and that toxic bacterial colonies resist the nematode grazing. Nematodes on agar plates were incubated with 96 colonies of E. coli EPI300-T1R and with 96 colonies of Pseudomonas aeruginosa PAO1, an organism with known toxicity against C. elegans (18). After 5 days, all colonies of the nontoxic E. coli were removed by grazing, whereas none or only a few of the toxic P. aeruginosa PAO1 colonies were grazed (Fig. 1). Next, an E. coli EPI300-T1R fosmid library with large DNA inserts (∼35 kb) of P. tunicata D2 (11) was screened for clones that resist the nematode grazing. Nine hundred clones of the E. coli library representing an ∼6-fold coverage of the P. tunicata D2 genome were exposed to predation by the worms. In order to identify bacterial toxins, we took advantage of the capacity of C. elegans to sense and behaviorally respond to bacterial metabolites (2, 51, 61). As a consequence, C. elegans has the ability to selectively graze nontoxic clones, whereas avoiding toxic clones of the E. coli host expressing heterologous DNA (see Video S1 in the supplemental material). After 5 days, two clones (designated AA11 and HG8) were observed not to be grazed, while all other clones of the library were removed by the nematodes (Fig. 2). Clone AA11, which displayed a bright yellow color, and clone HG8 were confirmed to fully resist nematode grazing on replicate plates, indicating that no false positives were scored using this method. The two nematode-resisting clones (AA11 and HG8) were selected for further studies.
FIG. 1.
Toxic colonies resist C. elegans grazing. The numbers of bacterial colonies (of a total of 96) of the nontoxic E. coli EPI300-T1R and the toxic P. aeruginosa PA01 grazed by C. elegans in 5 days are indicated. Each data point represents means ± the standard error of three replicates.
Fosmid DNA inserts of the P. tunicata D2 genome are responsible for killing of C. elegans.
Bacterial lawns of the two grazing resistant clones AA11 and HG8 were found to kill C. elegans. After 5 days, AA11 killed ca. 70% of the total number of worms, while HG8 caused death of 70% of the worms within the first 24 h (Fig. 3 b). Several eggs were found on the lawn of AA11, but no progeny were observed at day 5. On the HG8 lawn, few eggs and no young worms were present. No loss in viability of the nematode was found on a lawn of a randomly selected clone (BG6) from the E. coli library that did not show resistance to grazing in the original fosmid clone screen (Fig. 3b).
In order to exclude the possibility that the toxicity of the two active clones was caused by either a mutation in E. coli EPI300-T1R or an external contamination, the fosmids of these clones were retransformed into E. coli EPI300-T1R and subjected to the nematode assay. Both retransformants showed killing of the nematodes with the same kinetics as the original clones (data not shown). In addition, the killing was abolished when the copy-number of the fosmid was repressed by the omission of the inducer arabinose (see Materials and Methods). These results confirm that the two fosmid inserts of the P. tunicata D2 genome were responsible for the grazing resilience and killing of the worms.
The gene cluster producing the small molecule tambjamine is causing the slow killing of C. elegans.
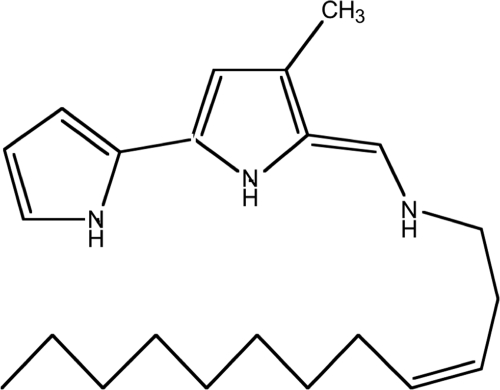
End sequencing of the slow-killing AA11 fosmid and mapping onto the P. tunicata D2 genome showed that the clone contains an insert of 39 kb, which includes a gene cluster involved in the production of a small yellow molecule, the antifungal compound tambjamine YP1 (11, 17). An AA11 transposon mutant library (96 clones) was screened for nematode killing, and 11 clones exhibiting an attenuated activity were identified. After sequencing from the transposon cassette 11 different genes were found to be disrupted. These genes were all located in the operon involved in the production of the P. tunicata D2 tambjamine compound YP1 (11, 15) (Fig. 4). Our results suggest that each gene in this operon is required for the expression of the slow-killing activity. All transposon mutants in the tambjamine gene cluster also displayed a loss of the yellow pigment, a finding characteristic of the AA11 clone. Moreover, a 1-h heat exposure at 65°C of the agar plate with the AA11 lawn, prior to addition of C. elegans, resulted in extended viability of the nematode by only 20 h (data not shown), further supporting the notion that the heat-stable tambjamine is involved in the killing of C. elegans. Evidence of the production of the tambjamine YP1 by the AA11 E. coli clone was demonstrated by Burke and coworkers in 2007 (11). These authors analyzed bacterial cell extracts of the AA11 E. coli clone by electrospray mass spectroscopy. The extract was shown to contain a distinct molecular mass of 356 Da (11), which is consistent with the mass of YP1 produced by P. tunicata D2 (17).
FIG. 4.
P. tunicata D2 tambjamine YP1.
Tambjamine compounds (Fig. 4) have been shown to possess antimicrobial (6, 11), antitumorigenic (31), immunosuppressive (22), antiproliferation (57), and ichtyodeterrent activities (35). To the best of our knowledge, the antinematode activity detected in the present study represents a novel property for this class of compounds. Tambjamine compounds were first isolated from marine invertebrates (35), but later studies revealed that they are produced by epibiotic bacteria on such marine living surfaces (11, 32), suggested to aid the host in protecting itself against predatory pressure (7).
A P. tunicata D2 gene coding for an unknown protein is involved in the fast killing of C. elegans.
The transposon mutant library of HG8 (insert size, 13.8 kb) revealed one insert-disrupted clone, H10 that had a lower rate of killing relative to the positive clone HG8 (Fig. 3c). Clone H10 was found to have a disruption in the P. tunicata D2 gene ZP_01132244 (NCBI locus), which encodes a protein of unknown function. The attenuated killing activity of the H10 clone was largely restored by complementation with a plasmid carrying the wild-type gene (Fig. 3c). In silico analysis showed that the gene product of ZP_01132244 contains a signal peptide and is therefore predicted to be excreted. More than 80% of the nematodes remained viable after a HG8 lawn was exposed to 65°C for 1 h, suggesting that the antinematode compound produced by this clone was heat sensitive.
The fast- and slow-killing activities display different modes of action.
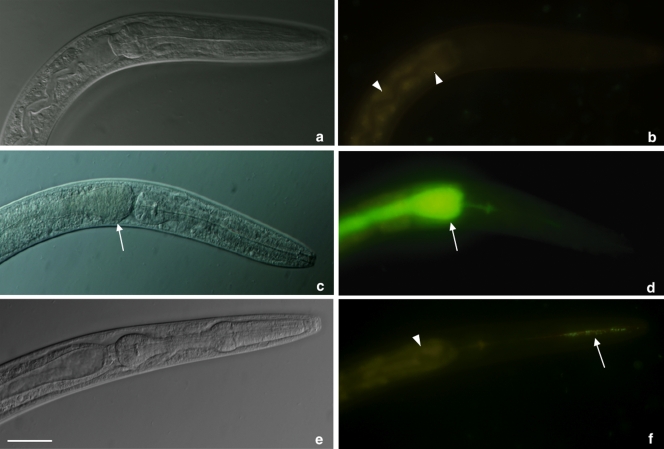
In order to gain insight into the mode of action, we exposed the worms to the two active bacterial clones labeled with the green fluorescent protein (GFP). Analysis by fluorescence microscopy of the negative control clone BG6 showed no bacterial colonization in the intestinal lumen (Fig. 5 b). The fast-killing clone HG8 displayed the same absence of colonization as did the negative control and only a small number of cells were present in the pharynx of worms (arrow in panel f). In contrast, worms that died in the presence of the slow-killing clone AA11 showed substantial colonization of the intestinal lumen (arrows in panels c and d). This result suggests that the tambjamine compound may play a role in facilitating bacterial colonization of the nematode's gut, while the fast-killing clone expressing the antinematode compound acts in a colonization-independent manner.
FIG. 5.
Visualization of E. coli clones GFP labeled inside the nematodes. Images show the pharynx and first part of the intestinal lumen of C. elegans by differential interference microscopy (a, c, and e), and under fluorescence microscopy (b, d, and f). No bacterial cells were detected in the intestinal lumen of worms exposed to the negative control BG6 (b). Similarly, nematodes killed by the fast-killing clone showed no colonization of the intestine and only few live cells were observed in the pharynx (arrow in panel f). Worms killed by the slow-killing clone AA11 show massive colonization of the intestinal lumen, which expanded (arrows in panels c and d). Autofluorescence of nematode intestinal cells is visible in panels b and f (arrowheads). Scale bar, 0.05 mm.
DISCUSSION
Recent studies of surface associated marine bacteria have led to the hypothesis that such microorganisms use a range of inhibitory bioactive compounds for the successful competition for nutrients and space and for protection against predators (16, 39). In silico and in vitro observations on such bacteria, including P. tunicata D2 (15, 44, 59), suggest an unexplored potential for the discovery of effective defense systems and toxic compounds against a range of eukaryotes. In order to explore the diversity of uncultured organisms, the first biologically active operons were cloned and screened for production of new antibiotic compounds approximately 10 years ago (8, 48), and several new bioactive compounds have been revealed using this approach (23, 36, 50). Contemporary high-throughput functional genomics and metagenomics, however, require the development of new and efficient screens for the discovery of novel molecules and activities (23, 36, 58). Using a simple, high-throughput screen, we have here identified bioactive compounds involved in the killing of the nematode C. elegans. This was achieved by the screening of a genomic library of E. coli containing large inserts of the P. tunicata D2 genome. To our knowledge, this is the first report of the successful application of such a screening approach.
Exhaustive studies have investigated the presence of toxic genes acting against the model organism C. elegans by screening bacterial mutant libraries for attenuated killing effects (see, for example, reference 53). Although this approach can provide a high-resolution detection of genes encoding toxic compounds, it usually requires a large number of mutants to be screened. For example, to identify the gene(s) responsible for the antinematode activity of P. tunicata D2, we estimate that at least 10,000 random transposon mutants must be screened for loss of activity in order to cover ∼4,500 protein-coding genes in the genome (59). Moreover, the transposon mutagenesis approach requires that the bacterial producers are readily cultivable, especially under the conditions that allow nematode growth. In the present study we developed a rapid, functional screen based on the selective nematode grazing of recombinant E. coli clones that can contain DNA from any source (including uncultured and/or pathogenic bacteria). Hence, our method harbors great potential for the exploration of unexplored biodiversity through metagenome-based screens.
In the natural environment the bacteriovorous C. elegans, encounters a great variety of microorganisms. Some of these bacteria are an ideal food source for the nematode, and some others are harmful; as a consequence, C. elegans has developed sophisticated strategies to distinguish and behaviorally respond to different food sources, including deterrence from noxious substances and attraction to nutrients or signals (51, 61). To date, at least two main mechanisms of grazing avoidance have been reported in C. elegans. The first relies on the ability of the nematode to detect repellent olfactory cues produced by microorganisms, which triggers an immediate avoidance behavior in C. elegans (45). The second mechanism involves the ingestion of pathogenic bacteria by the nematode, which induces a process of aversive learning leading to the ability to subsequently avoid the pathogen (62). In the present study we took advantage of the C. elegans' ability to behaviorally respond to bacterial metabolites, and we designed a screen for the genomic library that allows the worm to use both mechanisms of grazing avoidance. Our results demonstrate that C. elegans is able to distinguish between toxic and nontoxic bacterial colonies and preferentially feeds on the nonactive rather than on positive clones in the functional gene library (Fig. 2 and see Video S1 in the supplemental material). The screening method provided sufficient resolution to identify different types of nematode killing activity and to suggest modes of action. This is illustrated by the detection of a slow- and a fast-killing activity in the P. tunicata D2 library. The slow killing was based on the small molecule tambjamine, which appears to facilitate intestinal colonization of the E. coli host cells. The finding that the heat-killed slow-killing AA11 clone is still able to kill the worms, albeit after a longer period of time than for non-heat-killed cells, supports the role of the heat-stable tambjamine compound. The longer time period required for nematode killing is also consistent with the notion that the slow-killing phenotype reflects the combined effect of colonization of the worm's gut by live cells and the production of the tambjamine compound. A similar mode of action was also found to be involved in nematode killing by Burkholderia pseudomallei and B. cepacia (19, 33, 43). The fast-killing activity appears to act in a colonization-independent fashion, and the heat-dependent loss of activity by the HG8 clone is likely the result of denaturation of a heat-labile, antinematode activity. Together, these results demonstrate that gene-based screens of the kind presented here detect multiple activities with predicted mode of actions, a resolution that is not necessarily available when screening mutant libraries of bacterial isolates.
The method presented here has immediate applications for the identification and studies of bioactive compounds derived from bacteria in a range of settings. For example, as reported by Sifri et al. (53), 12 of 25 human pathogens investigated using the C. elegans model use toxins as major or synergistic determinants of virulence. We submit that our method has the potential to identify microbial toxins and thus facilitate the process of dissecting the interactions between host and pathogen. Moreover, bacterial toxins against C. elegans, such as the crystal toxin of Bacillus thuringiensis and the avermectin derivatives of Streptomyces avermitilis, have been developed into important commercial anthelmintic products in agriculture and human and animal health (10, 20, 21). The increased resistance by parasitic nematodes to currently used antihelminthics clearly necessitates the identification of new drug leads and targets, and the approach detailed here is likely to significantly facilitate the screening process in the early stage of drug discovery programs. Furthermore, in recent years bacterial toxins have emerged as an invaluable tool for the fields of molecular and cell biology, providing researchers with specific and efficient inhibitors of key cellular pathways (1, 30, 34, 49). New biological inhibitors with new molecular targets and mode of action are needed to explore eukaryotic cellular processes, and the method presented here has the specificity to reveal novel bioactive compounds with such characteristics. From an ecological point of view, the method described in the present study can be used to improve the current understanding of microbial interactions in the environment. In the majority of habitats, including marine systems, nematodes are effective bacterivorous predators, and their ability to selective graze microbes has a strong impact on the microbial community (5, 12-14, 41, 42); yet little is known about the mechanisms by which bacteria protect themselves from such predation. The present study uncovered two substances produced by the marine surface associated bacterium P. tunicata that are likely to be involved in predation protection. While most of the studies using C. elegans as a model for microbial virulence have focused on the investigation of known pathogens (53), to our knowledge ours is the first study that use C. elegans to investigate the inhibitory activity against nematodes and the potential defense mechanisms against predation of an environmentally relevant microorganism. More broadly, we submit that the methodology presented will be particularly useful for exploring environmental genomic (metagenomic) libraries representing uncultivable microbial communities in a range of ecological habitats that are likely to contain bacteria producing bioactive compounds.
Supplementary Material
Acknowledgments
This study was supported by the Australian Research Council and the Centre for Marine Bio-Innovation.
We thank the Caenorhabditis Genetics Center at the University of Minnesota for provision of C. elegans samples and the University International Postgraduate Award for supporting F.B.
Footnotes
Published ahead of print on 2 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aktories, K., E. A. Joseph, and R. P. Michel. 2006. Toxins as tools, p. 976-990. In The comprehensive sourcebook of bacterial protein toxins, 3rd ed. Academic Press, London, England.
- 2.Bargmann, C. I. 2006. Chemosensation in C. elegans. In The Caenorhabditis elegans research community. WormBook doi: 10.1895/wormbook.1.123.1.http://www.wormbook.org/chapters/www_chemosensation/chemosensation.html. [DOI] [PMC free article] [PubMed]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Bergarnasco, C., and P. Bazzicalupo. 2006. Chemical sensitivity in Caenorhabditis elegans. Cell. Mol. Life Sci. 63:1510-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanc, C., M. Sy, D. Djigal, A. Brauman, P. Normand, and C. Villenave. 2006. Nutrition on bacteria by bacterial-feeding nematodes and consequences on the structure of soil bacterial community. Eur. J. Soil Biol. 42:S70-S78. [Google Scholar]
- 6.Boger, D. L., and M. Patel. 1988. Total synthesis of prodigiosin, prodigiosene, and desmethoxyprodigiosin: Diels-Alder reactions of heterocyclic azadienes and development of an effective palladium(II)-promoted 2,2-bipyrrole coupling procedure. J. Organic Chem. 53:1405-1415. [Google Scholar]
- 7.Bowman, J. P. 2007. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 5:220-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brady, S. F., C. J. Chao, J. Handelsman, and J. Clardy. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Organic Lett. 3:1981-1984. [DOI] [PubMed] [Google Scholar]
- 9.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burg, R. W., B. M. Miller, E. E. Baker, J. Birnbaum, S. A. Currie, R. Hartman, Y. L. Kong, R. L. Monaghan, G. Olson, I. Putter, J. B. Tunac, H. Wallick, E. O. Stapley, R. Oiwa, and S. Omura. 1979. Avermectins, new family of potent anthelmintic agents producing organism and fermentation. Antimicrob. Agents Chemother. 15:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke, C., T. Thomas, S. Egan, and S. Kjelleberg. 2007. The use of functional genomics for the identification of a gene cluster encoding for the biosynthesis of an antifungal tambjamine in the marine bacterium Pseudoalteromonas tunicata. Environ. Microbiol. 9:814-818. [DOI] [PubMed] [Google Scholar]
- 12.De Mesel, I., S. Derycke, T. Moens, K. Van der Gucht, M. Vincx, and J. Swings. 2004. Top-down impact of bacterivorous nematodes on the bacterial community structure: a microcosm study. Environ. Microbiol. 6:733-744. [DOI] [PubMed] [Google Scholar]
- 13.De Mesel, I., S. Derycke, J. Swings, M. Vincx, and T. Moens. 2003. Influence of bacterivorous nematodes on the decomposition of cordgrass. J. Exp. Mar. Biol. Ecol. 296:227-242. [Google Scholar]
- 14.Djigal, D., A. Brauman, T. A. Diop, J. L. Chotte, and C. Villenave. 2004. Influence of bacterial-feeding nematodes (Cephalobidae) on soil microbial communities during maize growth. Soil Biol. Biochem. 36:323-331. [Google Scholar]
- 15.Egan, S., S. James, C. Holmstrom, and S. Kjelleberg. 2002. Correlation between pigmentation and antifouling compounds produced by Pseudoalteromonas tunicata. Environ. Microbiol. 4:433-442. [DOI] [PubMed] [Google Scholar]
- 16.Egan, S., T. Thomas, and S. Kjelleberg. 2008. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr. Opin. Microbiol. 11:219-225. [DOI] [PubMed] [Google Scholar]
- 17.Franks, A., P. Haywood, C. Holmstrom, S. Egan, S. Kjelleberg, and N. Kumar. 2005. Isolation and structure elucidation of a novel yellow pigment from the marine bacterium Pseudoalteromonas tunicata. Molecules 10:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan, Y. H., K. L. Chua, H. H. Chua, B. P. Liu, C. S. Hii, H. L. Chong, and P. Tan. 2002. Characterization of Burkholderia pseudomallei infection and identification of novel virulence factors using a Caenorhabditis elegans host system. Mol. Microbiol. 44:1185-1197. [DOI] [PubMed] [Google Scholar]
- 20.Griffitts, J. S., J. L. Whitacre, D. E. Stevens, and R. V. Aroian. 2001. Bt toxin resistance from loss of a putative carbohydrate-modifying enzyme. Science 293:860-864. [DOI] [PubMed] [Google Scholar]
- 21.Haber, C. L., C. L. Heckaman, G. P. Li, D. P. Thompson, H. A. Whaley, and V. H. Wiley. 1991. Development of a mechanism of action-based screen for anthelmintic microbial metabolites with avermectinlike activity and isolation of milbemycin-producing Streptomyces strains. Antimicrob. Agents Chemother. 35:1811-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han, S. B., H. M. Kim, Y. H. Kim, C. W. Lee, E. S. Jang, K. H. Son, S. U. Kim, and Y. K. Kim. 1998. T-cell specific immunosuppression by prodigiosin isolated from Serratia marcescens. Int. J. Immunopharmacol. 20:1-13. [DOI] [PubMed] [Google Scholar]
- 23.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvell, C. D., K. Kim, J. M. Burkholder, R. R. Colwell, P. R. Epstein, D. J. Grimes, E. E. Hofmann, E. K. Lipp, A. D. Osterhaus, R. M. Overstreet, J. W. Porter, G. W. Smith, and G. R. Vasta. 1999. Emerging marine diseases—climate links and anthropogenic factors. Science 285:1505-1510. [DOI] [PubMed] [Google Scholar]
- 25.Hilbi, H., S. S. Weber, C. Ragaz, Y. Nyfeler, and S. Urwyler. 2007. Environmental predators as models for bacterial pathogenesis. Environ. Microbiol. 9:563-575. [DOI] [PubMed] [Google Scholar]
- 26.Hilliard, M. A., C. I. Bargmann, and P. Bazzicalupo. 2002. C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr. Biol. 12:730-734. [DOI] [PubMed] [Google Scholar]
- 27.Hilliard, M. A., C. Bergamasco, S. Arbucci, R. H. A. Plasterk, and P. Bazzicalupo. 2004. Worms taste bitter: ASH neurons, QUI-1, GPA-3, and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. EMBO J. 23:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmstrom, C., S. Egan, A. Franks, S. McCloy, and S. Kjelleberg. 2002. Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol. Ecol. 41:47-58. [DOI] [PubMed] [Google Scholar]
- 29.Holmstrom, C., S. James, B. A. Neilan, D. C. White, and S. Kjelleberg. 1998. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 48:1205-1212. [DOI] [PubMed] [Google Scholar]
- 30.Iacovache, I., F. G. van der Goot, and L. Pernot. 2008. Pore formation: an ancient yet complex form of attack. Biochim. Biophys. Acta, Bioenerg. 1778:1611-1623. [DOI] [PubMed] [Google Scholar]
- 31.Kojiri, K., S. Nakajima, H. Suzuki, A. Okura, and H. Suda. 1993. A new antitumor substance, BE-18591, produced by a streptomycete. I. Fermentation, isolation, physico-chemical and biological properties. J. Antibiotics. 46:1799-1803. [DOI] [PubMed] [Google Scholar]
- 32.Konig, G. M., S. Kehraus, S. F. Seibert, A. Abdel-Lateff, and D. Muller. 2006. Natural products from marine organisms and their associated microbes. Chembiochem 7:229-238. [DOI] [PubMed] [Google Scholar]
- 33.Kothe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 34.Lalli, G., S. Bohnert, K. Deinhardt, C. Verastegui, and G. Schiavo. 2003. The journey of tetanus and botulinum neurotoxins in neurons. Trends Microbiol. 11:431-437. [DOI] [PubMed] [Google Scholar]
- 35.Lindquist, N., and W. Fenical. 1991. New tamjamine class alkaloids from the marine ascidian Atapozoa sp. and its nudibranch predators: origin of the tambjamines in Atapozoa. Experientia 47:504-506. [Google Scholar]
- 36.Lorenz, P., and J. Eck. 2005. Metagenomics and industrial applications. Nat. Rev. Microbiol. 3:510-516. [DOI] [PubMed] [Google Scholar]
- 37.Mårdén, P., A. Tunlid, K. Malmcrona-Friberg, G. Odham, and S. Kjelleberg. 1985. Physiological and morphological changes during short-term starvation of marine bacterial isolates. Arch. Microbiol. 142:326-332. [Google Scholar]
- 38.Markowitz, V. M., I. M. Chen, K. Palaniappan, K. Chu, E. Szeto, Y. Grechkin, A. Ratner, I. Anderson, A. Lykidis, K. Mavromatis, N. N. Ivanova, and N. C. Kyrpides. 2009. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38:382-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matz, C., J. S. Webb, P. J. Schupp, S. Y. Phang, A. Penesyan, S. Egan, P. Steinberg, and S. Kjelleberg. 2008. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS One 3:e2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 41.Moens, T., L. Verbeeck, A. de Maeyer, J. Swings, and M. Vincx. 1999. Selective attraction of marine bacterivorous nematodes to their bacterial food. Mar. Ecol. Prog. Ser. 176:165-178. [Google Scholar]
- 42.Newsham, K. K., J. Rolf, D. A. Pearce, and R. J. Strachan. 2004. Differing preferences of Antarctic soil nematodes for microbial prey. Eur. J. Soil Biol. 40:1-8. [Google Scholar]
- 43.O'Quinn, A. L., E. M. Wiegand, and J. A. Jeddeloh. 2001. Burkholderia pseudomallei kills the nematode Caenorhabditis elegans using an endotoxin-mediated paralysis. Cell Microbiol. 3:381-393. [DOI] [PubMed] [Google Scholar]
- 44.Persson, O. P., J. Pinhassi, L. Riemann, B.-I. Marklund, M. Rhen, S. Normark, J. M. Gonzalez, and A. Hagstrom. 2009. High abundance of virulence gene homologues in marine bacteria. Environ. Microbiol. 11:1348-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pradel, E., Y. Zhang, N. Pujol, T. Matsuyama, C. I. Bargmann, and J. J. Ewbank. 2007. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 104:2295-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purdy, A., F. Rohwer, R. Edwards, F. Azam, and D. H. Bartlett. 2005. A glimpse into the expanded genome content of Vibrio cholerae through identification of genes present in environmental strains. J. Bacteriol. 187:2992-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 48.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schiavo, G., and F. G. van der Goot. 2001. The bacterial toxin toolkit. Nat. Rev. Mol. Cell. Biol. 2:530-537. [DOI] [PubMed] [Google Scholar]
- 50.Schloss, P. D., and J. Handelsman. 2003. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14:303-310. [DOI] [PubMed] [Google Scholar]
- 51.Schulenburg, H., and J. J. Ewbank. 2007. The genetics of pathogen avoidance in Caenorhabditis elegans. Mol. Microbiol. 66:563-570. [DOI] [PubMed] [Google Scholar]
- 52.Schulenburg, H., and S. Muller. 2004. Natural variation in the response of Caenorhabditis elegans toward Bacillus thuringiensis. Parasitology 128:433-443. [DOI] [PubMed] [Google Scholar]
- 53.Sifri, C. D., J. Begun, and F. M. Ausubel. 2005. The worm has turned: microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13:119-127. [DOI] [PubMed] [Google Scholar]
- 54.Stretton, S., S. Techkarnjanaruk, A. M. McLennan, and A. E. Goodman. 1998. Use of green fluorescent protein to tag and investigate gene expression in marine bacteria. Appl. Environ. Microbiol. 64:2554-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Styer, K. L., G. W. Hopkins, S. S. Bartra, G. V. Plano, R. Frothingham, and A. Aballay. 2005. Yersinia pestis kills Caenorhabditis elegans by a biofilm-independent process that involves novel virulence factors. EMBO Rep. 6:992-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sulston, J., and J. Hodgkin. 1988. Methods, p. 587-606. In W. B. Wood (ed.), The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 57.Tanigaki, K., T. Sato, Y. Tanaka, T. Ochi, A. Nishikawa, K. Nagai, H. Kawashima, and S. Ohkuma. 2002. BE-18591 as a new H+/Cl− symport ionophore that inhibits immunoproliferation and gastritis. FEBS Lett. 524:37-42. [DOI] [PubMed] [Google Scholar]
- 58.Thomas, T., S. Egan, D. Burg, C. Ng, L. Ting, and R. Cavicchioli. 2007. The integration of genomics and proteomics into marine, microbial ecology. Mar. Ecol. Prog. Ser. 332:291-299. [Google Scholar]
- 59.Thomas, T., F. Evans, D. Schleheck, A. Mai-Prochnow, C. Burke, A. Penesyan, D. Dalisay, S. Stelzer-Braid, N. Saunders, J. Johnson, S. Ferriera, S. Kjelleberg, and S. Egan. 2008. Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated lifestyle in the marine environment. PLoS One 3:e3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, Y. 2008. Neuronal mechanisms of Caenorhabditis elegans and pathogenic bacteria interactions. Curr. Opin. Microbiol. 11:257-261. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., H. Lu, and C. I. Bargmann. 2005. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438:179-184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.