Abstract
Tap and shower water at two locations in the Netherlands was examined for the presence of rapidly growing nontuberculous mycobacteria. Cultures yielded Mycobacterium peregrinum, M. salmoniphilum, M. llatzerense, M. septicum, and three potentially novel species, a distribution different from that in clinical samples.
Nontuberculous mycobacteria (NTM) are omnipresent in our environment and cause opportunistic infections in humans after environmental exposure (4). Showers have recently been suggested as a source of NTM infection, because of their biofilms and aerosol formation (5). As in most environmental studies on NTM, the focus of the latter study was on the most common causative agents of human NTM disease, the Mycobacterium avium complex (MAC) bacteria. MAC bacteria have been detected frequently from water samples (4); molecular typing has even confirmed shower water as a source of MAC disease in a patient (3).
In recent years, the pathogenic potential of rapidly growing mycobacteria (RGM) has gained attention, though they are generally ignored in environmental studies (2, 12). In a study of swimming pool and whirlpool water in the Netherlands, RGM made up 30 to 40% of all isolates but were not identified to the species level (7). RGM cause pulmonary infections in patients with preexisting pulmonary disease and localized or disseminated skin disease after trauma or in the immunocompromised. RGM disease is a therapeutic challenge due to the extensive resistance of these mycobacteria to antimicrobial drugs (2). To investigate RGM exposure from home shower and tap water, we sampled water for Mycobacterium culture in two locations in the Netherlands.
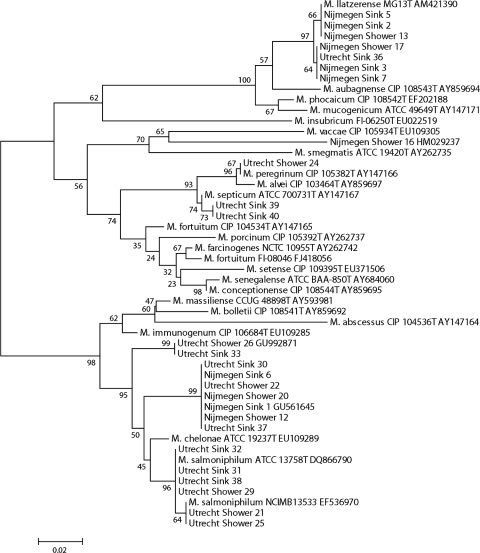
For this purpose, 4 liters of shower water and 4 liters of kitchen sink tap water were sampled from the homes of two of the investigators, in the cities of Nijmegen and Utrecht. In both cities, drinking water is produced from local groundwater sources. Since soil acts as a natural filter, no further disinfection treatments are performed during the production process. The cold water was collected after 10 s of flushing, applying a pulsating flow. All samples were decontaminated by a 30-min incubation with 0.005% cetylpyridinium chloride (CPC), and 1-liter aliquots were filtered through 0.4-μm filters. The filters were dissected, with one half applied directly on Middlebrook 7H10 and the other on antimicrobial-supplemented 7H11 agar sections of selective bi-plates (Becton & Dickinson, Erembodegem, Belgium), and incubated in duplicate at 30 and 37°C, as previously published (8). A control sample spiked with a clinical Mycobacterium fortuitum isolate was included. Media were checked for growth after 3, 4, and 7 days of incubation, without quantification. All colonies with distinct morphologies of presumed mycobacterial origin were picked, to a total of 10 per sampling site per city, and subcultured on Middlebrook 7H10 agar at 30°C, labeled by sampling location and with consecutive numbers, and identified by rpoB gene sequencing; species identifications were based on the 97% sequence similarity cutoff (1). Twenty-six of the 40 isolates were identified as Mycobacterium species. The rpoB gene sequences of these isolates were aligned with those of reference strains of the closest related mycobacteria by using Clustal X software (11). The resulting topology and tree, inferred by neighbor joining and visualized using the MEGA 4.0 software package (10), were evaluated by bootstrap analyses based on 1,000 resamplings (Fig. 1).
FIG. 1.
Phylogenetic tree based on rpoB gene sequences of the isolates from the water samples taken in the cities of Utrecht and Nijmegen and type strain sequences of related Mycobacterium species. A neighbor-joining tree was created, bootstrapped 1,000 times, and visualized with MEGA 4.0 (10). Bootstrap values are indicated at the nodes.
All water samples yielded growth of multiple colony morphologies within 7 days of incubation. Growth at 30°C was faster and yielded more-distinct morphologies; fungal contamination was frequent on plate sectors without antimicrobial supplement. From the Nijmegen kitchen sink water samples, we identified M. llatzerense and a novel M. chelonae-abscessus group member (GenBank accession no. GU561645); from the shower water samples, M. llatzerense, a novel RGM phylogenetically related to M. vaccae (GenBank accession no. HM029237), and the novel M. chelonae-abscessus group member (GenBank accession no. GU561645) were cultured. From the Utrecht shower samples, we identified M. salmoniphilum, M. peregrinum, and two groupings of novel M. chelonae-abscessus group members (GenBank accession no. GU561645 and GU992871), including the one also cultured from the Nijmegen samples (GenBank accession no. GU561645); from the Utrecht sink water sample, we identified M. septicum, M. llatzerense, M. salmoniphilum, and the two groupings of novel M. chelonae-abscessus group members (GenBank accession no. GU561645 and GU992871). Figure 1 represents a phylogenetic tree based on the rpoB sequences of the isolates from the water samples and those of type strains of the most closely related RGM species. Control samples correctly yielded M. fortuitum.
Although M. chelonae-abscessus group bacteria were frequent in the water samples, we did not find any true representatives of the species M. abscessus, M. chelonae, M. bolletii, or M. massiliense. This was surprising, as all isolates from a recent nationwide clinical study on 95 patients with M. chelonae-abscessus group isolates could be assigned to these four mentioned species based on >97% rpoB gene sequence homology (1, 13); these 95 patients included inhabitants of the cities of Utrecht and Nijmegen. In routine rpoB gene sequencing of clinical NTM isolates executed from January 2010 onwards, only M. abscessus, M. chelonae, and M. bolletii were identified (J. van Ingen and D. van Soolingen, personal communication). Thus, it seems there is a discrepancy between M. chelonae-abscessus group bacteria in these water samples and those found in clinical samples in the Netherlands. The absence of M. abscessus, the most common and notorious RGM causing human disease (2, 13), from the water samples is particularly striking. In addition, M. salmoniphilum, M. llatzerense, and two potentially novel M. chelonae-abscessus group species that were not previously detected in clinical samples in the Netherlands (13) were isolated from the water samples.
Both M. abscessus and M. chelonae have been identified in environmental samples (12). Yet, very few studies have applied rpoB gene sequencing for identification, which may partly explain discrepancies between environmental and clinical samples. By use of rpoB sequencing, M. chelonae but not M. abscessus was recently detected in water from a cooling tower in France (9). Our study is too small to draw strong conclusions, but our results suggest that M. abscessus disease in the Netherlands may be contracted from sources other than tap and shower water. A larger-scale study of clinical and environmental samples is needed to provide insight into this issue.
The M. fortuitum complex organisms are frequently isolated from environmental as well as clinical samples (2, 12). In the Utrecht samples, we detected M. septicum and M. peregrinum bacteria, both well-known opportunistic pathogens and members of the M. fortuitum complex (2). In the Nijmegen samples, no M. fortuitum complex members were isolated; an M. smegmatis group species and multiple strains of M. llatzerense were detected.
The M. llatzerense isolates are the first isolates of this species to be reported after the description of M. llatzerense as a separate species in the genus Mycobacterium (6). Recently, bacteria of this species were first isolated from a pulmonary specimen from a patient in the Netherlands. This isolate was considered clinically insignificant (J. van Ingen, personal communication).
In summary, multiple RGM species are present in tap and shower water in the Netherlands. We identified the well-known opportunistic pathogens M. peregrinum and M. septicum, though, strikingly, the most common RGM causing human disease, M. abscessus, was not cultured. We did, for the first time, isolate M. salmoniphilum and M. llatzerense in the Netherlands. A considerable number of RGM present in tap and shower water were novel species. The differences between RGM species distribution in these water samples and in clinical samples suggest that tap and shower water in the Netherlands may represent a potential source of human infection for some, but not all, RGM species. These preliminary results warrant a nationwide representative study.
Nucleotide sequence accession numbers.
The following nucleotide sequences have been deposited in GenBank: novel M. chelonae complex grouping 1, accession no. GU561645; novel M. chelonae complex grouping 2, accession no. GU992871; and novel M. smegmatis group member, accession no. HM029237.
Footnotes
Published ahead of print on 16 July 2010.
REFERENCES
- 1.Adékambi, T., P. Colson, and M. Drancourt. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Groote, M. A., and G. Huitt. 2006. Infections due to rapidly growing mycobacteria. Clin. Infect. Dis. 42:1756-1763. [DOI] [PubMed] [Google Scholar]
- 3.Falkinham, J. O., III, M. D. Iseman, P. de Haas, and D. van Soolingen. 2008. Mycobacterium avium in a shower linked to pulmonary disease. J. Water Health 6:209-213. [DOI] [PubMed] [Google Scholar]
- 4.Falkinham, J. O., III. 2009. Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J. Appl. Microbiol. 107:356-367. [DOI] [PubMed] [Google Scholar]
- 5.Feazel, L. M., L. K. Baumgartner, K. L. Peterson, D. N. Frank, J. K. Harris, and N. R. Pace. 2009. Opportunistic pathogens enriched in showerhead biofilms. Proc. Natl. Acad. Sci. U. S. A. 106:16393-16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomila, M., A. Ramirez, J. Gascó, and J. Lalucat. 2008. Mycobacterium llatzerense sp. nov., a facultatively autotrophic, hydrogen-oxidizing bacterium isolated from haemodialysis water. Int. J. Syst. Evol. Microbiol. 58:2769-2773. [DOI] [PubMed] [Google Scholar]
- 7.Havelaar, A. H., L. G. Berwald, D. G. Groothuis, and J. G. Baas. 1985. Mycobacteria in semi-public swimming-pools and whirlpools. Zentralbl. Bakteriol. Mikrobiol. Hyg. B 180:505-514. [PubMed] [Google Scholar]
- 8.Neumann, M., R. Schulze-Robbecke, C. Hagenau, and K. Behringer. 1997. Comparison of methods for isolation of mycobacteria from water. Appl. Environ. Microbiol. 63:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pagnier, I., M. Merchat, D. Raoult, and B. La Scola. 2009. Emerging Mycobacteria spp. in cooling towers. Emerg. Infect. Dis. 15:121-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 11.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Ingen, J., M. J. Boeree, P. N. R. Dekhuijzen, and D. van Soolingen. 2009. Environmental sources of rapid growing nontuberculous mycobacteria causing disease in humans. Clin. Microbiol. Infect. 15:888-893. [DOI] [PubMed] [Google Scholar]
- 13.van Ingen, J., R. de Zwaan, R. P. N. Dekhuijzen, M. J. Boeree, and D. van Soolingen. 2009. Clinical relevance of Mycobacterium chelonae-abscessus group isolation in 95 patients. J. Infect. 59:324-331. [DOI] [PubMed] [Google Scholar]