Abstract
A disease-like syndrome is currently affecting a large percentage of the Ianthella basta populations from the Great Barrier Reef and central Torres Strait. Symptoms of the syndrome include discolored, necrotic spots leading to tissue degradation, exposure of the skeletal fibers, and disruption of the choanocyte chambers. To ascertain the role of microbes in the disease process, a comprehensive comparison of bacteria, viruses, fungi, and other eukaryotes was performed in healthy and diseased sponges using multiple techniques. A low diversity of microbes was observed in both healthy and diseased sponge communities, with all sponges dominated by an Alphaproteobacteria, a Gammaproteobacteria, and a group I crenarchaeota. Bacterial cultivation, community analysis by denaturing gradient gel electrophoresis (Bacteria and Eukarya), sequencing of 16S rRNA clone libraries (Bacteria and Archaea), and direct visual assessment by electron microscopy failed to reveal any putative pathogens. In addition, infection assays could not establish the syndrome in healthy sponges even after direct physical contact with affected tissue. These results suggest that microbes are not responsible for the formation of brown spot lesions and necrosis in I. basta.
Sponges harbor a highly diverse range of microorganisms, including representatives from 28 bacterial phyla and both major lineages of the Archaea (reference 34 and references cited therein; 40). Microorganisms can comprise up to 40% of sponge biomass, although sponges with more developed aquiferous systems and looser mesohyl often have lower microbial abundances (35). Some sponge-microbe associations may be considered symbiotic (34), while others are nonspecific and may include potentially pathogenic microorganisms (7, 41).
Diseases of marine organisms have been attributed to bacteria, fungi, viruses, protozoans, and a variety of metazoan parasites (18). In sponges, bacteria and fungi are the most commonly reported pathogens, but the exact etiological agents are rarely identified, and little is known about the disease processes (38). In the past decade there has been an increase in reports of sponge disease around the globe, including the Caribbean, Panama, Papua New Guinea, and Slovenia (7, 14, 27, 29, 41, 44). Disease-like symptoms in sponges may also arise from environmental stressors (4, 17), physical damage (46), predation (20), or competitive interactions (22).
Since 2006, two studies have reported a disease-like syndrome in the sponge Ianthella basta, which is commonly distributed in Papua New Guinea (7) and along the Great Barrier Reef (24). A large percentage of I. basta sponges from the Torres Strait and the Palm Islands in the Great Barrier Reef were found to exhibit signs of disease, which included discolored, necrotic spots and exposed skeletal fibers (24). In sponges affected by this syndrome there was a high level of cellular degradation and debris within the remnants of the choanocyte chambers. In Papua New Guinea, I. basta exhibited high mortality between 1996 and 2000, with the affected sponges exhibiting mottled brown lesions, rotted tissue, and large holes (7). The etiological agent of disease in I. basta was not unequivocally ascertained in either study.
Previous research using 454 tag pyrosequencing has assessed the microbial community in I. basta and reported high diversity, with 1,099 operational taxonomic units (OTU) at 95% sequence similarity (40). However, most of this diversity was composed of rare organisms represented by only one or a few sequences. The community was dominated by the Alpha- and Gammaproteobacteria with a single Gammaproteobacteria OTU actually comprising 49% of all sequence tags (40). The rare microbial biosphere in I. basta included Acidobacteria, Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Cyanobacteria, Deinococcus-Thermus, Firmicutes, Gemmatimonadetes, Lentisphaerae, Nitrospira, Planctomyces, Poribacteria, Spirochaetes, TM7, Verrucomicrobia, and the beta, delta, and epsilon classes of the Proteobacteria (40).
With bacteria commonly implicated in sponge disease processes and shifts in microbial communities being used to detect putative pathogens in corals and sponges (3, 7, 33, 41), we sought to ascertain the role of microorganisms in the disease-like syndrome affecting I. basta and to determine how disease affects the symbiotic microbial population.
MATERIALS AND METHODS
Sample collection.
I. basta sponges that were displaying visible signs of necrosis and brown spot lesions were collected from two sites at Masig Island, central Torres Strait, Australia (9°44.260′S, 143°25.275′E and 9°44.779′S, 143°24.280′E) in June and November 2008. Three individual tissue samples were collected from each sponge comprising sections of (i) diseased tissue (D); (ii) the interface between diseased and apparently healthy tissue (M); and (iii) apparently healthy, nondiseased tissue (ND). In addition, sections from healthy control sponges were collected for microbial comparison (H). In all, 13 healthy and 32 diseased samples were collected. Immediately after collection, tissue samples were fixed in 1.5 ml of 100% ethanol and stored at −20°C prior to molecular analysis.
DNA extraction and DGGE.
All samples were extracted by using two different extraction protocols to maximize the amplification of a broad range of microbes. DNA was extracted according to the manufacturer's protocol with a Power Plant DNA isolation kit (MoBio Laboratories, Carlsbad, CA) and using a modified version of (43). Briefly, 0.5 g of sponge tissue was added to 0.5 ml of grinding buffer (100 mM Tris [pH 9, 100 mM EDTA, 1% sodium dodecyl sulfate, 100 mM NaCl, and Milli-Q water). Tubes were immersed in liquid nitrogen and ground with a sterile plastic pestle before the addition of an another 0.25 ml of grinding buffer and 18.75 μl of 20.3-mg ml−1 proteinase K. The samples were incubated at 65°C for 60 min with gentle rotation before the addition of 187.5 μl of 5 M potassium acetate. Samples were incubated on ice for 30 min and centrifuged at 8,000 × g for 15 min at room temperature. DNA in the supernatant was precipitated with 0.8 volumes of isopropanol, washed twice with 70% ethanol, resuspended overnight at 4°C in Milli-Q water, and stored at −20°C.
The 16S and 18S rRNA gene of samples from each extraction method were amplified by PCR with bacterial primers (1055f, 5′-ATGGCTGTCGTCAGCT-3′; 1392r, 5′-ACGGGCGGTGTGTRC-3′) (12) and eukaryotic primers (NS1f, 5′-GTAGTCATATGCTTGTCTC-3′; NS2r, 5′-GGCTGCTGGCACCAGACTTGC-3′) (42). Both reverse primers were modified to contain a 40-bp GC clamp (26). PCRs contained 5 μl of deoxynucleoside triphosphate (2.5 mM), 5 μl of 10× OptiBuffer, 0.15 μl of each primer (100 pmol μl−1), 0.4 μl of bovine serum albumin (BSA; 10 mg ml−1), 3 μl of MgCl2 (50 mM), 0.5 μl of Bio-X-ACT Taq polymerase (Bioline, London, United Kingdom), and 1 μl of DNA template. Reactions were made up to 50 μl of total volume with Milli-Q water. The PCR conditions for the 16S primers were as follows: 1 cycle at 95°C for 5 min; 30 cycles at 95°C for 30 s, 55°C for 1 min, and 70°C for 1 min; and a final elongation at 70°C for 10 min. The PCR conditions for the 18S primers were as follows: 1 cycle at 95°C for 3 min; 30 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final elongation at 72°C for 7 min. PCR products from both extraction methods were pooled, and 20 μl of each sample was added to an 8% (wt/vol) polyacrylamide gel containing either a 50 to 70% (Bacteria) or 35 to 70% (Eukarya) denaturing gradient of formamide and urea. The gels were run at 60°C for 16 h in 1× TAE buffer at 75 V using the Ingeny D-Code system, stained with 1× SYBR gold for 10 min, visualized under UV illumination, and photographed with the Vilber Lourmat ChemiSmart 3000 system. Reference bands from healthy and diseased samples, as well as bands that were present in diseased but absent from healthy samples, were excised, reamplified with PCR, and checked for correct mobility on a 50 to 70% or 35 to 70% denaturing gradient gel electrophoresis (DGGE) gel, respectively. PCR products were sequenced by Macrogen, Inc. (Seoul, Korea), using the forward primer 1055f.
Cloning and sequencing.
The 16S rRNA gene from three healthy, middle, and diseased samples was amplified by PCR with the universal bacterial primers 63f (5′-CAGGCCTAACACATGCAAGTC-3′) (25) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) (21) and the archaeal primers 21f (5′-TTCCGGTTGATCCYGCCGGA-3′) (32) and 958r (5′-TCCGGCGTTGAMTCCAATT-3′) (9). PCRs contained 5 μl of deoxynucleoside triphosphate (2.5 mM), 5 μl of 10× OptiBuffer, 0.15 μl of each primer (100 pmol μl−1), 0.4 μl of BSA (10 mg ml−1), 3 μl of MgCl2 (50 mM), 0.2 μl of Hot Star Taq DNA polymerase (Qiagen, Inc., Chatsworth, CA), and 1 μl of DNA. Reactions were made up to a 50-μl total volume with Milli-Q water. The PCR conditions were as follows: 1 cycle at 95°C for 5 min; 32 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min; and a final elongation at 72°C for 10 min. The triplicate PCR products were pooled to create a single clone library for each of the healthy, middle, and diseased samples. PCR products were cloned with a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Plasmids were checked for inserts by PCR amplification using M13 forward and reverse primers. Restriction digests using HhaI and HaeIII (New England Biolabs, Inc.) were performed to determine the OTU for each library. Duplicates from each representative OTU in each library were sent to Magrogen for sequencing using 63f and 1492r (bacteria) and 21f and 958r (archaea) as the sequencing primers.
Phylogenetic analysis.
DGGE and clone sequences were compared to available databases by using the Basic Local Alignment Search Tool (BLAST) (2) to determine the nearest relatives and the percent similarity. Only unique clone sequences (irrespective of the library they originated from) were submitted to GenBank under accession numbers GU784983 to GU784990, and the DGGE sequences were submitted to GenBank under accession numbers GU784991 to GU785007. Sequences were checked for chimera formation by using Greengenes (10) and Bellerophon (16), and all chimeric sequences were removed before tree construction. Sequences were compiled, automatically aligned, and manually edited in the ARB software package (http://www.arb-home.de [23]). Initially, trees were calculated with almost complete 16S rRNA (1,400-bp) sequences for all close relatives of target sequences by using the neighbor-joining and maximum-parsimony methods in ARB. Partial sequences were subsequently imported into the tree without changing the branch topology by using the ARB parsimony-interactive method. The robustness of inferred tree topologies was evaluated after 1,000 bootstrap resamplings of the neighbor-joining data in the PHYLIP program (11). Escherichia coli and Thermotoga maritima were used as outgroups for the bacterial and archaeal trees, respectively.
Bacterial cultivation.
Aseptic techniques were used to isolate bacteria from small portions (∼1 cm2) of tissue from five healthy and ten diseased sponges. Sponge tissue was rinsed briefly in 70% ethanol and rapidly transferred to sterile artificial seawater (ASW). The tissue was then removed, cut into small pieces using a sterile scalpel, and finely ground by using a mortar and pestle. This slurry was suspended in 9 ml of sterile ASW. Tenfold serial dilutions of the suspension were prepared to a dilution of 10−3, and 100 μl of each dilution was plated in triplicate on Bacto Marine Agar 2216 (Difco Laboratories, Detroit, MA). All plates were incubated at 28°C for up to 7 days. Three distinct colony morphotypes were identified, and these appeared to be the same in both diseased and healthy sponges. Five representatives of each morphotype were selected from both healthy and diseased sponges and serially streaked onto Marine Agar 2216 to obtain pure cultures. Total bacterial counts were recorded from each plate as CFU ml−1 of sponge tissue, and the results were averaged. DNA from I. basta isolates was extracted by using a Wizard genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's protocol. The 16S rRNA gene was amplified by PCR with universal bacterial primers 63f and 1492r, following the same conditions described above. PCR products were sent to Macrogen, where they were subsequently purified and sequenced using 63f as the sequencing primer. Sequences from I. basta isolates were submitted to GenBank under the accession numbers GU817016 to GU817018.
TEM.
In order to visualize potential pathogens in situ, small I. basta tissue sections (∼1 mm in diameter) from five healthy sponges and seven sponges displaying visible signs of necrosis and brown spot lesions were collected for observation using transmission electron microscopy (TEM). Morphotype enumeration was conducted on 10 fields of view per sample to assess variation in morphotype abundance between healthy and diseased sponges. Samples were fixed for 24 h in 2.5% glutaraldehyde in fresh filtered seawater, subsequently placed in filtered artificial seawater, and stored at 4°C until further processing. Fixed tissue was placed in a 1% osmium tetroxide solution (prepared in 0.1 M cacodylate buffer) for 3.5 h and subsequently dehydrated in a graded acetone series (50, 60, 70, 80, 90, and 100%). Tissue was then embedded in Epon resin, sectioned with an ultramicrotome, and stained with 5% uranyl acetate in 50% methanol, followed by Reynolds lead citrate. Ultrathin sections of 60 nm were mounted on Formvar-coated copper grids and viewed using a JEOL 1010 operated at 80 kV. Images were captured by using a SoftImaging Megaview III digital camera.
Pilot infection trial.
To assess disease transmission, a small-scale pilot study was undertaken to determine whether healthy sponges could be infected by a sponge showing visible signs of necrosis. Healthy sponges were placed in a 40-liter tank at a distance of 30 cm from a necrotic sponge for 8 days. The tank was placed inside a 1,000-liter flowthrough, 70% daylight-shaded outdoor aquarium. Sponges were monitored daily for the onset of brown spot lesions and/or necrosis. After 8 days, the sponges were showing no visible signs of disease, so healthy sponges were placed in direct contact with diseased tissue and left for a further 5 days.
Data analyses.
PAST statistical software (15) was used to carry out a nonmetric multidimensional scaling (nMDS) analysis, using the Raup-Crick measure of similarity, to analyze microbial community composition using a presence (scored as 1)/absence (scored as 0) matrix for each of the DGGE bands.
RESULTS
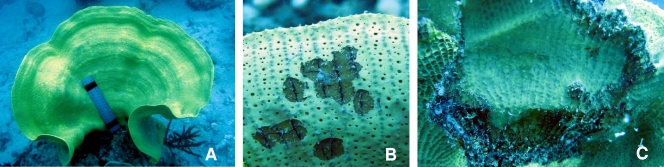
A total of 13 healthy control I. basta (Fig. 1 A) and 32 sponges displaying signs of disease were collected from Masig Island over the two sampling periods. Visible signs of disease included brown necrotic spots in which sponge skeletal fibers were apparent. In addition, some I. basta exhibited high levels of tissue degradation with completely exposed skeletal fibers (Fig. 1B and C).
FIG. 1.
Healthy and diseased I. basta specimens. (A) Healthy control; (B) diseased specimen with brown spot lesions; (C) diseased specimen with exposed skeletal fibers.
Bacterial community analysis.
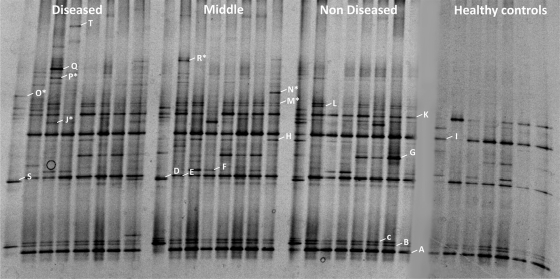
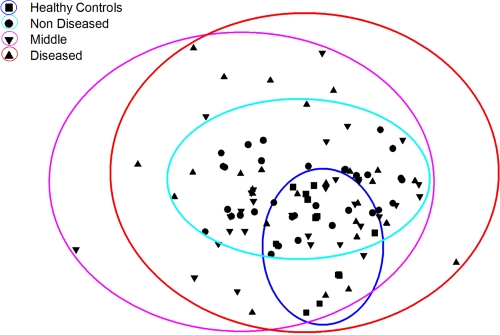
DGGE analysis of I. basta samples revealed a relatively low diversity of microbes with a total of 31 bands detected (Fig. 2). Comparison of the DGGE profiles in healthy and diseased I. basta revealed remarkably similar banding patterns, regardless of the health status of the sponge (Fig. 2). Healthy control samples, as well as non-necrotic portions of diseased sponges, showed similar DGGE banding patterns and little variation between samples. In contrast, middle and diseased samples showed much larger variation, possessing bands that were missing in healthy control and nondiseased samples (e.g., bands Q, R, and T). However, these bands were not consistently present in all diseased samples (Fig. 2). The nMDS shows a cluster of healthy control sponges in the center of the nMDS plot, and there is little separation between healthy control sponges and several data points depicting diseased samples (Fig. 3). The variability associated with middle and diseased samples in the DGGE (Fig. 2) is depicted by the wide separation of many of these samples in the nMDS analysis.
FIG. 2.
Representative DGGE gel image of 16S rRNA-defined bacterial populations from healthy control I. basta, unaffected tissue from diseased I. basta (nondiseased), tissue from the interface between the lesion and healthy tissue (middle), and lesion tissue (diseased). Individual bands excised for sequencing are labeled on the right hand side of the band, and asterisks (*) denote bands that yielded low sequence quality.
FIG. 3.
Nonmetric multidimensional scaling (nMDS) analysis of sponge bacterial community composition using DGGE banding pattern data to construct a similarity matrix. Circles were manually drawn to assist differentiation of the disease categories.
16S rRNA sequencing of excised DGGE bands revealed relatively low bacterial diversity restricted to the Alpha and Gammaproteobacteria (Table 1). In addition, three bands present in all samples, had highest sequence similarity (95 to 96%) to an uncultured Crenarchaeota from an ascidian. Overall, DGGE bands from I. basta had low similarity to sequences in the NCBI database, with band H being the only sequence with a similarity of >96%. The phylogenetic affiliation of six bands (labeled with an asterisk) could not be determined due to low sequence quality (Fig. 2).
TABLE 1.
Sequence similarity in excised 16S rRNA DGGE bands from I. basta as determined by using BLAST
| Band | Accession no. | % Similarity | Description |
|---|---|---|---|
| A | EU373876 | 90 | Alphaproteobacteria (Mediterranean sediment) |
| B | EF173334 | 88 | Alphaproteobacteria (soil) |
| C | EU373879 | 90 | Alphaproteobacteria, (Mediterranean sediment) |
| D | FM242455 | 94 | Gammaproteobacteria (unidentified environmental sample) |
| E | FM242456 | 95 | Gammaproteobacteria (unidentified environmental sample) |
| F | FM242457 | 96 | Gammaproteobacteria (unidentified environmental sample) |
| G | EU117208 | 93 | Pseudomonas viridiflava |
| H | FJ005061 | 99 | Pseudomonas oryzihabitans |
| I | EU283427 | 96 | Crenarchaeote (ascidian) |
| K | EU283423 | 96 | Crenarchaeote (ascidian) |
| L | EU283427 | 95 | Crenarchaeote (ascidian) |
| Q | FJ231146 | 95 | Unidentified bacterium (high altitude lake in Chile) |
| S | FM242455 | 94 | Gammaproteobacteria (unidentified environmental sample) |
| T | AY492079 | 90 | Fucus vesiculosus mitochondrion |
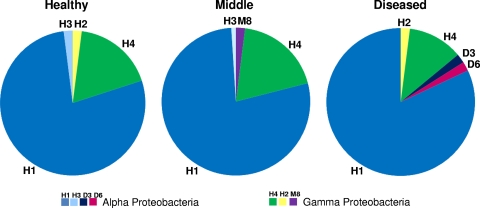
Clone library analysis of I. basta samples was consistent with the DGGE results revealing low microbial diversity and high similarity between each of the three libraries (Fig. 4). All clone sequences fell into only two classes, the Alpha- and Gammaproteobacteria, with all three libraries dominated by the single Alphaproteobacteria sequence (H1). This Alphaproteobacteria accounted for 78% of the healthy and middle libraries and 82% of the diseased library. The other dominant sequence in all libraries was a Gammaproteobacteria, clone H4, which comprised 18, 19, and 12% of the healthy, middle, and diseased libraries, respectively. Two closely related Alphaproteobacteria sequences (D3 and D6) were only detected in the library from diseased sponges, but each sequence comprised only 2% of the library. Two other Gammaproteobacteria sequences were detected, with one of them (M8) only detected in the middle library and the other (H2) present in both the healthy and the diseased libraries. These sequences made up only a small percentage of each of the libraries (2%) (Fig. 4).
FIG. 4.
Pie charts showing the differences in bacterial community composition in I. basta between healthy control sponges, the interface between the lesion and healthy tissue (middle), and lesion tissue (diseased). The graphs were constructed based on the frequency of clones from each of the three libraries. Clones given the same label in each of the three libraries had identical 16S rRNA sequences.
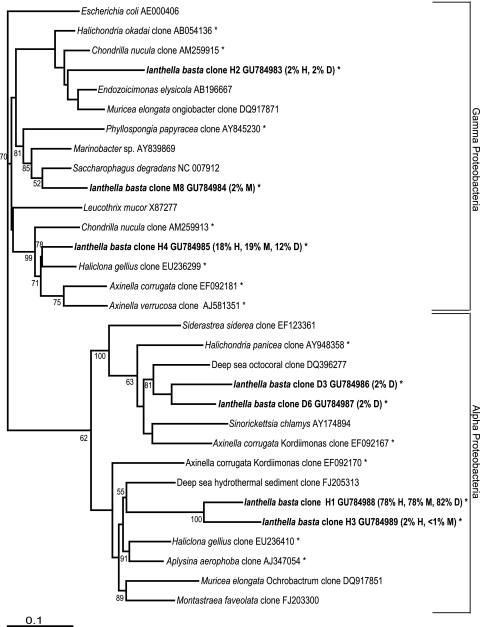
Phylogenetic analysis showed that clone H1 was only distantly related (86% sequence similarity) to its closest relatives, which includes a sequence retrieved from a deep-sea hydrothermal vent and numerous other sponge and coral-derived sequences (Fig. 5). Clone H4 was most closely related to symbionts previously reported from the sponges Haliclona gellius, Chondrilla nucula, and Axinella spp. Although clones D3 and D6 showed some similarity to a sequence from black band diseased corals (EF123361), they were most closely affiliated to uncultured symbionts of a soft coral (DQ396277) and sponges (AY948358 and EF092167). I. basta clone H3, present in both the healthy and middle libraries, was closely related to the dominant Alphaproteobacteria sequence H1.
FIG. 5.
Maximum-likelihood phylogenetic tree from analysis of all 16S rRNA gene sequences retrieved from clone library analysis. I. basta sequences are indicated by boldfacing, with the percentage of each library it comprised listed in parentheses afterward, and asterisks (*) indicate clones isolated from marine sponges. The numbers at the nodes are percentages, indicating the levels of bootstrap support based on analysis of 1,000 resampled data sets. Only values >50% are shown. The scale bar represents 0.1 substitutions per nucleotide position.
Culture-based analysis isolated only three unique morphotypes, and these were cultured from both healthy and diseased I. basta. Five replicates of each morphotype were sequenced from both diseased and healthy sponges, and in all instances 16S rRNA sequences were identical for the individual morphotypes. No single isolate was present in diseased samples and absent from healthy samples, nor were any of the dominant cultures more abundant in diseased samples. However, the mean CFU for the diseased samples was higher than healthy samples (83 ± 12 CFU ml−1 versus 57 ± 8 CFU ml−1). Two of the I. basta isolates (GU817016 and GU817017) revealed high similarities with marine Alphaproteobacteria. I. basta isolate CLCM was identical to an Alphaproteobacteria isolated from the mucus of a Red Sea coral (DQ107390). Similarly, isolate CMSM1 was identical to a sponge-associated Alphaproteobacteria (EF513634). Although I. basta isolate CMSM2 was identical to a Bacillus thuringiensis strain (GU384894), it was also 99% similar to a Bacillus strain previously cultured from I. basta in Papua New Guinea (DQ323748). None of the cultured isolates were detected by using molecular techniques.
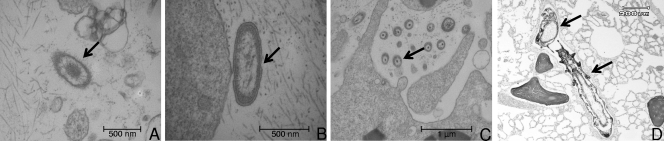
Consistent with molecular and culture-based methods, low bacterial diversity and abundance was also visualized by TEM in both healthy and diseased samples (Fig. 6). Three dominant bacterial morphotypes identified in all samples were defined as furry (Fig. 6A), full (Fig. 6B), and filament (Fig. 6C). The trans-section of the filament morphotype showed a mean size of 158 nm with the mean sizes of the furry and full morphotypes being 259 and 301 nm, respectively. Both the outer and the inner cell membranes of the full morph were clearly defined, indicating a Gram-negative bacterium. None of the morphotypes showed variation in abundance in healthy (furry, n = 1.1 ± 0.33/field of view; full, n = 0.8 ± 0.28; and filament, n = 7.6 ± 2.4) or diseased tissue (furry, n = 1.1 ± 0.34/field of view; full, n = 0.6 ± 0.20; filament, n = 8.1 ± 2.5). There was also no particular association of these morphotypes with any individual sponge cell type. No viruslike particles were detected in any samples. Preliminary histological analysis of I. basta tissue revealed the consistent presence of a polychaete worm within the sponge tissue (Fig. 6D). However, TEM and subsequent histology failed to reveal any differences in the abundance of this worm between diseased and healthy sponge tissue.
FIG. 6.
Three dominant bacterial morphotypes of I. basta identified by using TEM in both healthy (n = 5) and diseased (n = 7) tissue, and a light microscope image depicting the polychaete worm associated with both healthy (n = 6) and diseased (n = 11) tissue. (A) Fuzzy; (B) full; (C) filament; (D) polychaete worm.
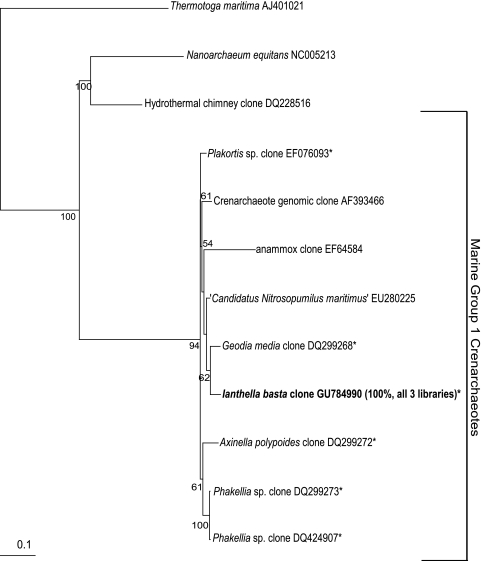
Only a single archaeal sequence was detected in clone libraries from healthy, middle and diseased I. basta libraries. The sequence was a marine group 1 crenarchaeota, related to other sponge-specific Archaea (95%) (Fig. 7).
FIG. 7.
Maximum-likelihood phylogenetic tree from analysis of the 16S rRNA gene sequence retrieved from archaeal clones. The I. basta sequence is indicated by boldfacing, and asterisks (*) indicate clones isolated from other marine sponges. The numbers at the nodes are percentages indicating the levels of bootstrap support based on analysis of 1,000 resampled data sets. Only values >50% are shown. Scale bar represents 0.1 substitutions per nucleotide position.
Eukaryotic community analysis.
DGGE banding patterns were highly similar regardless of the health status of the sponge. No bands were consistently present in diseased samples and absent from healthy controls (see Fig. S1 in the supplemental material). All but two of the sequenced bands came from the sponge DNA (97% sequence similarity to Hexadella pruvoti), and the other bands corresponded to the polychaete worm. No microbial eukaryotic sequences were recovered with these universal primers.
Infection trial.
Transmission of disease symptoms was unsuccessful after the 10-day pilot infection trial. Healthy sponges showed no evidence of brown spot lesions or necrosis following passive infection or direct contact with lesion tissue.
DISCUSSION
Widespread necrosis, cellular disruption, and degradation of choanocyte chambers occur in I. basta with brown spot lesions (24). Despite these disease-like symptoms; bacterial cultivation, molecular community analysis (Bacteria and Eukarya) and electron microscopy undertaken in the present study failed to reveal any putative pathogens. Regardless of the technique used, shifts in microbial community structure between healthy control and diseased I. basta were never observed, providing substantial evidence that bacteria are likely not the etiological agent of the disease-like syndrome. In addition, no eukaryotic organisms (e.g., fungi, protozoa, and worms) could be implicated in the formation of brown spot lesions and necrosis. Further support for the absence of a primary pathogen in I. basta was provided by the infection assay where lesions could not be induced in healthy sponges even after direct contact with infected tissue. Although TEM did not reveal the presence of any viruslike particles, future research would be required to unequivocally confirm the absence of viruses.
Of the three isolates cultivated from I. basta, none were detected by using molecular techniques, suggesting that they are not a major component of the microbial community. Interestingly, one of the isolates from the present study (GU817018) had 99% sequence similarity to a Bacillus strain isolated from a diseased I. basta from Papua New Guinea (PNG) (7). In the study of I. basta from PNG, affected sponges were more abundant close to shore, and a correlation was found between the disease and the cultivation of five Bacillus and Pseudomonas isolates. The authors of that study speculate that bacterial pesticide pollution from agriculture may have introduced new bacteria that were pathogenic to sponges. Subsequent research with sponges and corals showed that addition of up to 5,000 μg of the pesticides VectoBac G (containing B. thuringiensis israelensis) and VectoLex G (containing B. sphaericus) per liter could not induce symptoms of disease in I. basta (28). In addition, the Bacillus strain from the present study was readily cultured from both healthy and diseased I. basta in similar densities, suggesting that it is an unlikely primary pathogen.
It is now well established that marine sponges can host a large diversity of microorganisms (34, 40). Although a previous study detected 1,099 different bacterial OTU from I. basta on the Great Barrier Reef using 454 tag pyrosequencing (40), the present study demonstrated low microbial diversity using cultivation, microcopy, and sequencing of 16SrRNA clone libraries. Excluding the rare biosphere of microbes detected by deep 454 tag pyrosequencing, the two studies were consistent in describing the composition of the microbial community, with the bulk of sequences falling in the Alpha- and Gammaproteobacteria. Two bacterial sequences (comprising at least 90% of the total bacterial community) and a single archaeal sequence were detected with both DGGE and 16S rRNA clone sequencing in all healthy and diseased sponges. The consistently low diversity in diseased sponges contrasts with previous studies where an increase in microbial diversity is detected in response to disease or environmental stress in sponges and corals (8, 30, 31, 39, 41). It has been hypothesized that the increased bacterial diversity in diseased animals may be due to elevated nutrients associated with decaying cells and/or a breakdown in the host defense mechanisms (41). The inability of opportunistic seawater-derived microbes to proliferate in “diseased” I. basta suggests that the sponges' chemical defense and/or immune system are still functional even at an advanced stage of tissue degradation.
All clone sequences from I. basta had low similarity to sequences in the NCBI database, indicating that I. basta hosts a novel microbial community. Similarly, Webster and coworkers (28) found that ca. 25% of the 16S rRNA-V6 tag sequences from I. basta were below the threshold for reliable assignment and may in fact represent previously unknown microbes. While I. basta clones had low similarity to other known sequences, it is worth noting that all clones clustered with other sequences retrieved from marine sponges.
The term “disease” can have various meanings, depending on the source and context it is used in. Oxford's English dictionary defines disease as “a disorder of structure or function in a human, animal, or plant, especially one that produces specific symptoms.” Alternatively, Webster's dictionary defines disease as “an impairment of health or condition of abnormal functioning.” These definitions also state that many medical terms describing symptoms are called “diseases,” especially when the cause of the symptom is unknown. This is particularly relevant for marine organisms, where causative agents are yet to be identified for many of the disease-like syndromes. For instance, despite the fact that no etiological agents can be identified in many of the coral diseases (reference 33 and references cited therein) and that no bacteria, protozoans or viruses could be implicated in the formation of soft tunics in the ascidian Halocynthia roretzi, these syndromes are still referred to as “diseases” (19). Regardless of the specific interpretation of “disease,” we have illustrated here that care should be taken not to assume disease-like symptoms come from an infectious pathogen.
Considering that no infectious agent could be implicated, an environmental origin of the disease-like syndrome should be considered. Previous studies have shown tissue necrosis to arise from various stimuli such as: physical damage due to abiotic factors (6, 46), predation (20), and chemical defense (22). While small feeding scars have been described on Antarctic and temperate sponges due to opistobranchs and nudibranchs, the damage is normally minimal and localized (45). Visual monitoring of I. basta over the course of different seasons and times of day, spanning a 2-year period, has failed to detect any potential predators or parasites that could be responsible for the extensive necrosis observed in I. basta throughout Torres Strait and the Great Barrier Reef. Sponge necrosis and mortality have previously been attributed to elevated temperature (5, 36), high salinity (37), low water exchange (1), and pollution (13). Considering the widespread incidence of disease in I. basta, it is unlikely that localized environmental factors such as pollution or low water flow are responsible.
Our results suggest that microorganisms are not responsible for the disease that is affecting large numbers of I. basta on the Great Barrier Reef and Torres Strait. Although molecular evidence revealed no difference in the microbial communities residing within healthy and diseased sponges, it is still possible that the disease is caused by microbes present in extremely low abundance (hence not detected by molecular methods) or existing symbionts switching on virulence mechanisms (despite no change in their abundance or distribution). These scenarios should be explored further to eliminate microbes as causative agents of disease. Future research will also investigate the role of elevated seawater temperature, high sedimentation, and autoimmune dysfunction in the onset of brown spot lesions and necrosis.
Supplementary Material
Acknowledgments
This study was supported by an AIMS@JCU and MTSRF postgraduate award to H.M.L. We also acknowledge the technical, scientific, and financial assistance from the AMMRF.
We thank R. de Nys for his support and editing contributions and C. Wolff, L. Evans-Illidge, M. Jonker, and J. Morris for diving and fieldwork.
Footnotes
Published ahead of print on 9 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allemand-Martin, A. 1914. Contribution à l'étude de la culture des éponges. C. R. Assoc. Advancement Sci. Tunis 42:375-377. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourne, D. G. 2005. Microbiological assessment of a disease outbreak on corals from Magnetic Island (Great Barrier Reef, Australia). Coral Reefs 24:304-312. [Google Scholar]
- 4.Bruno, J. F., L. E. Petes, C. D. Harvell, and A. Hettinger. 2003. Nutrient enrichment can increase the severity of coral diseases. Ecol. Lett. 6:1056-1061. [Google Scholar]
- 5.Cerrano, C., A. Arillo, G. Bavestrello, B. Calcinai, R. Cattaneo-Vietti, A. Penna, M. Sarà, and C. Totti. 2000. Diatom invasion in the Antarctic hexactinellid sponge Scolymastra joubini. Polar Biol. 23:441-444. [Google Scholar]
- 6.Cerrano, C., G. Magnino, and A. Sarà. 2001. Necrosis in a population of Petrosia ficiformis (Porifera, Demospongiae) in relation with environmental stress. Ital. J. Zool. 68:131-136. [Google Scholar]
- 7.Cervino, J. M., K. Winiarski-Cervino, S. W. Polson, T. Goreau, and G. W. Smith. 2006. Identification of bacteria associated with a disease affecting the marine sponge Ianthella basta in New Britain, Papua New Guinea. Mar. Ecol. Prog. Ser. 324:139-150. [Google Scholar]
- 8.Cooney, R. P., O. Pantos, M. D. Le Tissier, M. R. Barer, A. G. O'Donnell, and J. C. Bythell. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401-413. [DOI] [PubMed] [Google Scholar]
- 9.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A. 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 172:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1993. PHYLIP (Phylogenetic Inference Package) version 3.5c. Department of Genetics, University of Washington, Seattle.
- 12.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gashout, S. F., D. A. Haddud, A. A. El-Zintani, and R. M. A. Elbare. 1989. Evidence for infection of Libyan sponge grounds. International Seminar on the Combat of Pollution and the Conservation of Marine Wealth in the Mediterranean Sea, Gulf of Sirte, Libya.
- 14.Gochfeld, D. J., C. Schloeder, and R. W. Thacker. 2007. Sponge community structure and disease prevalence on coral reefs in Bocas del Toro, Panama, p. 335-343. In M. Custódio, G. Lôbo-Hajdu, E. Hajdu, and G. Muricy (ed.), Museu Nacional serie livros. Museu Nacional-Univ Federal Do Rio De Janeiro, Rio de Janeiro, Brazil.
- 15.Hammer, Ø., D. A. T. Harper, and P. D. Ryan. 2001. PAST: statistics for paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9. [Google Scholar]
- 16.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 17.Jones, R. J., J. Bowyer, O. Hoegh-Gulberg, and L. L. Blackall. 2004. Dynamics of a temperature-related coral disease outbreak. Mar. Ecol. Prog. Ser. 281:63-77. [Google Scholar]
- 18.Kinne, O. 1980. Diseases of marine animals: general aspects, p. 13-73. In O. Kinne (ed.), Diseases of marine animals, vol. I. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 19.Kitamura, S. I., S. I. Ohtake, J. Y. Song, S. J. Jung, M. J. Oh, B. D. Choi, K. Azumi, and E. Hirose. 2010. Tunic morphology and viral surveillance in diseased Korean ascidians: soft tunic syndrome in the edible ascidian, Halocynthia roretzi (Drasche), in aquaculture. J. Fish Dis. 33:153-160. [DOI] [PubMed] [Google Scholar]
- 20.Knowlton, A. L., and R. C. Highsmith. 2005. Nudibranch-sponge feeding dynamics: benefits of symbiont-containing sponge to Archidoris montereyensis (Cooper, 1862) and recovery of nudibranch feeding scars by Halichondria panicea (Pallas, 1766). J. Exp. Mar. Biol. Ecol. 237:36-46. [Google Scholar]
- 21.Lane, D. J. 1991. 16S rRNA sequencing, p. 115-148. In E. Stackerbrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 22.Lopez-Victoria, M., S. Zea, and E. Weil. 2006. Competition for space between encrusting excavating Caribbean sponges and other coral reef organisms. Mar. Ecol. Prog. Ser. 312:113-121. [Google Scholar]
- 23.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luter, H. M., S. Whalan, and N. S. Webster. 2010. Prevalence of tissue necrosis and brown spot lesions in a common marine sponge. Mar. Freshw. Res. 61:484-489. [Google Scholar]
- 25.Marchesi, J. R., T. Sata, A. J. Weightman, T. A. Martine, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagelkerken, I., L. Aerts, and L. Pors. 2000. Barrel sponge bows out, p. 14-15. In Reef encounter. International Society for Reef Studies, Topeka, KS.
- 28.Negri, A. P., R. M. Soo, F. Flores, and N. S. Webster. 2009. Bacillus insecticides are not acutely harmful to corals and sponges. Mar. Ecol. Prog. Ser. 381:157-165. [Google Scholar]
- 29.Olson, J. B., D. J. Gochfeld, and M. Slattery. 2006. Aplysina red band syndrome: a new threat to Caribbean sponges. Dis. Aquat. Organisms 71:163-168. [DOI] [PubMed] [Google Scholar]
- 30.Pantos, O., and J. C. Bythell. 2006. Bacterial community structure associated with white band disease in the elkhorn coral Acropora palmata determined using culture independent 16S rRNA techniques. Dis. Aquat. Organisms 69:79-88. [DOI] [PubMed] [Google Scholar]
- 31.Pantos, O., R. P. Cooney, M. D. A. Le Tissier, M. R. Barer, A. G. O'Donnell, and J. C. Bythell. 2003. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ. Microbiol. 5:370-382. [DOI] [PubMed] [Google Scholar]
- 32.Reysenbach, A., L. J. Giver, G. S. Wickham, and N. R. Page. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sussman, M., B. L. Willis, S. Victor, and D. G. Bourne. 2008. Coral pathogens identified for White Syndrome (WS) epizootics in the Indo-Pacific. PLoS One 3:e2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, M. W., R. Radax, D. Steger, and M. Wagner. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vacelet, J., and C. Donadey. 1977. Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30:301-314. [Google Scholar]
- 36.Vicente, V. P. 1989. Regional commercial sponge extinction in the West Indies: are recent climatic changes responsible? Mar. Ecol. Prog. Ser. 10:179-191. [Google Scholar]
- 37.Walton Smith, F. G. 1939. Sponge mortality at British Honduras. Nature 143:785. [Google Scholar]
- 38.Webster, N. S. 2007. Sponge disease: a global threat? Environ. Microbiol. 9:1363-1375. [DOI] [PubMed] [Google Scholar]
- 39.Webster, N. S., R. E. Cobb, and A. P. Negri. 2008. Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2:830-842. [DOI] [PubMed] [Google Scholar]
- 40.Webster, N. S., M. W. Taylor, F. Behnam, S. Lücker, T. Rattel, S. Whalan, M. Horn, and M. Wagner. 2009. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed]
- 41.Webster, N. S., J. R. Xavier, M. Freckelton, C. A. Motti, and R. Cobb. 2008. Shifts in microbial and chemical patterns within the marine sponge Aplysina aerophoba during a disease outbreak. Environ. Microbiol. 10:3366-3376. [DOI] [PubMed] [Google Scholar]
- 42.White, T. J., T. Burns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal rRNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), Methods and applications. Academic Press, Inc., New York, NY.
- 43.Wilson, K., L. Yutao, V. Whan, S. Lenhnert, K. Byrne, S. Moore, S. Pongsomboon, A. Tassanakajon, G. Rosenberg, E. Ballment, Z. Fayazi, J. Swan, M. Kenway, and J. Benzie. 2002. Genetic mapping of the black tiger shrimp Panaeus monodon with amplified fragment length polymorphism. Aquaculture 204:297-309. [Google Scholar]
- 44.Wulff, J. L. 2007. Disease prevalence and population density over time in three common Caribbean coral reef sponge species. J. Mar. Biol. Assoc. U.K. 87:1715-1720. [Google Scholar]
- 45.Wulff, J. L. 2006. Rapid diversity and abundance decline in a Caribbean coral reef sponge community. Biol. Conserv. 127:167-176. [Google Scholar]
- 46.Wulff, J. L. 2006. Resistance versus recovery: morphological strategies of coral reef sponges. Funct. Ecol. 20:699-708. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.