Abstract
Waste lagoons of swine operations are a source of Cryptosporidium oocysts. Few studies, however, have reported on oocyst concentrations in swine waste lagoons; none have reported on oocyst viability status, nor has there been a systematic assessment of species/genotype distributions across different types of swine facilities. Ten swine waste lagoons associated with farrowing, nursery, finishing, and gestation operations were each sampled once a month for a year. Oocysts were extracted from triplicate 900-ml effluent samples, enumerated by microscopy, and assessed for viability by dye exclusion/vital stain assay. DNA was extracted from processed samples, and 18S ribosomal DNA (rDNA) genes were amplified by PCR and sequenced for species and genotype identification. Oocysts were observed at each sampling time at each lagoon. Annual mean concentrations of total oocysts and viable oocysts ranged between 24 and 51 and between 0.6 and 12 oocysts ml−1 effluent, respectively. The species and genotype distributions were dominated (95 to 100%) by Cryptosporidium suis and Cryptosporidium pig genotype II, the latter of which was found at eight of the lagoons. The lagoon at the gestation facility was dominated by Cryptosporidium muris (90%), and one farrowing facility showed a mix of pig genotypes, Cryptosporidium muris, and various genotypes of C. parvum. The zoonotic C. parvum bovine genotype was observed five times out of 407 18S rDNA sequences analyzed. Our results indicate that pigs can have mixed Cryptosporidium infections, but infection with C. suis is likely to be dominant.
Over the last few decades, pork production in North America has undergone significant growth and centralization into large concentrated swine (Sus scrofa) operations with more animals on fewer farms (18). A consequence of the increase in numbers of swine per facility is a concomitant increased concentration of swine waste. Present housing facilities for swine are designed to collect feces and urine in wastewater lagoons, in which the waste undergoes anaerobic transformations. One of several public health concerns over swine lagoons is the potential presence of infectious bacteria, viruses, and protozoa (4). Because of the notoriety given to swine waste lagoon spills in the coastal flood plain of North Carolina that were associated with a series of hurricanes in 1998 and 1999 (21), large-scale swine operations have become a focus of environmental and public health concerns.
The cause of the massive outbreak of cryptosporidiosis in Milwaukee, WI, in 1993 was afterwards determined to be Cryptosporidium hominis, the human genotype of C. parvum and an obligate parasite of humans (33, 44). At the time, however, it was thought to be caused by C. parvum (22). Because of this initial misidentification of the cryptosporidial source of the outbreak, the connection between C. parvum and large-scale confined livestock operations has become a focused area of research. Although manure-associated outbreaks of C. parvum have implicated bovine sources, a Canadian study found that the prevalence of Cryptosporidium in swine lagoons was greater than that in dairy liquid manure (9). Olson et al. (24) also reported the presence of Cryptosporidium oocysts of undetermined genotype at four of six hog operations in Canada. Atwill et al. (2) observed C. parvum oocysts in feces of feral pigs. Hutchison et al. (13) observed C. parvum oocysts of undetermined genotype in 5 and 13% of fresh and stored fecal samples, respectively, from pigs of undeclared age. Guselle et al. (10) followed the course of a naturally occurring C. parvum infection in 33 weaned pigs. Following the protocol of the genetic analysis of Morgan et al. (23), Guselle et al. (10) identified this C. parvum genotype as being adapted to pigs. At the time, the zoonotic potential of this C. parvum pig-adapted genotype was considered uncertain (23).
Recently, two genotypes of Cryptosporidium have been recognized as host adapted to swine: Cryptosporidium suis (formerly Cryptosporidium pig genotype I) and Cryptosporidium pig genotype II (28, 29). Xiao et al. (37) reported on an immunocompromised person who was infected with a Cryptosporidium pig genotype and thus implicated Cryptosporidium from swine as potentially zoonotic and a public health concern. Before molecular methods were developed to differentiate pig genotypes of Cryptosporidium from other species, C. parvum was thought to infect 152 species of mammals and consist of several cryptic species (6). An extensive survey of swine effluent from swine finishing operations in Ireland indicated a prevalence of both C. suis and Cryptosporidium pig genotype II (39). Hamnes et al. (11) reported prevalence of both C. suis and Cryptosporidium pig genotype II in feces of suckling pigs across Norway and thus implicated farrowing operations as sources of this parasite.
Other than the prevalence of Cryptosporidium in feces of young pigs and effluent lagoons of older pigs in finishing operations, little comprehensive data on oocyst concentrations, viability of oocysts, and distributions of Cryptosporidium species and genotypes have been reported. No systematic study of swine lagoon effluents from large-scale facilities has been reported for the four separate stages of swine development, (i) breeding and gestation, (ii) farrowing (parturition), (iii) nursery (in which weaned piglets are kept until 8 to 9 weeks of age), and (iv) finishing (in which 8- to 9-week-old pigs are kept to market weight). The objective of this investigation was to determine for 1 year the frequencies, concentrations, viability statuses, and distributions of Cryptosporidium species and genotypes in lagoons associated with the four types of swine operations in the Southern Piedmont and in coastal plain watersheds of Georgia.
MATERIALS AND METHODS
Sampling sites.
Ten swine lagoons were sampled. Seven lagoons were located in the Southern Piedmont and three were located in the coastal plain of Georgia. Three lagoons were on farrowing operations, four were on finishing operations, one was on a nursery operation, one was on a gestation operation, and one was on a lagoon that received effluent from a farrowing and nursery operation (designated Far/Nur). Each lagoon was sampled once a month from June 2007 to May 2008.
Lagoon sampling protocol.
The sampling protocol that was developed considered all safety aspects. Since small boats were launched on lagoons to take samples, all personnel were trained in small boat safety. Three 1-liter surface samples were taken at three different designated locations across each lagoon, one of which was taken near the effluent source pipe. The samples were contained in one-liter Nalgene bottles. Samples were stored on ice while in transit to the laboratory.
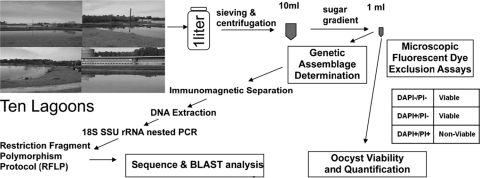
The percentage of solids in each lagoon effluent was determined. Ten- to 50-ml subsamples were filtered through tared Whatman no. 2 filter paper. The filter papers were then weighed before and after drying in an oven at 105°C for 24 h. Percentages of solids were determined by dividing the difference between the weight of the dried filter and the tared filter weight by the volume of sample filtered. After subsampling for percentages of solids, the samples were sent on ice by Federal Express overnight to Cornell University for oocyst extraction and further analysis as depicted in Fig. 1. Enumerations and viability assays were performed at the ARS laboratory in Georgia, and BLAST analyses and sequence determinations were performed at Cornell University.
FIG. 1.
Flow diagram of sampling, processing, and analysis protocol. SSU, small subunit.
Sample preparation.
Of the three effluent samples collected from each of the 10 lagoons, 900 ml of each was filtered through U.S. sieves (no. 200 and 500, with pore sizes of 75 and 25 μm, respectively; Seedburo Equipment Company), and the filtrate was collected in five 250-ml conical tubes. These tubes were centrifuged at 1,500 × g for 10 min. The supernatant was gently vacuumed off, and the pellets were combined. The centrifugation, vacuuming, and combining of pellets continued until the total volume fit into a 50-ml tube. The 50-ml tube was centrifuged at 1,500 × g for 10 min. The resulting pellet was <10 ml. Particularly dense lagoon samples were first floated with 1.3-specific gravity (sp. gr.) sugar before filtration in 50-ml centrifuge tubes and centrifuged at 1,000 × g for 15 min. The top 15 ml was collected and poured over U.S. sieves no. 200 and 500. The resulting filtrate was processed as stated above for less-dense samples.
To further purify any Cryptosporidium oocysts present in the sieved and centrifuged processed pellet, the pellet was placed on a discontinuous sugar gradient of 1.05, 1.08, and 1.10 sp. gr. and concentrated by ultracentrifugation (55,000 × g) in a swinging-bucket rotor for 15 min at 8°C. Oocysts were extracted in and just above the 1.08-sp. gr. layer. After the oocysts were extracted, deionized water was added to bring the volume to three times the extracted volume to reduce the specific gravity to 1.0. This tube was centrifuged at 1,500 × g for 10 min and the supernatant discarded by vacuuming. The pellet was washed into a smaller centrifuge tube. The centrifugation, vacuuming, and placing into a smaller centrifuge tube continued until the total volume was reduced to 1 ml. This 1-ml suspension was divided in half; one half was used for enumeration and viability assessment, and the other was used for genotyping.
Dye exclusion/vital stain assay.
The details of this assay have been described previously (1, 3, 14). Briefly, 10 μl of 4′,6-diamidino-2-phenylindole (DAPI; 2 mg ml−1 in methanol) and 10 μl of propidium iodide (PI; 1 mg ml−1 in 0.1× phosphate-buffered saline, pH 7.2) were added to a 100-μl suspension of extracted oocysts in a 1.5-ml microcentrifuge tube. After incubation of the suspension in the dark at room temperature for 1 h, 10 μl of Merifluor fluorescent antibody (Meridian Bioscience Inc., Cincinnati, OH) was added and mixed, and the suspension was incubated in the dark at room temperature for 0.5 h (1, 14). Similar to the result reported by Anguish and Ghiorse (1), oocysts were initially identified with the fluorescent antibody. Further details of its applicability as an indicator of viability or potential for animal infectivity and thus the capability of reproduction under appropriate conditions have been previously discussed (27). As explained in the reply to Robertson et al. (27), to assay the viability of oocysts extracted from an environmental matrix, such as soil, the dye permeability assay is treated explicitly as an indirect assessment of potential infectivity. As such, this dye permeability method (14) was adapted to the U.S. EPA's method 1622 in a study assessing viability of oocysts extracted from surface waters (31). All phases of dye permeability and impermeability have been illustrated in color micrographs (1) and exemplified in images of nonviable oocysts extracted from lagoon effluent (Fig. 2). Viable oocysts were the sum of DAPI-negative (DAPI−), PI-negative (PI−) oocysts and DAPI-positive (DAPI+) PI− oocysts; DAPI+ PI+ oocysts were considered inactivated. Empty oocyst shells were also counted as inactivated in this assay. The number of viable oocysts was determined as the percentage of total oocysts enumerated.
FIG. 2.

Image of oocysts from the lagoon at the nursery is a combination of images of the immunofluorescent stain and DAPI and PI dyes. The oocyst near the center is DAPI+ PI− and is considered viable and potentially infective. The oocysts at 11 and 5 o'clock have excysted and are not viable; the oocysts at 12 and 6 o'clock are DAPI+ PI+ and are not viable. Bar with internal subdivisions, 5.085 μm.
Microscopy and quantification of oocysts.
For enumeration and determination of the viability status of oocysts from each processed sample, duplicate 10-μl aliquots of a stained oocyst suspension were each placed on an agar-coated microscope slide (1% Noble agar air dried on an ethanol-cleaned slide) and each mounted with a 22-mm2 coverslip. The slides were examined with a Leica DMR fluorescent microscope with a 100×/1.40-0.7 (magnification/numerical aperture) oil PL APO differential interference contrast (DIC) objective and 10× eyepieces and with filters to observe oocysts stained with the fluorescent antibody and the two nucleic acid dyes DAPI and PI. Images of each microscopic field were captured with SimplePCI imaging software (Compix Inc., Sewickley, PA) and examined for oocysts. Twenty random fields (out of >105 possible fields) were brought into view, and oocysts, if present, were counted and their viabilities were assessed for each duplicate subsample of each replicate processed effluent sample. Total counts were determined by the following equation: cells ml−1 = [(N × Ac)/(Af × Vu)] × (Vc/Vs) (where N is the mean cell count, Ac is the area under the coverslip, Af is the area of the microscopic field of view, Vu is the volume of sample under the coverslip, Vc is the volume of final oocyst concentrate, and Vs is the volume of effluent from which the oocysts were concentrated).
Genotyping.
From the sample preparation, a 0.5-ml portion was used for genotyping as published by Xiao et al. (36, 38). In brief, the genotyping procedure consisted of immunomagnetic separation (Dynabeads anti-Cryptosporidium kit; Invitrogen Dynal), with the resulting bead-oocyst complex being used directly in the DNA extraction procedure (QIAamp DNA minikit; Qiagen). Several volumes of DNA, 1, 3, 4, and 6 μl, were used in the PCR. Primers used in the analysis are as delineated by Xiao et al. (36). Positive PCR products were identified by electrophoresis on a 2% agarose gel. In the interest of time, a limited number of positive PCR products were used for the restriction fragment length polymorphism (RFLP) protocol. Instead, all amplified PCR products, including those that were identified by RFLP, were sequenced (Applied Biosystems automated 3730xl DNA analyzer; Biotechnology Resource Center, Cornell University). The resulting sequences were analyzed using the Basic Local Alignment Search Tool (National Center for Biotechnology Information).
Data analysis.
Because data like those for the raw oocyst counts have been shown to fit a Poisson distribution (16), the count data were transformed for statistical analysis with the equation X′ = (X + 0.5)1/2 (43) and transformed means were determined. The transformed means were then back transformed into numbers of oocysts ml−1 of effluent for presentation. To make the data behave as if they were normally distributed, these oocyst densities were transformed into natural log numbers to conduct an analysis of variance with lagoons as fixed effects and replicate samples as a random effect in analysis of variance while comparing means for significant differences (at a P value of ≤0.05) with Proc Mixed, version 9.2 (30). Significant differences were assigned to tests of transformed means, which are presented in untransformed units of concentration. Analysis of variance was also conducted on the data for percent solids with Proc Mixed (30). Regression relationships between concentrations of total oocysts, viable oocysts, and percent solids from lagoon effluent were analyzed with Proc Reg (30). A chi-square analysis was also performed on the frequency distribution of observed Cryptosporidium genotypes with Proc Freq (30), and the logistic regression procedure of SAS (30) was performed on normalized (frequency of genotype/total number of genotypes per lagoon) frequency data.
RESULTS
Concentration and viability assessment.
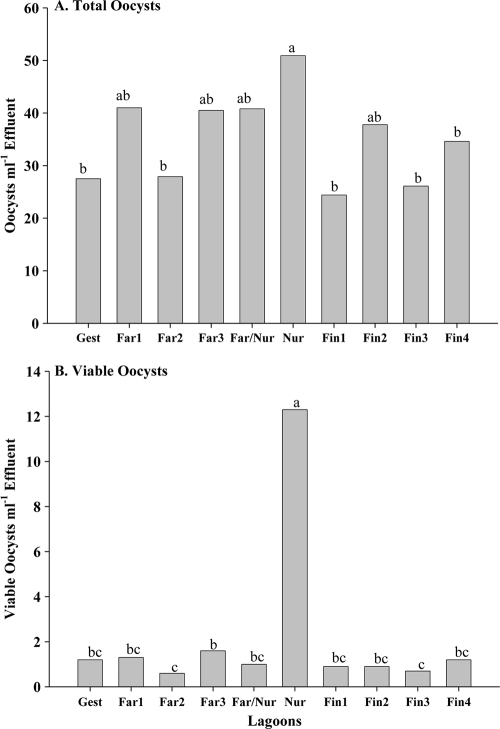
Cryptosporidium oocysts were recovered from every lagoon and on each sampling date. Mean oocyst concentrations ranged between 11.0 and 215.7 ml−1 of lagoon effluent for finishing facilities, between 11.0 and 210.8 ml−1 lagoon effluent for the farrowing facilities, between 11.0 and 354.1 ml−1 for the nursery facilities, and between 11.0 and 126.8 ml−1 lagoon effluent for the one gestation facility sampled. The annual mean concentration of oocysts over the 12 months of sampling by the lagoon indicated that the nursery had a greater concentration of oocysts than three of the finishing facilities, one farrowing facility, and the gestation facility (Fig. 3 A). The annual mean concentration of viable oocysts in lagoon effluent was greater at the nursery than at the lagoons of the other facilities (Fig. 3B). Of the mean total annual oocysts at the nursery, 24.2% were observed to be viable; the percentages of viable oocysts among mean annual total oocysts for the other swine facilities ranged between 2.4 and 4.4%. Regression analysis indicated no correlation between total oocyst concentrations and viable oocyst concentrations. The annual mean percentage of lagoon effluent solids was significantly greater for the lagoon receiving effluent from the gestation facility than the other lagoons; differences in annual mean percentages of solids between the other lagoons were not observed (data not shown). Regression analysis indicated no correlation between percentages of solids and total oocyst concentrations.
FIG. 3.
Annual mean concentrations of total oocysts (A) and viable oocysts (B) at each swine facility. Gest, gestation; Far, farrowing; Nur, nursery; Fin, finishing. Different letters above the bars indicate differences at a P value of ≤0.05.
Distribution of species and genotypes.
Nine species and genotypes of Cryptosporidium oocysts were observed in the effluent of the 10 lagoons sampled over the 12-month sampling period: C. suis, Cryptosporidium pig genotype II, C. muris, C. parvum rat genotypes, C. parvum mouse genotypes, C. parvum bovine genotypes, Cryptosporidium meleagridis, Cryptosporidium felis, and one unknown C. parvum genotype. Genetic analysis indicated that C. suis and pig genotype II (Fig. 4) were observed in nearly all of the lagoons. The distributions of Cryptosporidium oocyst species and genotypes (Table 1) were dominated (73.9%) by C. suis, which was observed in every lagoon except for the lagoon at the gestation facility. Cryptosporidium pig genotype II was observed in lagoons of eight of the 10 facilities; C. muris was dominant in the lagoon of the gestation facility and was observed in one finishing facility and one farrowing facility lagoon, as well as in the lagoon receiving effluent from both a nursery and farrowing facility. Of the less frequently observed species and genotypes of Cryptosporidium oocysts, the C. parvum rat genotype (GenBank accession number FJ205699) was observed eight times, C. parvum mouse genotypes (GenBank accession numbers AF112571, AF108863, and EU553589) were observed seven times, a C. parvum bovine genotype (GenBank accession number AF308600) was observed two times, and C. meleagridis (GenBank accession number AF112574) was observed once over the 12 sampling times at the farrowing facility designated Far1. Two C. parvum bovine genotypes (GenBank accession numbers DQ656354 and AF308600) were observed in the lagoon of the farrowing facility designated Far2. An unknown C. parvum genotype (GenBank accession number EF489037) (45) was observed in the nursery lagoon, and a C. parvum bovine genotype (GenBank accession number AF308600) was observed in the lagoon receiving effluent from both a nursery and farrowing facility. Cryptosporidium felis was observed once in the lagoon effluent of the gestation facility. The viability status was unknown for the oocysts from which the DNA was extracted for genetic analysis.
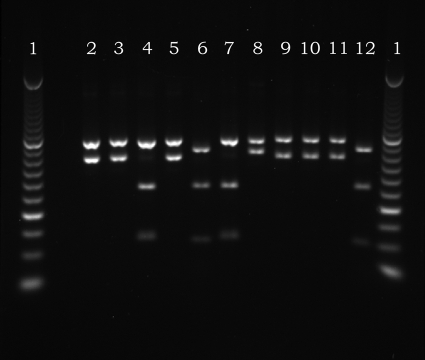
FIG. 4.
RFLP patterns of the dominant Cryptosporidium spp. from the different swine lagoons. Lane 1, 50-bp ladder; lane 2, Nur, C. suis; lane 3, Nur, C. suis; lane 4, C. parvum bovine genotype (control); lane 5, Nur, C. suis; lane 6, Far3, pig genotype II; lane 7, C. parvum bovine genotype (control); lane 8, unsequenced isolate; lane 9, Far/Nur, C. suis; lane 10, Far/Nur, C. suis; lane 11, Fin4, C. suis; lane 12, Fin4, pig genotype II.
TABLE 1.
Distribution of Cryptosporidium genotypes among the 10 swine lagoons examineda
| Facility | No. (%) of isolates of indicated species/genotype |
|||
|---|---|---|---|---|
| C. suis | Pig genotype II | C. muris | Other | |
| Fin1 | 54 (94.7) | 3 (5.3) | 0 | 0 |
| Fin2 | 41 (100) | 0 | 0 | 0 |
| Fin3 | 44 (81.5) | 4 (7.4) | 6 (11.1) | 0 |
| Fin4 | 21 (63.6) | 12 (36.4) | 0 | 0 |
| Far1 | 9 (26.5) | 2 (5.9) | 5 (14.7) | 18 (52.9) |
| Far2 | 33 (93.9) | 0 | 0 | 2 (6.1) |
| Far3 | 17 (85.0) | 2 (15.0) | 0 | 0 |
| Nur | 42 (80.8) | 7 (13.5) | 0 | 1 (5.7) |
| Far/Nur | 40 (90.9) | 2 (4.5) | 1 (2.3) | 1 (2.3) |
| Gestation | 0 | 3 (7.5) | 36 (90.0) | 1 (2.5) |
| Total | 301 (73.9) | 35 (8.6) | 48 (11.8) | 23 (5.6) |
Gest, gestation; Far, farrowing; Nur, nursery; Fin, finishing.
For the purpose of performing a chi-square analysis, all of the less frequently observed genotypes of C. parvum, Cryptosporidium meleagridis, and Cryptosporidium felis and the one unknown C. parvum genotype were considered other genotypes (Table 1). The chi-square analysis (Table 2) indicated that the observations were not similarly distributed across the 10 swine operations (P < 0.0001). When the facilities were analyzed by production type (finishing, farrowing, nursery, and gestation), the distributions of the species and genotypes of Cryptosporidium oocysts were still not similar among the facility types (P < 0.001) and were likely skewed by there being only one gestation facility in which no C. suis was observed. When the chi-square analysis was run with C. suis and the other species and genotypes were truncated into a single set, the distributions were still not similar among production facilities (P < 0.0001). If data for the gestation facility and farrowing facility Far1 were removed from the data set, there was still significant variation among the remaining eight operations when testing for the proportion of C. suis in positive samples (P < 0.0001). However, when the facility types excluding the gestation facility were analyzed, no significant variation in the proportions of C. suis was observed (P = 0.9653).
TABLE 2.
Results of chi-square analysis of the four sets of Cryptosporidium species and genotypes across the 10 locationsa
| Analysis type | df | Chi-square | Probability |
|---|---|---|---|
| Location by CrypType | 27 | 480.458 | <0.0001 |
| FacType by CrypType | 9 | 328.266 | <0.0001 |
| Location by CrypType (C. suis/other)b | 9 | 202.809 | <0.0001 |
| FacType by CrypType (C. suis/other)c | 3 | 139.490 | <0.0001 |
| Location by CrypType (C. suis/other, without Far1/Gest)d | 7 | 30.805 | <0.0001 |
| FacType by CrypType (C. suis/other, without Gest)e | 2 | 0.0706 | 0.9653 |
Results of chi-square analysis of the four sets of Cryptosporidium species and genotypes (CrypType; i.e., C. suis, Cryptosporidium pig genotype II, C. muris, and the other less frequently observed species and genotypes [as delineated in Table 1]) across the 10 locations and by type of facility (FacType).
Analysis of the distribution of C. suis and all other species and genotypes across locations.
Analysis of the distribution of C. suis and all other species and genotypes across facilities.
Analysis of the distribution of C. suis and other species and genotypes across locations with Far1 and the gestation facility (Gest) removed.
Analysis of the distribution of C. suis and other species and genotypes across facilities with only Gest removed.
Results of a logistic regression analysis (Table 3) indicated that the odds of observing C. suis in the lagoon effluent of the swine facilities sampled would be 71.6 times greater than the odds of observing C. parvum genotypes. Although not statistically significant, the odds of observing Cryptosporidium pig genotype II would be 2.4 times the odds of observing C. parvum genotypes; the odds of observing C. muris would be 0.8 times the odds of observing C. parvum genotypes.
TABLE 3.
Odds ratio estimates of the distribution of Cryptosporidium genotypes/species across the 10 swine lagoonsa
| Genotype/species | Odds ratiob | 95% Wald confidence interval |
|---|---|---|
| C. suis | 71.616*** | 8.830-531.355 |
| Pig genotype II | 2.382 | 0.487-9.049 |
| C. muris | 0.766** | 0.168-4.504 |
The distribution of each of the shown genotypes/species is compared to the distributions of the other less frequently observed species and genotypes, such as the zoonotic bovine C. parvum type II, as well as others.
**, significant at a P of ≤0.01; ***, significant at a P of ≤0.001.
DISCUSSION
Viability assessment and distribution of Cryptosporidium genotypes.
Cryptosporidium oocysts were detected and quantified in all the effluent lagoons on each sampling occasion. In contrast to our results, previous prevalence studies on the distribution of Cryptosporidium oocysts in swine effluent lagoons have reported observation frequencies less than 100%. Reinoso and Becares (26) reported detecting Cryptosporidium oocysts in 53% of the effluent samples taken from 13 intensive swine operations in Spain. Hutchison et al. (13) reported detecting oocysts in only 3 of 58 swine effluent lagoons sampled in the United Kingdom. Cote et al. (5) reported detecting oocysts in only one operation out of 32 finishing swine operations sampled in Quebec, Canada. Xiao et al. (39) reported detecting Cryptosporidium in effluent from 14 of 33 finishing facilities in Ireland. The difference between our frequency of observations and those of Reinoso and Becares (26), Hutchison et al. (13), and Xiao et al. (39) may be attributed to differences in sampling scheme, sample size, and methods of oocyst extraction and detection. The concentrations that we have reported (Fig. 3) are in the ranges that Reinoso et al. (26) and Hutchison et al. (13) reported, 16 to 233 and 140 to 310 oocysts ml−1 effluent, respectively. Assessment of viability was not reported in any of these previous studies.
Our observation that the nursery operation, which has two houses and a 0.3-ha lagoon, compared to the finishing operations with four houses and 0.6-ha lagoons, had the greatest concentration of oocysts is in agreement with the observation of Vitovec et al. (34). They reported that the greatest prevalence of Cryptosporidium oocysts in porcine fecal samples was associated with postweaning piglets between 6 and 8 weeks old. The greatest proportion of viable oocysts was also associated with the nursery operation (Fig. 3). The nursery's greater frequency of viable oocysts may be attributable to the greater influx and load of oocysts in the effluent lagoon. However, the observation that the viability assessment for the lagoon receiving effluent from a farrowing and nursery operation was not different from those of the other lagoons implies that our observation of the elevated oocyst viability at the nursery operation may be anomalous. Apart from the nursery operation we sampled, the mean concentration of viable oocysts in the other lagoons was less than 10% of the mean total. Our results indicate that there appears to be a continuous input of oocysts into the lagoons from their respective housing facilities. Based on infection prevalences of 16, 31, and 100% in sows, piglets, and postweaning piglets (20), respectively, we can infer that the apparent inflow of oocysts into the lagoon indicates that some of the pigs in the houses were shedding oocysts in their feces at any given time. The apparent die-off of oocysts may, in part, be attributed to known ammonia emissions from lagoons (12) and deleterious effects of ammonia on oocyst viability (15).
Our determination of the mean concentration of oocysts between 25,000 and 500,000 oocysts dm−3 is indicative of their persistence in effluent lagoons for all four types of swine operations and expands on the observations of Xiao et al. (39) that oocysts of unknown viability appeared to persist in swine effluent from finishing operations in Ireland. We observed that a small fraction of the oocysts appeared to be viable and potentially infective. Thus, an application of swine effluent to spray fields would include a load of potentially infective Cryptosporidium oocysts. Given that 500 m3 of swine effluent may be applied to a spray field as a source of N for plant growth (32) and given that the concentration range of viable oocysts for any of the lagoons in our study was between 1,000 and 12,000 dm−3, the load of viable oocysts applied to a spray field could range between 5 × 108 and 6 × 109 viable oocysts.
The distribution of Cryptosporidium oocysts among the lagoons of the swine facilities sampled indicated that the frequency of observing C. suis would be significantly greater than that for C. parvum genotypes, at a ratio of >10:1. Combining C. suis and Cryptosporidium pig genotype II further increases the ratio of swine-adapted genotypes to C. parvum genotypes. Whereas Langkjaer et al. (19) reported that pig genotype II was most prevalent for weaners, we observed that C. suis dominated the nursery lagoon. Except in the gestation facility, the ratio of C. suis to Cryptosporidium pig genotype II favored the former and ranged from 20:1 to 7:4. The yearlong sampling of the lagoon receiving effluent from the gestation facility proved to be anomalous: 90% of the samples were identified as C. muris, no C. suis was observed, a small percentage of Cryptosporidium pig genotype II was identified, and C. felis was observed once. Cryptosporidium felis has been known to infect cows and both immunocompromised and immunocompetent humans (7). Because this observation of C. felis was associated with an effluent lagoon (which could average a volume of 20,000 m3) into which fecal waste from gestating sows was collected, its presence appears to implicate swine as a source, which would be, to the best of our knowledge, a first.
The observation of the C. parvum rat genotype at farrowing facility Far1 may be a first for swine facilities in the United States. This C. parvum rat genotype matched one that Feng et al. (8) reported isolating from a wastewater treatment plant in Shanghai, China, and suggests a worldwide distribution of this genotype. The source of this rat genotype was likely a known population of rats inhabiting the houses. The C. meleagridis isolate from Far1 matched one isolated from a turkey (36). In addition to C. muris being observed in the lagoon at Far1, several isolates of a C. parvum mouse genotype that matched those identified in three different studies were observed (23, 25, 36). The source of this genotype was likely a population of mice inhabiting the operation. An unknown “other” Cryptosporidium genotype observed at the nursery matched an isolate observed in Ireland (45) and suggests an isolate with a continental distribution.
Our observations over the yearlong period of sampling the lagoons of the 10 swine facilities support previous observations that pigs can be infected with C. suis, Cryptosporidium pig genotype II, and C. parvum (29) as well as C. muris (39, 45). We emphasize that the percentages of C. parvum genotypes observed ranged from 0 to 2% except for that at farrowing facility Far1. This observation is similar to the prevalence study that Zintl et al. (45) reported. The low frequency of C. parvum genotypes that we observed also supports the prevalence study that Johnson et al. (17) reported, in which they maintained that domestic pigs do not appear to pose a significant public health risk.
Because the morphologies of C. suis and C. parvum oocysts are indistinguishable (29), observations by microscopy of these oocysts in swine feces and lagoon effluent may have been assumed to be C. parvum. The reports in the literature that pigs and lagoons of swine facilities may be a significant source of C. parvum (2, 9, 13, 24) may actually refer to C. suis. The weaner pigs infected with the C. parvum pig genotype that Guselle et al. (10) reported were likely infected by C. suis, as Ryan et al. (29) later described. As Zintl et al. (45) observed, mixed infections in pigs can be common, and our data support this view. Cryptosporidium suis is, however, likely to be the dominant genotype, and the frequency of observing C. parvum genotypes in pigs has been low (42). Based on recent studies, the zoonotic potential of C. suis is not great. Most human cases of cryptosporidiosis in the United States and worldwide (over 90%) are associated with C. hominis and C. parvum. A smaller proportion of cases have been associated with C. meleagridis, C. felis, and C. canis (40, 41). Although C. suis and C. suis-like organisms have been associated with sporadic cases of cryptosporidiosis, these reports number in the single digits (35, 40). Cryptosporidium suis appears not to be an important cause of zoonotic disease in humans; however, immunocompromised individuals, known to be at greater risk of infection with Cryptosporidium spp., should take special precautions to prevent exposure to these parasites.
Acknowledgments
We express our appreciation to Gregory Surratt, Michael Thornton, and Shaheen Humayoun for their expert technical assistance.
This project was supported in part by the USDA-CSREES-National Research Initiative Competitive Grants Program.
Footnotes
Published ahead of print on 16 July 2010.
REFERENCES
- 1.Anguish, L. J., and W. C. Ghiorse. 1997. Computer-assisted laser scanning and video microscopy for analysis of Cryptosporidium parvum oocysts in soil, sediment, and feces. Appl. Environ. Microbiol. 63:724-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atwill, E. R., R. A. Sweitzer, M. D. G. C. Pereira, I. A. Gardner, D. Van Vuren, and W. M. Boyce. 1997. Prevalence of and associated risk factors for shedding Cryptosporidium parvum oocysts and Giardia cysts within feral pig populations in California. Appl. Environ. Microbiol. 63:3946-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, A. T., L. J. Robertson, and H. V. Smith. 1992. Viability of Cryptosporidium parvum oocysts: correlation of in vitro excystation with inclusion of exclusion of fluorogenic vital dyes. Appl. Environ. Microbiol. 58:3488-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, D., L. Todd, and S. Wing. 2000. Concentrated swine feeding operations and public health: a review of occupational and community health effects. Environ. Health Perspect. 108:685-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cote, C., A. Villeneuve, L. Lessard, and S. Quessy. 2006. Fate of pathogenic and nonpathogenic microorganisms during storage of liquid hog manure in Quebec. Livestock Sci. 102:204-210. [Google Scholar]
- 6.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection, and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 7.Fayer, R., M. Santin, J. M. Trout, and J. P. Dubey. 2006. Detection of Cryptosporidium felis and Giardia duodenalis assemblage F in a cat colony. Vet. Parasitol. 140:44-53. [DOI] [PubMed] [Google Scholar]
- 8.Feng, Y., N. Li, L. Duan, and L. Xiao. 2009. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J. Clin. Microbiol. 47:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan, T. Y., and R. A. Holley. 2003. Pathogen survival in swine manure environments and transmission of human enteric illness—a review. J. Environ. Qual. 32:383-392. [DOI] [PubMed] [Google Scholar]
- 10.Guselle, N. J., A. J. Applebee, and M. E. Olson. 2003. Biology of Cryptosporidium parvum in pigs: from weaning to market. Vet. Parasitol. 113:7-18. [DOI] [PubMed] [Google Scholar]
- 11.Hamnes, I. S., B. K. Gjerde, T. Forberg, and L. J. Robertson. 2007. Occurrence of Cryptosporidium and Giardia in suckling piglets in Norway. Vet. Parasitol. 144:222-233. [DOI] [PubMed] [Google Scholar]
- 12.Harper, L. H., R. R. Sharpe, T. B. Parkin, A. D. Visscher, O. van Cleemput, and F. M. Byers. 2002. Nitrogen cycling though swine production systems: ammonia, dinitrogen, and nitrous oxide emissions. J. Environ. Qual. 33:1189-1201. [DOI] [PubMed] [Google Scholar]
- 13.Hutchison, M. L., L. D. Walters, S. M. Avery, B. A. Synge, and A. Moore. 2004. Levels of zoonotic agents in British livestock manures. Lett. Appl. Microbiol. 39:207-214. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins, M. B., L. J. Anguish, D. D. Bowman, M. J. Walker, and W. C. Ghiorse. 1997. Assessment of a dye permeability assay for determination of inactivation rates of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 63:3844-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins, M. B., D. D. Bowman, and W. C. Ghiorse. 1998. Inactivation of Cryptosporidium parvum oocysts by ammonia. Appl. Environ. Microbiol. 64:784-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins, M. B., A. J. Franzluebbers, and S. B. Humayoun. 2006. Assessing short-term responses of prokaryotic communities in bulk and rhizosphere soils to tall fescue endophyte infection. Plant Soil 289:309-320. [Google Scholar]
- 17.Johnson, J., R. Buddle, S. Reid, A. Armson, and U. M. Ryan. 2008. Prevalence of Cryptosporidium genotypes in pre- and post-weaned pigs in Australia. Exp. Parasitol. 119:418-421. [DOI] [PubMed] [Google Scholar]
- 18.Key, N., and W. McBride. 2007. The changing economics of U.S. hog production. ERR-52. United States Department of Agriculture, Economic Research Service, Washington, DC. http://www.ers.usda.gov/Publications/ERR52/. Accessed 10 June 2010.
- 19.Langkjaer, R. B., H. Vigre, H. L. Enemark, and C. Maddox-Hyttel. 2007. Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology 134:339-350. [DOI] [PubMed] [Google Scholar]
- 20.Maddox-Hyttel, C., R. B. Langkjaer, H. L. Enemark, and H. Vigre. 2006. Cryptosporidium and Giardia in different age groups of cattle and pigs—occurrence and management associated risk factors. Vet. Parasitol. 141:48-59. [DOI] [PubMed] [Google Scholar]
- 21.Mallin, M. A. 2000. Impacts of industrial animal production on rivers and estuaries. Am. Sci. 88:26-37. [Google Scholar]
- 22.McDonald, A. C., W. R. MacKenzie, D. G. Addiss, M. S. Gradus, G. Linke, E. Zembrowski, M. R. Hurd, M. J. Arrowood, P. J. Lammie, and J. W. Priest. 2001. Cryptosporidium parvum-specific antibody responses among children residing in Milwaukee during the 1993 waterborne outbreak. J. Infect. Dis. 183:1373-1379. [DOI] [PubMed] [Google Scholar]
- 23.Morgan, U. M., L. Xiao, R. Fayer, T. K. Graczik, A. A. Lal, P. Deplazes, and R. C. A. Thompson. 1999. Phylogenetic analysis of Cryptosporidium isolates from captive reptiles using 18S rDNA sequence data and random amplified polymorphic DNA analysis. J. Parasitol. 85:525-530. [PubMed] [Google Scholar]
- 24.Olson, M. E., C. L. Thorlakson, L. Deselliers, D. W. Morck, and T. A. McAllister. 1997. Giardia and Cryptosporidium in Canadian farm animals. Vet. Parasitol. 68:375-381. [DOI] [PubMed] [Google Scholar]
- 25.Pedraza-Díaz, S., L. M. Ortega-Mora, B. A. Carrion, V. Navarro, and M. Gomez-Bautista. 2009. Molecular characterization of Cryptosporidium isolates from pet reptiles. Vet. Parasitol. 160:204-210. [DOI] [PubMed] [Google Scholar]
- 26.Reinoso, R., and E. Becares. 2008. The occurrence of intestinal parasites in swine slurry and their removal in activated sludge plants. Bioresour. Technol. 99:6661-6665. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, L. J., A. T. Campbell, and H. V. Smith. 1998. Viability of Cryptosporidium parvum oocysts: assessment by the dye permeability assay. Appl. Environ. Microbiol. 64:3544-3545. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan, U. M., B. Samarasinghe, C. Read, J. R. Buddle, I. D. Robertson, and R. C. A. Thompson. 2003. Identification of a novel Cryptosporidium genotype in pigs. Appl. Environ. Microbiol. 69:3970-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan, U. M., P. Monis, H. L. Enemark, I. Sulaiman, B. Samarasinghe, C. Read, R. Buddle, I Robertson, L. Zhou, R. C. A. Thompson, and L. Xiao. 2004. Cryptosporidium suis n. sp. (Apicomplexa: Cryptosporidiidae) in pigs (Sus scrofa). J. Parasitol. 90:769-773. [DOI] [PubMed] [Google Scholar]
- 30.SAS Institute. 2004. SAS/STAT 9.1 user's guide. SAS Institute, Cary, NC.
- 31.Simmons, O. D., III, M. D. Sobsey, C. D. Heaney, F. W. Schaefer III, and D. S. Francy. 2001. Concentration and detection of Cryptosporidium oocysts in surface water samples by method 1622 using ultrafiltration and capsule filtration. Appl. Environ. Microbiol. 67:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sistani, K. R., M. R. McLaughlin, and G. E. Brink. 2008. Soil nutrient evaluation from swine effluent application to five forage-system practices. Nutr. Cycl. Agroecosyst. 82:265-271. [Google Scholar]
- 33.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptospordium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitovec, J., K. Hamadejova, L. Landova, M. Kvac, D. Kvetonova, and B. Sak. 2006. Prevalence and pathogenicity of Cryptosporidium suis in pre- and post-weaned pigs. J. Vet. Med. B 53:239-243. [DOI] [PubMed] [Google Scholar]
- 35.Wang, J. C., L. X. Zhang, C. S. Ning, F. C. Jian, and M. F. Sun. 2007. Identification and phylogenetic analysis of a Cryptosporidium isolate from a patient admitted in hospital. Chin. J. Zoon. 23:997-1000. [Google Scholar]
- 36.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. A. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao, L., C. Bern, M. Arrowood, I. Sulaiman, L. Zhou, V. Kawai, A. Vivar, A. A. Lal, and R. H. Gilman. 2002. Identification of the Cryptosporidium pig genotype in a human patient. J. Infect. Dis. 185:1845-1848. [DOI] [PubMed] [Google Scholar]
- 38.Xiao, L. L., A. A. Lal, and J. Jiang. 2004. Detection and differentiation of Cryptosporidium oocysts in water by PCR-RFLP. Methods Mol. Biol. 268:163-176. [DOI] [PubMed] [Google Scholar]
- 39.Xiao, L., J. E. Moore, U. Ukoh, W. Gatei, C. J. Lowery, T. M. Murphy, J. S. G. Dooley, B. C. Miller, P. J. Rooney, and J. R. Rao. 2006. Prevalence and identity of Cryptosporidium spp. in pig slurry. Appl. Environ. Microbiol. 72:4461-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao, L. L., and Y. Y. Feng. 2008. Zoonotic cryptosporidiosis. FEMS Immunol. Med. Microbiol. 52:309-323. [DOI] [PubMed] [Google Scholar]
- 41.Xiao, L. 2009. Molecular epidemiology of human cryptosporidiosis in developing countries, p. 25-30. In M. G. Ortega-Pierres, S. Caccio, R. Fayer, T. Mank, and R. C. A. Thompson (ed.), Giardia and Cryptosporidium: from molecules to disease. CABI, Wallingford, United Kingdom.
- 42.Xiao, L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80-89. [DOI] [PubMed] [Google Scholar]
- 43.Zar, J. H. 1999. Biostatistical analysis. Prentice-Hall, Upper Saddle River, NJ.
- 44.Zhou, A., A. Singh, J. Jiang, and L. Xiao. 2003. Molecular surveillance of Cryptosporidium spp. in raw waste water in Milwaukee: implications for understanding outbreak occurrence and transmission dynamics. J. Clin. Microbiol. 41:5254-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zintl, A., D. Neville, D. Maguire, S. Fanning, G. Mulcahy, H. V. Smith, and T. De Waal. 2007. Prevalence of Cryptosporidium species in intensively farmed pigs in Ireland. Parasitology 134:1575-1582. [DOI] [PubMed] [Google Scholar]