Abstract
The oral biofilm community consists of >800 microbial species, among which Streptococcus mutans is considered a primary pathogen for dental caries. The genomic island TnSmu2 of S. mutans comprises >2% of the genome. In this study, we demonstrate that TnSmu2 harbors a gene cluster encoding nonribosomal peptide synthetases (NRPS), polyketide synthases (PKS), and accessory proteins and regulators involved in nonribosomal peptide (NRP) and polyketide (PK) biosynthesis. Interestingly, the sequences of these genes and their genomic organizations and locations are highly divergent among different S. mutans strains, yet each TnSmu2 region encodes NRPS/PKS and accessory proteins. Mutagenesis of the structural genes and putative regulatory genes in strains UA159, UA140, and MT4653 resulted in colonies that were devoid of their yellow pigmentation (for strains UA140 and MT4653). In addition, these mutant strains also displayed retarded growth under aerobic conditions and in the presence of H2O2. High-performance liquid chromatography profiling of cell surface extracts identified unique peaks that were missing in the mutant strains, and partial characterization of the purified product from UA159 demonstrated that it is indeed a hybrid NRP/PK, as predicted. A genomic survey of 94 clinical S. mutans isolates suggests that the TnSmu2 gene cluster may be more prevalent than previously recognized.
The human oral cavity harbors >800 microbial species, and each individual likely hosts at least 100 different species (1, 30, 31). Dental biofilm formation is a sequential process. After a new tooth emerges or a new surface is cleaned, the surface is colonized by a group of bacteria termed the “pioneer colonizers,” comprised mostly of the mitis group streptococci (i.e., Streptococcus gordonii, S. sanguinis, S. mitis, etc.). Later, early colonizers, such as Streptococcus mutans, Veillonella, and bridge species such as fusobacteria, join the community through interactions with the pioneer colonizers or with available sites on the tooth surface. Growth of the early colonizers then modifies the local environment, making it favorable for the growth of late colonizers, which consist mostly of Gram-negative, strictly anaerobic bacteria. Through further cell growth and coadhesion, this will eventually result in a mature biofilm (for a review, see reference 21).
The mutans group streptococci (mainly S. mutans) are considered primary pathogens for the development of dental caries (tooth decay) (24), and the mitis group streptococci are oral commensals (5, 6). The two groups of streptococci are known competitors, with S. mutans producing mutacins to kill the mitis group streptococci and the mitis group streptococci producing H2O2 to kill S. mutans (22). Since ∼60 to 80% of the pioneer colonizers are the mitis group streptococci, an intriguing question arises: how does S. mutans gain entry into this hostile environment and persist in the early biofilm community? By searching the genome of S. mutans, evidence for such a niche-specific adaptation may be revealed in a large genomic island (GI) termed TnSmu2 (2). GIs are ubiquitous in prokaryotes. They are characterized by the presence of insertion elements on either end of a large DNA segment (10 to 200 kb) and typically exhibit a G+C percentage that is distinct from that of the rest of the genome. Many GIs carry genes that confer a selective advantage to the bearer strain under a particular set of environmental conditions but are otherwise dispensable. GIs can encode a tremendous variety of functions, including host adherence, toxin production, type III and type IV secretion, iron acquisition, and host modulation (refer to references 9, 15, and 19 for reviews).
In this communication, we report the characterization of the function of the TnSmu2 elements from 3 strains of S. mutans. We demonstrate that these GIs all carry a gene cluster encoding nonribosomal peptide synthetases (NRPS), polyketide synthases (PKS), accessory proteins, transporters, and transcription regulators, although the sequences of the genes vary widely among different strains. Furthermore, we present evidence that these gene clusters are responsible for the biosynthesis of a hybrid NRP/PK pigment that we refer to as mutanobactin, which is likely involved in tolerance to oxygen and H2O2 stresses. We have also shown that this gene cluster is more prevalent among S. mutans clinical isolates than was previously recognized.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. mutans strains used in this study are listed in Tables S2 and S4 in the supplemental material. All S. mutans strains were routinely grown on brain heart infusion (BHI; Difco, Sparks, MD) agar plates or on BHI agar supplemented with either spectinomycin (800 μg ml−1) or kanamycin (Kan; 800 μg ml−1). A chemically defined medium (CDM) was used in growth assays under aerobic and anaerobic conditions as well as in luciferase reporter assays. The composition of the CDM has been described previously (16).
Bioinformatic analysis.
BLASTP programs (http://www.ncbi.nlm.nih.gov/blast/) were used to analyze individual open reading frames (ORFs) of the TnSmu2 gene cluster to identify homologs in the database. Comparisons of the mutanobactin gene clusters in strains UA140 and MT4653 were performed using BLAST 2 software. The NRPS-PKS program (http://www.nii.res.in/nrps-pks.html), developed by Ansari et al. (3), was used to predict the modules and domains of NRPS/PKS and to determine the substrate specificity of the A domains.
Construction of mutant strains.
Primers used for mutagenesis are listed in Table S1 in the supplemental material. To delete the whole NRPS-PKS gene cluster, in-frame allelic exchange of the NRPS-PKS genes with the Kan cassette was performed by using a three-step direct PCR ligation strategy according to the method of Doran et al. (10). Briefly, for UA159, primers UA159-ABC-upF and UA159-ABC-upR were used to amplify a 2,873-bp fragment from downstream of SMU.1346c, and UA159-mefE-dnF and UA159-mefE-dnR were used to amplify a 2,685-bp fragment from upstream of SMU.1338c (Fig. 1 A). For UA140 and MT4653, UA140-RRKK-upF and UA140-RRKK-upR were used to amplify a 2,380-bp fragment from upstream of ORF3, and UA140-13-15dnF and UA140-13-15dnR were used to amplify a 2,169-bp fragment from downstream of ORF12 (Fig. 1B). The UA159-ABC-upR and UA140-RRKK-upR primers and the UA159-mefE-dnF and UA140-13-15dnF primers were designed with 25-bp extensions corresponding to the 5′ and 3′ ends of the Kan cassette, respectively. The Kan cassette, aphAIII, was amplified from pGNaa3 (29) with primers kan-F and kan-R. After purification, the upstream and downstream fragments of UA140 and UA159 were mixed with the Kan cassette fragment in equal amounts for a PCR using Phusion DNA polymerase (NEB). The initial denaturation was performed at 94°C for 2 min, followed by 10 cycles without primers to allow annealing and elongation of the overlapping fragments (94°C for 10 s, 55°C for 15 s, and 72°C for 7 min). After the addition of external primers (UA159-ABC-upF and UA159-mefE-dnR for UA159 and UA140-RRKK-upF and UA140-13-15dnR for UA140), the program continued, with 30 cycles (94°C for 10 s, 55°C for 15 s, and 72°C for 7 min) and then a final 10-min extension at 72°C. The resulting PCR products were used directly to transform UA159, UA140, and MT4653. The transformants were selected on Kan plates, and the deletion of the NRPS-PKS gene cluster on the chromosome was confirmed by PCR.
FIG. 1.
TnSmu2 gene cluster and predicted modules of NRPS gene products. (A) Genomic organization in strain UA159. SMU.1345c, -1342c, -1341c, -1340c, and -1339c encode NRPS, and SMU.1344c, -1343c, -1336c, and -1335c encode PKS. SMU.1349 is the transcription activator of the operon. Numbers above the genes are gene numbers in the database (http://www.oralgen.lanl.gov). Names below the genes were designated in this study. (B) Genomic organization in strain UA140. ORFs 3, 4, and 11 encode NRPS, and ORFs 5 to 8 encode PKS. ORF1 and ORF2 encode a two-component system required for the transcription of the entire gene cluster. Numbers above the genes are ORF numbers, and names below the genes were assigned in this study. Predicted domains for NRPS and PKS are listed below the gene names. Domain names for NRPS: A, adenylation domain, which selects the cognate amino acid, activates it as an amino acyl adenylate, and transfers it to the T domain (subscript letters denote the specified amino acid of each A domain); T, T domain, also known as a peptidyl carrier protein (PCP), where a thioester bond is formed; C, condensation domain, responsible for peptide bond formation between the amino acid present on the T domain of the same module and the peptidyl intermediate bound to the T domain of the preceding module; E, epimerization domain; TE, termination domain, containing a thioesterase module. Domain names for PKS: AT, acyltransferase domain, which selects an appropriate extender unit (usually malonyl-coenzyme A [malonyl-CoA] or methylmalonyl-CoA) and transfers it to the ACP domain, where a thioester bond is formed; ACP, acyl carrier protein domain; KS, ketosynthase domain, responsible for decarboxylative condensation between the extender unit present on the ACP domain of the same module and the polyketide intermediate bound to the ACP domain of the preceding module; KR, ketoreductase domain; ppt, phosphopantetheine attachment site. At the phosphopantetheine attachment site, a 4′-phosphopantetheine prosthetic group is attached through a serine. This prosthetic group acts as a swinging arm for the attachment of activated fatty acid and amino acid groups. The domain structures were predicted using a Web-based program (http://www.nii.res.in/nrps-pks.html).
To delete the possible regulator SMU.1349 of UA159 and the two-component system histidine kinase-encoding ORF1 of UA140 and MT4653, a double-crossover strategy was used. Briefly, for each construct, two fragments were generated by PCR, corresponding to approximately 1 kb of upstream and downstream sequence relative to the target gene. To facilitate ligation, an XbaI restriction site and an XhoI restriction site were incorporated into the 3′ end of the upstream fragment and the 5′ end of the downstream fragment, respectively. The PCR product was digested with XbaI and XhoI and ligated with the Kan cassette released from pGNaa3 digested with the same enzymes. The ligation mixture was diluted 200 times and used as the DNA template for PCR. For UA159, primers 1349up-F and 1349dn-R were used, and for UA140 and MT4653, primers 140HKup-F and 140HKdn-R were used to amplify the ligated fragment. The PCR products were directly transformed into UA159, UA140, and MT4653. The transformants were selected on Kan plates, and the deletion was confirmed by PCR. A complete list of plasmids and strains constructed in this study is provided in Table S2 in the supplemental material.
Construction of luciferase reporter strains.
To construct a luciferase transcriptional reporter fusion to the NRPS-PKS operon in UA159, a 400-bp fragment containing the promoter region upstream of SMU.1348c (IGR1065) was amplified by PCR and ligated into the transcription fusion vector pFW5-luc to generate the plasmid pFW-IGR1065-luc. To construct the same reporter gene fusion in UA140, a 400-bp fragment between ORF4 and ORF5 (Fig. 1B) was amplified by PCR and cloned into pFW5-luc to generate the plasmid pFW-orf4-luc. These plasmids were integrated into the chromosomes of S. mutans UA159 and UA140 via single-crossover homologous recombination. The resulting strains were confirmed by PCR.
Effects of oxygen on growth.
To determine the effect of oxygen on the growth of the wild-type (WT) and mutanobactin mutant strains of S. mutans, cells were grown to stationary phase in CDM at 37°C anaerobically. The stationary-phase cultures of WT and mutant strains were normalized to an optical density at 600 nm (OD600) of 1.0 and then inoculated at a 1:100 dilution into fresh CDM. Two hundred microliters of this cell suspension was added to each well of a Bioscreen system plate, and cell growth was monitored by measuring the OD600, using a Bioscreen C lab system (Helsinki, Finland). To create aerobic conditions, the system was set to shake for 10 s every 30 min. For the anaerobic control culture, 200 μl of the same cell suspension was added to each well of a 96-well plate, which was then reduced in an anaerobic chamber and sealed within the chamber with TopSeal (TopSeal was not compatible with the Bioscreen system plate). Cell growth was monitored using a FluoStar Optima plate reader (BMG Lab Technologies).
Effects of H2O2 on growth.
Cell suspensions were prepared as described above. To the UA140 culture, H2O2 was added to a final concentration of 0.1 mM, and to the UA159 and MT4653 cultures, H2O2 was added to a final concentration of 0.6 mM. These concentrations were previously determined to be inhibiting but not lethal to the respective cultures. To each well of a 96-well plate, 150-μl aliquots of the cell suspension were added inside an anaerobic chamber, and the plate was sealed by using TopSeal. The sealed plate was then placed inside a FluoStar Optima plate reader, and the cell growth was monitored by measuring the OD600. Both WT and mutant cultures without H2O2 addition were used as controls. Wells containing medium only were used as blank controls, and all samples were tested in replicate series (n ≥ 3).
Biofilm formation assay.
Biofilm formation of the wild-type and mutant strains was conducted under both anaerobic and aerobic conditions in 6-well plates on a rocker platform in a 37°C incubator. Cell cultures were grown and diluted in the same fashion as that described above, except that the CDM contained 0.5% sucrose instead of glucose. To create anaerobic conditions, the plate was reduced in an anaerobic chamber and sealed with TopSeal. The biofilm was inspected microscopically after 24 h of growth.
Luciferase assay.
Luciferase assays were performed as previously described (25). Briefly, 20 μl of 1 mM d-luciferin (Sigma, St. Louis, MO) suspended in 100 mM citrate buffer, pH 6, was added to 100 μl of the cell culture. To ensure sufficient levels of the intracellular ATP pool, cells were recharged with 1% glucose for 10 min prior to luciferin addition. Luciferase activity was measured by using a 20/20 luminometer (Turner Biosystems, Sunnyvale, CA). Usually, two parallel cultures were measured at each time point and the average value was recorded. Each experiment was repeated at least three times.
Mutanobactin extraction and HPLC profiling.
S. mutans WT and mutant strains were grown overnight in BHI broth, and the overnight cultures were painted onto BHI agar plates. The plates were incubated in an anaerobic chamber at 37°C for 24 h, and the cells were collected from the plate by scraping with a cotton tip and then were resuspended in phosphate-buffered saline (PBS) in a 1.5-ml Eppendorf tube. Cells were concentrated by centrifugation, and the pellet was extracted 3 times with 200 μl of methanol (ACS grade; Sigma). After removal of the extraction solvent in vacuo, the remaining organic residues (the dry weight yield was ∼10 mg per plate for each S. mutans strain) were redissolved in methanol and passed through a C18 SPE cartridge prior to high-performance liquid chromatography (HPLC). The extracts were analyzed using a C18 Synergi Hydro-RP column under gradient elution conditions (0 to 5 min, 10% acetonitrile; 5 to 25 min, 10 to 100% acetonitrile; and 25 to 40 min, hold at 100% acetonitrile), using a variable-wavelength detector.
RNA extraction and real-time PCR.
Overnight cultures of WT S. mutans UA140 and UA159 and their corresponding regulator mutants were grown in CDM to an OD600 of 0.8 at 37°C in an anaerobic chamber, and cells were collected by centrifugation. The cell pellets were resuspended in 1 ml of ice-cold Trizol and disrupted with 300 μl of 0.1-mm zirconia beads, using a Fastprep kit (MP Biomedical). After centrifugation, the supernatant was collected and RNA purified using an Ambion RiboPure Bacteria kit according to the manufacturer's protocol. The isolated RNA was treated with DNase I (Ambion) to remove traces of chromosomal DNA. After the treatment, RNA samples were cleaned with a Qiagen RNeasy MinElute cleanup kit. Total RNA (300 ng) was used for cDNA synthesis, using StrataScript reverse transcriptase (RT) (Stratagene) according to the manufacturer's protocol. Real-time PCR primers used in the study are listed in Table S1 in the supplemental material. Real-time PCR was performed using an Applied Biosystems 7300 machine, and the reaction mixtures were prepared using Applied Biosystems SYBR green PCR master mix. Changes in gene expression were calculated automatically by the Applied Biosystems 7300 system software, using the ΔΔCT method, and are briefly described as follows: ΔCT = threshold cycle of the target [CT (target)] − CT (housekeeping gene) and ΔΔCT = ΔCT1 − ΔCT2. Fold changes were calculated as 2−ΔΔCT. The 16S rRNA gene was used as the housekeeping gene reference, and all samples included a no-RT control to assess genomic DNA contamination in the reaction mixtures.
PCR analysis of distribution of the NRPS-PKS gene cluster.
Primers used to detect the NRPS-PKS genes are listed in Table S3 in the supplemental material. Primers UA140-orf5-F and UA140-orf5-R were designed to amplify a 515-bp fragment from ORF5 of the UA140 mutanobactin gene cluster. Primers UA159-BacC-F and UA159-BacC-R were designed to amplify a 214-bp fragment from SMU1339c of UA159. Primers MT-orf1-F and MT-orf1-R were designed to amplify a 364-bp fragment from ORF1 of contig 608 of MT4653 (see Table 1). Primers 16S-F and 16S-R were designed to amplify a 120-bp fragment of the 16S rRNA gene of S. mutans. The four pairs of primers were mixed to make a primer cocktail, which included 10 μM (each) MT-orf1-F and MT-orf1-R, 0.5 μM (each) UA140-orf5-F and UA140-orf5-R, 1.25 μM (each) UA159-BacC-F and UA159-BacC-R, and 2.5 μM (each) 16S-F and 16S-R.
TABLE 1.
NRPS/PKS genes in S. mutans strains UA159, UA140, and MT4653
| Strain, contig | Gene or ORF | Length (bp) | Putative function and/or annotation | Best match | GenBank accession no. |
|---|---|---|---|---|---|
| UA159 | 1334C | 683 | sfp; phosphopantetheinyl transferase | Bacillus licheniformis | YP_077646.1 |
| 1335C | 984 | fabK; enoyl(acyl-carrier protein) reductase | Clostridium tetani | NP_780843.1 | |
| 1336C | 939 | pksD; malonyl-coenzyme A(acyl-carrier protein) transacylase | Bacillus amyloliquefaciens | CAG23951.1 | |
| 1337C | 750 | α/β-Hydrolase | Desulfitobacterium hafnieuse | YP_520330.1 | |
| 1338C | 1,155 | mefE; ABC transporter, macrolide permease | Pyrococcus abyssi | NP_126768.1 | |
| 1339C | 4,368 | bacC; bacitracin synthetase | Brevibacillus texasporus | AAY29583.1 | |
| 1340C | 4,887 | bacA; bacitracin synthetase 1 | B. licheniformis | AAC06346.1 | |
| 1341C | 3,690 | grs; gramicidin S synthetase 2 | Aneurinibacillus migulanus | PoC063 | |
| 1342C | 8,175 | bacA; bacitracin synthetase 1 | B. licheniformis | AAC06346.1 | |
| 1343C | 3,315 | pksC; hybrid NRPS/PKS | Clostridium kluyveri | YP_001395738.1 | |
| 1344C | 1,218 | fabD; acyl-carrier protein S-malonyl transferase | Streptococcus pneumoniae | NY_357974.1 | |
| 1345C | 1,902 | ituA; peptide synthetase similar to MycA | Bacillus subtilis | BAB69698.1 | |
| 1346C | 720 | bacT; thioesterase II-like protein | B. licheniformis | BAA36683.1 | |
| 1347C | 2,343 | ymbB; ABC transporter permease | D. hafnieuse | ZP_01370240.1 | |
| 1348C | 702 | pasA; ABC transporter ATP binding | Clostridium acetobutylicum | NP_347458.1 | |
| 1349 | 576 | Transcription regulator of TetR family | Streptococcus sanguinis | YP_001034156.1 | |
| UA140 | 1 | 1,400 | Two-component histidine kinase | Streptococcus equi | YP_002747386.1 |
| 2 | 458 | Response regulator of LuxR family | Streptococcus equi | YP_002124238.1 | |
| 3 | 7,350 | NRPS; NpsA | Lactobacillus plantarum | NP_784351.1 | |
| 4 | 9,834 | NRPS; bacitracin synthetase 3 | B. licheniformis | O68008 | |
| 5 | 1,266 | PKS | C. kluyveri | YP_001395122.1 | |
| 6 | 4,524 | PKS | Clostridium cellulolyticum | ZP_01574699.1 | |
| 7 | 3,321 | PKS | C. cellulolyticum | ZP_01574700.1 | |
| 8 | 1,374 | PKS | C. cellulolyticum | ZP_01574699.1 | |
| 9 | 884 | ABC transporter ATPase | Leuconostoc citreum | YP_001728676.1 | |
| 10 | 737 | ABC transporter permease | Leuconostoc citreum | YP_001728675.1 | |
| 11 | 6,153 | Hybrid PKS/NRPS | C. kluyveri | YP_001395123.1 | |
| 12 | 695 | Thioesterase II-like protein | B. licheniformis | BAA36683.1 | |
| 13 | 521 | Phosphopantetheinyl transferase | C. kluyveri | YP_001395126.1 | |
| 14 | 962 | Avirulence factor D; avrD | Vibrio cholerae | EDN13368.1 | |
| MT4653, contig 608a | 1 | 318 (partial) | PKS1 | B. amyloliquefaciens | YP_0012421029.1 |
| 2 | 9,075 | PKS2 | Kordia algicida | ZP_02163865.1 | |
| 3 | 2,673 | PKS3 | K. algicida | ZP_02163862.1 | |
| MT4653, contig 613a | 1 | 3,627 (partial) | Mixed type I NRPS/PKS | Symbiont bacterium of Paederus fuscipes (beetle) | AAS47562.1 |
| 2 | 3,582 | PKS4 | K. algicida | ZP_02163862.1 | |
| 3 | 7,047 | PKS5 | C. cellulolyticum | ZP_01574364.1 |
Another contig, contig 629, is nearly identical to the UA140 gene cluster. Therefore, MT4653 contains at least two gene clusters, with one identical to that of UA140 and another containing extra genes encoding PKS and a hybrid NRPS/PKS.
S. mutans clinical isolates.
A total of 94 previously identified S. mutans clinical isolates were randomly selected from an S. mutans sample bank located at the New York University College of Dentistry. Fifty-four (57.4%) of the isolates were obtained from individuals with active caries (caries-active [CA] individuals), and 40 (42.6%) were from those without active caries (caries-inactive [CI] individuals). Caries status was measured using the dmft/DMFT index (number of decayed, missing, and filled teeth [lowercase for primary teeth and uppercase for permanent teeth]). Individuals with dmft/DMFT scores of ≥6 were categorized in the caries-active group. Individuals with dmft/DMFT scores of ≤1 were categorized in the caries-inactive group.
RESULTS
Diversity of the cargo genes in TnSmu2 islands among S. mutans isolates.
The genome sequence of S. mutans strain UA159 was completed in 2002, and a large genomic island (56,013 bp, or 2.76% of the genome) was annotated TnSmu2 (2) or GI 12 in the database (http://www.oralgen.lanl.gov/). This region contains genes homologous to those for gramicidin and bacitracin (nonribosomal peptide antibiotics) synthetases. Some of these genes are also the largest (8,175 bp) in the S. mutans genome. Surprisingly, despite their unusually large size, these genes have remained uncharacterized. Our interest in this region began after we recently sequenced 4 additional strains of S. mutans (UA140, MT4653, AF199, and 19) (see Table S4 in the supplemental material) in order to assess the interstrain diversity among S. mutans clinical isolates. A genomic analysis of these strains revealed large contigs containing sequences that were not present in strain UA159. A BLASTX search revealed that these contigs encoded proteins homologous to NRPS, PKS, and accessory proteins as well as to transporters and transcription regulators. These findings prompted us to revisit the TnSmu2 region of UA159. The locus consists of at least 16 genes directly related to NRP and PK biosynthesis (Table 1 and Fig. 1A). Among these genes, four encode NRPS, with the highest homology being to bacitracin and gramicidin synthetases (SMU.1339c to -1342c). One encodes a peptide synthetase homologous to MycA (SMU.1345c), two encode PKS (SMU.1343c and -1344c), one codes for a conserved hypothetical protein with a beta-ketoacyl synthase domain (SMU.1336c), and one encodes a putative enoyl-acyl carrier protein (ACP) reductase (SMU.1335c). If all of these proteins function as a multienzyme complex, then the product is likely to be a hybrid NRP/PK (11). In addition, accessory proteins required for NRP/PK synthesis, such as phosphopantetheinyl transferase, thioesterase, ABC transporters, and permeases, are also encoded by the same gene cluster. A putative transcription regulator (SMU.1349) shares the same intergenic region with the NRPS-PKS gene cluster but is transcribed in the opposite direction (Fig. 1A).
Compared with that in UA159, the NRPS-PKS locus in UA140 has a completely different gene composition and organization (Table 1; Fig. 1B). This locus contains 15 genes distributed on 5 putative operons transcribed in the same orientation. The first putative operon includes genes for a two-component system with a histidine kinase (HK) (ORF1) and a luxR family response regulator. The second includes two large NRPS genes, with the first being homologous to the NpsA gene of Lactobacillus plantarum (ORF3) and the second being homologous to the bacitracin synthetase 3 gene of Bacillus licheniformis (ORF4). The third operon includes 6 genes (ORFs 5 to 10), with 5 PKS genes and 1 ABC transporter gene. The fourth operon encodes a hybrid PK/NRPS (ORF12) with homology to that of Clostridium kluyveri and also encodes a thioesterase II-like protein. The fifth putative operon contains a phosphopantetheinyl transferase gene and a homolog of the avirulence factor D (avrD) gene primarily found in Pseudomonas syringae.
The organization of the NRPS-PKS gene cluster in MT4653 appears to be more complex. In addition to a gene cluster identical to that of UA140, there are currently two additional contigs with sequences that do not match either the UA140 or UA159 sequence. A BLASTX search of these contigs found that one (contig 608) contains at least 3 PKS genes, and the other (contig 618) contains at least 2 PKS genes and 1 type I hybrid PKS/NRPS gene. Our PCR amplification analysis indicated that these two contigs are joined by a 12-kb stretch of DNA (data not shown). However, for the time being, we have yet to determine the sequence of the gap or whether these contigs are adjacent to the first NRPS-PKS gene cluster or located elsewhere. Nonetheless, these results suggest that MT4653 probably has two gene clusters involved in NRP/PK biosynthesis. Analysis of strains AF199 and 19 revealed that they contain the same gene cluster as UA140. Therefore, in the rest of the study, we focused on three strains, UA159, UA140, and MT4653.
Domain analysis of NRPS suggests possible amino acid compositions of NRP products.
NRPS catalyze the activation and condensation of amino acids to yield a peptide product (12). Each amino acid is activated by the acetylation module (A) of an NRPS, which usually encompasses about 500 amino acids. The specificity of the amino acid is determined by the amino acid residues located within the binding center of the A module, while the occurrence and specific order of the modules in the NRPS complex dictate the number and sequence of the amino acid residues in the nonribosomal peptide product (7). In general, the structure of the A modules is conserved among different NRPS across both the prokaryotic and eukaryotic kingdoms. Based on the crystal structure of gramicidin synthetase A (GrsA/PheA), eight amino acids which constitute the binding pocket have been identified (A236, W239, T278, I299, A301, A322, I330, and C331) (8). By aligning PheA with the A modules of other NRPS whose substrates are known, one can predict the specific amino acid in a particular NRP based on the sequence of the 8 amino acid residues (also termed the specificity-conferring sequence) (8, 33). We analyzed the NRPS genes in UA159 and UA140 by using an online NRPS-PKS prediction tool (http://www.nii.res.in/nrps-pks.html). In UA159, a total of 7 A modules were identified, with SMU.1345 harboring 1, SMU.1342 harboring 2, SMU.1341 harboring 1, SMU.1340 harboring 2, and SMU.1339 harboring 1. The amino acids specified by these A modules were predicted to be Asp, Leu, Ser, Pro, Val, Cys, and Gly, respectively (Fig. 1A). In UA140, a total of 6 A modules were identified, with ORF3 containing 2, ORF4 containing 3, and ORF11 containing 1. The specific amino acids specified were Phe, Phe, Asn, Asn, Gly, and Asn, respectively (Fig. 1B). The specificity-conferring sequences of these A modules are presented in Table S5 in the supplemental material.
Mutations of putative NRPS-PKS gene clusters result in colony discoloration.
To determine whether a hybrid NRP/PK product was made, we first deleted the first NRPS gene in each strain by allelic replacement with a terminatorless kanamycin resistance gene cassette. To our surprise, mutant colonies of UA140 and MT4653 were white, whereas the WT colonies of both strains were yellow (see Fig. S1 in the supplemental material), suggesting that the deleted genes are involved in pigment synthesis. Since the wild-type colonies of UA159 are normally white, we could not visually determine whether the genes in UA159 are also involved in white pigment synthesis. Using colony color as an indicator, we then made deletions in each of the genes in the putative NRPS-PKS gene cluster in UA140. All genes were required for pigment synthesis, except for the putative ABC transporter gene (ORF10) (Fig. 1 and data not shown).
HPLC profiling of putative NRP/PK products.
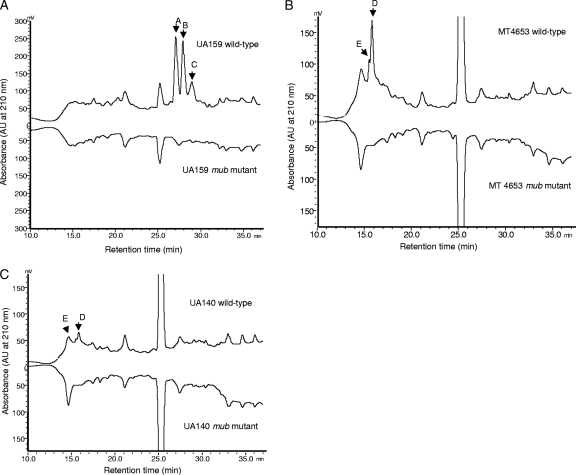
To confirm that the NRPS-PKS genes were involved in pigment biosynthesis in all strains, we conducted HPLC profiling of cell extracts, using a C18 Synergi Hydro-RP column. To exclude possible complications resulting from the accumulation of intermediate products if deletions of single NRPS genes were carried out, we created complete operon deletion mutants for each strain. We hypothesized that if the entire gene cluster in each strain was responsible for the biosynthesis of a single biomolecule, then peaks should be detectable for the WT strains that were not present for the mutants. Both WT and mutant cells were collected from plate cultures, extracted with methanol, and subjected to HPLC profiling. As shown in Fig. 2, there were conspicuous peaks that were observed for the WT strains but missing for the mutants. Of particular interest was the observation that strains UA159 and UA140 had different profiles, consistent with the diversity of their NRPS-PKS gene clusters and predicted amino acid compositions (Fig. 1). Interestingly, the profile of MT4653 was nearly identical to that of UA140, despite its having a second NRPS-PKS gene cluster. This suggested that the second gene cluster was probably not involved in pigment synthesis or was not extractable using our protocol. It should be noted that deletion of the second gene cluster did not result in a white colony phenotype, which suggests that the second gene cluster is not involved in yellow pigment synthesis (data not shown). It is also notable that during revision of the manuscript, we purified the putative NRP/PK product from UA159. Preliminary characterization of the molecule by nuclear magnetic resonance analysis determined the sequence to be Asp-PK-Ser-Pro-Val-Cys-Gly (data will be published elsewhere), which is identical to the sequence predicted from the A domains (Fig. 1). We also confirmed that the two major peaks in the UA159 profile (Fig. 2A) are isomers of the same product (data not shown).
FIG. 2.
HPLC profiles of methanol extracts of wild-type and TnSmu2 deletion mutants (mub) of strains UA159 (A), MT4653 (B), and UA140 (C). Cells were scraped from overnight cultures grown anaerobically on BHI agar plates and were resuspended in PBS. PBS was removed by centrifugation, and the cell surface molecules were extracted with methanol. Arrowheads indicate peaks that are missing for the mutants. AU, absorbance units.
Based on the genomic organizations of the NRPS-PKS gene clusters, the HPLC profiles of the WT and mutant strains, and the nature of the product (a putative hybrid NRP/PK), we named the product mutanobactin and the NRPS-PKS gene cluster the mub locus. Accordingly, the NRPS genes were named mubA to mubE in UA159 and mubA to mubC in UA140 and MT4653; the PKS and related genes were named mubG to mubJ in both UA159 and UA140; the thioesterase and phosphopantetheinyl transferase genes in both UA159 and UA140 were named mubT and mubP, respectively; and the ABC transporters were named mubX to mubZ. In addition, the α/β-superfamily hydrolase gene in UA159 (SMU.1337c) was named mubM (Fig. 1).
The mub gene cluster is involved in oxygen tolerance.
With the identification of the NRPS-PKS gene products responsible for pigment synthesis, we next investigated the biological function of this gene cluster. In the process, we fortuitously discovered that yellow colonies on the WT plate turned white after being exposed to air for 24 h (see Fig. S2 in the supplemental material). This suggested that the pigment probably reacted with oxygen. From this observation, the following question arose: could this pigment serve as a mechanism for oxygen tolerance in S. mutans? To answer this question, we performed a time course study of WT and mub mutant strains in chemically defined medium under both anaerobic and aerobic conditions. While no significant difference was observed between the WT and mutant strains under anaerobic conditions (see Fig. 4A to C), there were conspicuous differences in both growth rate and terminal OD between the WT and mutant strains under aerobic conditions (Fig. 3 A to C). In the presence of oxygen, the lag phase for all mutants was increased by 3 to 4 h, and the growth rate was reduced 66%, 20%, and 46% for UA140, UA159, and MT4653, respectively. In addition, the terminal OD was reduced ∼10% in UA159 and MT4653. These results suggested that these NRPS-PKS gene clusters and their products (the pigments) were involved in oxygen tolerance.
FIG. 4.
Effect of H2O2 on cell growth of strains UA159 (A), UA140 (B), and MT4653 (C). Cells were grown anaerobically in chemically defined medium, with and without H2O2. Squares, WT; triangles, mutant; closed symbols, no H2O2 was added; open symbols, sublethal concentrations of H2O2 were added. The growth rate was calculated only for the WT and mutant strains grown in the presence of H2O2. The experiment was repeated at least three times, with representative results shown.
FIG. 3.
Effect of oxygen on cell growth of strains UA159 (A), UA140 (B), and MT4653 (C). Cells were grown aerobically in chemically defined medium by use of a Bioscreen system. Squares, WT; triangles, mutant. Growth rate was calculated using the formula μ = ln2/g, where g is the doubling time at log phase. The experiment was repeated at least three times, with representative results shown.
Mutation of the mub locus reduces H2O2 resistance of S. mutans.
Having determined the role of the mub locus in oxygen tolerance of S. mutans, our next question was whether this locus was also involved in tolerance to other oxidative stresses normally encountered in the oral cavity, such as H2O2. To test this, WT cells of UA140 were first collected from plate cultures and suspended in PBS. The cells were divided into 2 tubes, and H2O2 was added to 1 tube at a final concentration of 3 mM. Cells were pelleted by centrifugation after 24 h of incubation in an anaerobic chamber. As expected, the H2O2-treated cells lost their yellow color, while the untreated cells remained yellow (see Fig. S3 in the supplemental material). This suggested that the pigment also reacted with H2O2. We next tested the growth of the WT and mutant cells in the presence of sublethal concentrations of H2O2. Cells were grown anaerobically in the presence or absence of 0.1 mM (for UA140) or 0.6 mM (for UA159 and MT4653) H2O2, and cell growth was monitored in a FluoStar plate reader. As shown in Fig. 4 A to C, the growth of the mub mutants was severely impaired in the presence of H2O2, especially for UA159 and MT4653. The lag time was prolonged for all three mutant strains compared with their respective WT strains. Moreover, the growth rate was reduced for all mub mutants compared with their respective WT strains (Fig. 4A to C). These results suggest that the mub locus is also required for H2O2 resistance of S. mutans.
mub mutants hinder biofilm formation under aerobic conditions.
It is commonly accepted that the oxygen concentration within a biofilm should be lower than that on the surface. Since S. mutans exists mainly in biofilms on the tooth's surface, we asked the question of whether the function of the mub locus is relevant to the natural habitat of S. mutans. To test this, biofilm assays were conducted with UA140 and UA159 WT and mub mutant strains. While no difference was noticeable between the WT and mutant strains under anaerobic conditions (data not shown), conspicuous differences were observed under aerobic conditions (Fig. 5). For both UA140 and UA159, the WT cells formed mature biofilms with well-developed three-dimensional structures; however, the mutant cells formed only a single layer on the surface of the polystyrene, despite having equal numbers in inocula to those of the WT (data not shown). These results indicate that the mub locus is important not only for planktonic growth but also for biofilm formation under aerobic conditions.
FIG. 5.
Effect of mub locus on biofilm formation. WT and mutant cells were grown in CDM containing 0.5% sucrose under aerobic conditions for 24 h. Biofilms were inspected by microscopy after washing off the loosely attached cells. 140, UA140; 159, UA159.
Transcription analysis of the mub gene cluster under normal growth conditions.
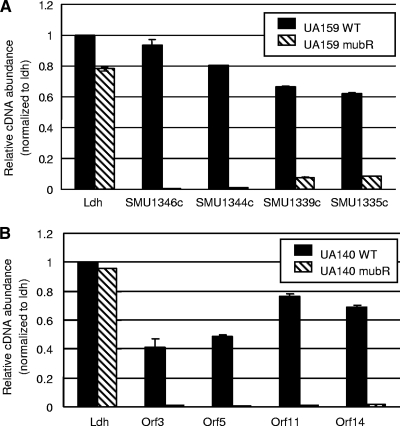
Given the importance of the mub locus in oxygen and H2O2 resistance and in biofilm formation, we hypothesized that the mub genes should be expressed during growth. To test this, we first analyzed a microarray data set generated from a separate study with UA159 cells grown to mid-log phase and found that the signals of the mub genes were comparable to those of numerous highly expressed housekeeping genes (see Table S6 in the supplemental material). To further confirm the microarray results, real-time RT-PCR was used to measure the expression of SMU.1346c, using the constitutively expressed lactate dehydrogenase (ldh) gene as a control. As shown in Fig. 6 A, the expression level of SMU.1346c was as high as that of ldh. Other genes in the locus were also analyzed, and similar results were obtained (data not shown). Compared to that in UA159, the expression level of ORF3 in UA140 was slightly lower, at approximately 40% of the ldh level (Fig. 6B). To obtain transcription profiles for the mub gene clusters, a luciferase reporter was inserted into both UA159 and UA140 by transcriptional fusion of luc with the IGR1065 and P2 promoters, respectively (Fig. 1). Luciferase activity was measured over the course of cell growth. As shown in Fig. 6C and D, expression of these promoters followed the growth curve, with peak expression levels at the late log/early stationary phase. After the early stationary phase, expression levels from both promoters declined dramatically.
FIG. 6.
Level and pattern of mub operon expression. (A and B) Real-time RT-PCR measurements of the expression levels of the first structural genes of the mub operons in UA159 (A) and UA140 (B) relative to the ldh gene expression level. Total RNA was extracted from cells grown anaerobically to late log phase and was used as a template in an RT reaction mix. (C and D) Expression patterns of mub gene operons in UA159 (C) and UA140 (D), as measured by luciferase reporter activity. Closed symbols, growth curve; open symbols, gene expression levels. RLU, relative light units. Experiments were repeated at least three times, with representative results shown.
Identification of transcription regulators of the mub gene cluster.
To further investigate the regulation of mub gene expression, we used functional genomics to identify the transcription regulator(s) of the mub gene cluster. For UA159, two putative transcription regulator genes in the TnSmu2 GI are annotated in the database (SMU.1349 and SMU.1361). To see which gene regulated mub gene expression, both genes were inactivated via an insertional inactivation, and the expression levels of genes located in different regions of the putative mub operon were measured by real-time RT-PCR. While deletion of SMU.1361 did not affect gene expression of the mub locus (data not shown), deletion of SMU.1349 abolished expression of all genes in the mub operon (Fig. 7 A), indicating that SMU.1349 is a major transcription activator of the mub genes in UA159. Therefore, SMU.1349 was renamed mubR to reflect this function (Fig. 1).
FIG. 7.
Expression levels of mub genes in WT and regulatory gene mutant strains of UA159 (A) and UA140 (B), as measured by real-time RT-PCR. The expression level of the ldh gene in the WT strain was arbitrarily assigned as 1.
In contrast to the case for UA159, genes encoding a two-component system exist upstream of the mub gene cluster in UA140 and MT4653 (Fig. 1). To see whether this system regulates mub gene expression, similar mutations to those described above were made in the histidine kinase gene (ORF1). The mutant strains lost their yellow pigment, suggesting that pigment production was abolished in the mutants (data not shown). To further confirm this, real-time RT-PCR was performed on one gene of each putative operon of the mub locus in UA140. As expected, none of these genes showed expression in the ORF1 mutant (Fig. 7B), suggesting that the two-component system activates transcription of all the mub genes in UA140 and MT4653, despite their presence in different putative operons. Based on this result, ORF1 and ORF2 were named mubK and mubR, respectively (Fig. 1).
Distribution of the mub gene cluster among S. mutans clinical isolates.
Given the important role of mutanobactin in oxygen and oxidative stress resistance and biofilm formation of S. mutans, we hypothesized that the mub gene cluster is ubiquitous in the S. mutans population. To test this, we designed three sets of primers, based on the unique sequences of UA159 SMU.1339, UA140 mubA, and MT4653 PKS1 (Table 1; see Table S3 in the supplemental material). PCR amplification of the chromosomal DNA would produce one product (214 bp) from UA159, one product (500 bp) from UA140, and two products (500 bp and 364 bp) from MT4653 (Fig. 8). As a control, a set of primers specific to the 16S rRNA gene of S. mutans was designed, which would generate a 120-bp PCR product from all strains of S. mutans (Fig. 8). When a cocktail was made from all 4 sets of primers and used in a PCR with the three target strains, each expected PCR amplicon was generated (Fig. 8). This primer cocktail was then used in PCRs with clinical isolates as templates. Among the 94 clinical isolates, 18.1% contained the same gene cluster as UA159, 17% contained the same gene cluster as UA140, 3.2% were positive for the MT4653 gene cluster (Table 2), and 61.7% were negative for any of the mub probes, although they were positive for the S. mutans-specific 16S rRNA gene probe (data not shown). Interestingly, all isolates that were PCR positive with the UA140 and MT4653 primers also had yellow colonies on the plate (data not shown). Furthermore, there was no obvious correlation between the presence of the mub gene cluster and patient caries status.
FIG. 8.
Specificity of the PCR primer mixture for detection of the specific mutanobactin genes in UA140, UA159, and MT4653. A band of 500 bp is indicative of the presence of the same gene cluster as that in UA140, a band of 214 bp indicates the presence of the same gene cluster as that in UA159, and two bands, of 500 bp and 364 bp, indicate the presence of the two gene clusters in MT4653. See Materials and Methods for details.
TABLE 2.
Distribution of NRPS-PKS gene clusters among S. mutans clinical isolates from patients with different caries statuses
| Primers and PCR result | No. (%) of isolates with PCR result | No. (%) of isolates with PCR result, by caries status |
Exact significance (1-sided) | |
|---|---|---|---|---|
| CA (n = 54) | CI (n = 40) | |||
| UA159 primers | ||||
| Positive | 17 (18.1) | 11 (20.4) | 6 (18.1) | 0.349 |
| Negative | 77 (81.9) | 43 (79.6) | 34 (81.9) | |
| UA140 primers | ||||
| Positive | 16 (17.0) | 10 (18.5) | 6 (15.0) | 0.436 |
| Negative | 78 (83.0) | 44 (81.5) | 34 (85.0) | |
| MT4653 primers | ||||
| Positive | 3 (3.2) | 1 (1.9) | 2 (5.0) | 0.388 |
| Negative | 91 (96.8) | 53 (98.1) | 38 (95.0) | |
DISCUSSION
In this study, we characterized the cargo genes carried on the TnSmu2 GI of S. mutans UA159 and the similar GIs of strains UA140 and MT4653. Our data suggest that this GI carries genes coding for nonribosomal peptide synthetases, polyketide synthases, and accessory proteins which are responsible for the biosynthesis of the pigment (named mutanobactin) carried by S. mutans. Mutational analysis further demonstrated that this gene cluster (named the mub locus) is involved in oxygen tolerance, H2O2 resistance, and biofilm formation of S. mutans. Furthermore, transcriptional analysis showed that the mub genes are highly expressed during growth. HPLC profiling of cell surface extracts identified unique peaks only in the WT strains, and preliminary characterization of the product from UA159 demonstrated that it is a hybrid NRP/PK compound. Based on these results, we concluded that the mub locus is responsible for the biosynthesis of a hybrid NRP/PK pigment involved in oxygen and oxidative stress tolerance of S. mutans. Inspection of other sequenced genomes of oral microbes did not find similar gene clusters (data not shown), suggesting that S. mutans is so far the only oral bacterium harboring such a gene cluster. This finding has significant implications for understanding the ecology of S. mutans in the oral cavity, as discussed below.
The oral streptococci are anaerobic bacteria but reside in the supragingival plaque (biofilm above the gum line). Yet the oral cavity is an oxygen-rich environment, especially for organisms living above the gum line. In addition, the supragingival plaque is bathed in saliva, which contains H2O2 produced by salivary enzymes and innate immune cells (4). Likewise, the pioneer colonizers, such as the mitis group streptococci, produce copious amounts of H2O2 as a defense mechanism to ward off competitors (4, 22, 23). How do these species cope with oxygen and H2O2 stress? It appears that all oral streptococci have evolved common as well as unique mechanisms. For example, all oral streptococci, including S. mutans, have superoxide dismutase (SOD), two NADH oxidases (Nox), and a Dpr homolog, which help them to grow under aerobic conditions (18). In addition, the mitis group streptococci (e.g., S. gordonii) avoid H2O2 autotoxicity by utilizing Mn2+ in place of Fe2+ as a cofactor of their enzymes and by limiting their dependence on [4Fe-4S] cluster proteins (20). Since H2O2 damage is caused by the generation of hydroxyl radicals through the Fenton reaction, involving the oxidation of Fe2+ to Fe3+, this strategy essentially renders the cell more tolerant to H2O2. S. mutans appears to use both Mn2+ and Fe2+ as cofactors of its SOD (26), and its pyruvate formate-lyase is more sensitive to oxygen than that of S. sanguinis (37). Therefore, alternative strategies must be employed to cope with the oxygen and H2O2 challenges imposed by the host and the competing species of mitis group streptococci. Here we show that in addition to the basic mechanisms shared by all oral streptococci (28, 38-40), the mutanobactin gene cluster carried on the TnSmu2 GI provides another layer of protection against oxygen and H2O2 stress for S. mutans. Since the mutanobactin gene cluster does not exist in other oral streptococci, this unique feature reflects a specific adaptation of S. mutans to the environment created by the pioneer colonizers (the mitis group streptococci) in the oral cavity.
The importance of the mutanobactin gene cluster in niche adaptation of S. mutans is further illustrated by its prevalence in all 6 of the sequenced S. mutans strains (UA159, UA140, MT4653, 19, AF199, and NN2025) and in ∼38% of the clinical isolates we analyzed (Table 2). However, this proportion may be underestimated given the diversity of the cargo genes in the TnSmu2 GI. In fact, in the recently sequenced S. mutans strain NN2025 (27), two gene clusters (regions 24 and 25) encoding NRPS/PKS were found, but none of the genes share significant sequence homology with the cargo genes in TnSmu2 of UA159 or any of the strains we sequenced, indicating that these genes are diversified yet may perform similar functions. Furthermore, according to the report of Maruyama et al. (27), regions 24 and 25 were found in 92.8% and 77.3%, respectively, of the Japanese and Finnish strains they tested. Based on these observations, we predict that as more S. mutans strains are sequenced, more varieties of the mutanobactin gene cluster will be found, and it is highly likely that this gene cluster exists in most or all S. mutans strains. The reason that it was not found in previous studies is probably that the sequences are too diverse to be detected by probes based on the UA159 sequence (36).
In addition to sequence diversity, the location of the mutanobactin gene cluster appears to be divergent as well. In a recent study, Waterhouse and Russell (36) showed that only 6 of 40 S. mutans isolates contained the same TnSmu2 GI as UA159, while 11 did not give a PCR product with primers encompassing the borders of the TnSmu2 locus and the rest gave a 4.5- to 5.5-kb PCR product suggestive of the absence of the TnSmu2 gene cluster. Interestingly, strain MT4653 gave a PCR product of 5.5 kb, yet it contains the same gene cluster as UA140. This suggests that the gene cluster in MT4653 is located elsewhere on the chromosome. In fact, in the contig containing this gene cluster in MT4653, we could find only one flanking gene that was the same as those in UA159. Similarly, strain 19 did not yield a PCR product in Waterhouse and Russell's study, but it has the same gene cluster as UA140, indicating that this strain harbors the TnSmu2 GI at the same location as UA159, although its cargo genes have completely different sequences. Furthermore, in the other sequenced strain, NN2025, the two NRPS-PKS gene clusters are found at different locations from that of TnSmu2 in UA159.
From the gene cluster organization and sequence, we predicted that the TnSmu2 cargo genes are responsible for the biosynthesis of a hybrid NRP/PK pigment that we named mutanobactin. This supposition was supported first by our mutagenesis studies of the individual genes in the NRPS-PKS gene cluster in strains UA140 and MT4653, in which we found that deletion of any individual gene (except for the putative ABC transporter gene) resulted in a white colony phenotype. Second, the HPLC profiles of the wild-type and mutant strains of UA159, UA140, and MT4653 demonstrated that unique peaks were missing for the TnSmu2 cargo gene deletion strains (Fig. 2). In addition, bioinformatic analysis of the NRPS protein sequence predicted that the mutanobactin molecule produced by UA159 would contain 7 amino acids, while those of UA140 and MT4653 would have 6 amino acids. Recently, the amino acid composition and partial structure of the mutanobactin molecule from UA159 were solved and showed an identical amino acid sequence to that predicted (Fig. 1), with a PK moiety attached between the first and second amino acid residues (data not shown). Interestingly, a recent report showed that Streptococcus equi also carries an NRPS gene cluster, on an integrative conjugative element which is responsible for the biosynthesis of a siderophore similar to the yersiniabactin produced by Yersinia species (17).
It is important that based on our results, we renamed the cargo genes of TnSmu2 the mub genes in strains UA159, UA140, and MT4653 (Fig. 1). This nomenclature is based on the function of the pigment (mutanobactin) synthesized by the gene products, whereas the nomenclature in the Los Alamos database is based on sequence homology with known genes. We believe that this homology-based nomenclature could be misleading, especially with regard to the functions of these genes. For example, SMU.1339c, -1340c, and -1342c are annotated as bacitracin synthetase genes. Bacitracin is a nonribosomal peptide antibiotic produced by Bacillus species (14, 32), and S. mutans is relatively more resistant to bacitracin than other streptococci (13, 34, 35). Thus, the existence of genes homologous to bacitracin synthetase genes in S. mutans was assumed to be an indication of bacitracin production by S. mutans (27). The results presented here indicate that the product synthesized by these bacitracin synthetase-like proteins is not bacitracin but, instead, a hybrid NRP/PK pigment. Furthermore, we tested methanol extracts of UA159, UA140, and MT4653 against a panel of oral bacterial species, both Gram positive and Gram negative, and did not detect any antibacterial activity.
Based upon our transcriptional studies of the TnSmu2 locus, we demonstrated that a single regulatory protein, SMU.1349, is required for gene expression of the mub gene cluster in UA159, while the mub operons in UA140, MT4653, and possibly AF199 and 19 are activated by a novel two-component system that is not present in the UA159 genome. These gene clusters are highly expressed in all strains, and even more so in UA159, with levels as high as that of the housekeeping gene ldh (Fig. 6). This high-level gene expression is consistent with their role in oxygen and oxidative stress tolerance; however, unlike that of the other oxidative stress-induced genes, such as sod, nox-1, and dpr (28, 38-40), mub gene cluster expression is not induced by oxygen or H2O2 (C. Wu and F. Qi, unpublished observation). This suggests that either these genes are induced at extremely low oxygen or H2O2 concentrations or they are constitutively expressed during growth as a defensive measure against any sudden changes in oxygen or H2O2 levels.
In summary, we have demonstrated that the TnSmu2 cargo genes are much more prevalent than previously recognized and that their sequence and genomic location are diverse as well. Yet their prominent high-level expression and unique presence in S. mutans suggest that they play a significant role in ensuring survival of S. mutans in the multispecies biofilm environment of the oral cavity.
Supplementary Material
Acknowledgments
We thank Fares Najar for sequencing the S. mutans strains.
This work was supported in part by NIH grants R01-DE014757 and R21-DE017349 to F.Q. and by COBRE grant P20-RR018741-05 to J.M. and J.F.
Footnotes
Published ahead of print on 16 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari, M. Z., G. Yadav, R. S. Gokhale, and D. Mohanty. 2004. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32:W405-W413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashby, M. T., J. Kreth, M. Soundarajan, and L. S. Sivuilu. 2009. Influence of a model human defensive peroxidase system on oral streptococcal antagonism. Microbiology 155:3691-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caufield, P. W., A. P. Dasanayake, Y. Li, Y. Pan, J. Hsu, and J. M. Hardin. 2000. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect. Immun. 68:4018-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 8.Conti, E., T. Stachelhaus, M. A. Marahiel, and P. Brick. 1997. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16:4174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 10.Doran, K. S., E. J. Engelson, A. Khosravi, H. C. Maisey, I. Fedtke, O. Equils, K. S. Michelsen, M. Arditi, A. Peschel, and V. Nizet. 2005. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J. Clin. Invest. 115:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du, L., C. Sanchez, and B. Shen. 2001. Hybrid peptide-polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab. Eng. 3:78-95. [DOI] [PubMed] [Google Scholar]
- 12.Fischbach, M. A., and C. T. Walsh. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106:3468-3496. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez de Annan, S., R. E. Ruiz de Valladares, and I. L. Benito de Cardenas. 1997. Mitis salivarius-bacitracin 10% sacarose agar for oral streptococci and Streptococcus mutans counts. Acta Odontol. Latinoam. 10:47-53. [PubMed] [Google Scholar]
- 14.Haavik, H. I. 1975. Bacitracin production by the neotype; Bacillus licheniformis ATCC 14580. Acta Pathol. Microbiol. Scand. 83(Suppl.):534-540. [DOI] [PubMed] [Google Scholar]
- 15.Hacker, J., B. Hochhut, B. Middendorf, G. Schneider, C. Buchrieser, G. Gottschalk, and U. Dobrindt. 2004. Pathogenomics of mobile genetic elements of toxigenic bacteria. Int. J. Med. Microbiol. 293:453-461. [DOI] [PubMed] [Google Scholar]
- 16.He, X., C. Wu, D. Yarbrough, L. Sim, G. Niu, J. Merritt, W. Shi, and F. Qi. 2008. The cia operon of Streptococcus mutans encodes a unique component required for calcium-mediated autoregulation. Mol. Microbiol. 70:112-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heather, Z., M. T. Holden, K. F. Steward, J. Parkhill, L. Song, G. L. Challis, C. Robinson, N. Davis-Poynter, and A. S. Waller. 2008. A novel streptococcal integrative conjugative element involved in iron acquisition. Mol. Microbiol. 70:1274-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi, M., Y. Yamamoto, and Y. Kamio. 2000. Molecular biology of oxygen tolerance in lactic acid bacteria: functions of NADH oxidases and Dpr in oxidative stress. J. Biosci. Bioeng. 90:484-493. [PubMed] [Google Scholar]
- 19.Hochhut, B., U. Dobrindt, and J. Hacker. 2005. Pathogenicity islands and their role in bacterial virulence and survival. Contrib. Microbiol. 12:234-254. [DOI] [PubMed] [Google Scholar]
- 20.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2002. Oxidative stress tolerance is manganese (Mn2+) regulated in Streptococcus gordonii. Microbiology 148:3255-3263. [DOI] [PubMed] [Google Scholar]
- 21.Kolenbrander, P. E., R. J. Palmer, Jr., A. H. Rickard, N. S. Jakubovics, N. I. Chalmers, and P. I. Diaz. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42:47-79. [DOI] [PubMed] [Google Scholar]
- 22.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187:7193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreth, J., Y. Zhang, and M. C. Herzberg. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J. Bacteriol. 190:4632-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loimaranta, V., J. Tenovuo, L. Koivisto, and M. Karp. 1998. Generation of bioluminescent Streptococcus mutans and its usage in rapid analysis of the efficacy of antimicrobial compounds. Antimicrob. Agents Chemother. 42:1906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, M. E., B. R. Byers, M. O. Olson, M. L. Salin, J. E. Arceneaux, and C. Tolbert. 1986. A Streptococcus mutans superoxide dismutase that is active with either manganese or iron as a cofactor. J. Biol. Chem. 261:9361-9367. [PubMed] [Google Scholar]
- 27.Maruyama, F., M. Kobata, K. Kurokawa, K. Nishida, A. Sakurai, K. Nakano, R. Nomura, S. Kawabata, T. Ooshima, K. Nakai, M. Hattori, S. Hamada, and I. Nakagawa. 2009. Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content. BMC Genomics 10:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama, K. 1992. Nucleotide sequence of Streptococcus mutans superoxide dismutase gene and isolation of insertion mutants. J. Bacteriol. 174:4928-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu, G., T. Okinaga, L. Zhu, J. Banas, F. Qi, and J. Merritt. 2008. Characterization of irvR, a novel regulator of the irvA-dependent pathway required for genetic competence and dextran-dependent aggregation in Streptococcus mutans. J. Bacteriol. 190:7268-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paster, B. J., I. Olsen, J. A. Aas, and F. E. Dewhirst. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 42:80-87. [DOI] [PubMed] [Google Scholar]
- 32.Podlesek, Z., and M. Grabnar. 1987. Genetic mapping of the bacitracin synthetase gene(s) in Bacillus licheniformis. J. Gen. Microbiol. 133:3093-3097. [DOI] [PubMed] [Google Scholar]
- 33.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 34.Tanzer, J. M., A. C. Borjesson, L. Laskowski, A. B. Kurasz, and M. Testa. 1984. Glucose-sucrose-potassium tellurite-bacitracin agar, an alternative to mitis salivarius-bacitracin agar for enumeration of Streptococcus mutans. J. Clin. Microbiol. 20:653-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuda, H., Y. Yamashita, Y. Shibata, Y. Nakano, and T. Koga. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 46:3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterhouse, J. C., and R. R. Russell. 2006. Dispensable genes and foreign DNA in Streptococcus mutans. Microbiology 152:1777-1788. [DOI] [PubMed] [Google Scholar]
- 37.Yamada, T., S. Takahashi-Abbe, and K. Abbe. 1985. Effects of oxygen on pyruvate formate-lyase in situ and sugar metabolism of Streptococcus mutans and Streptococcus sanguis. Infect. Immun. 47:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto, Y., K. Fukui, N. Koujin, H. Ohya, K. Kimura, and Y. Kamio. 2004. Regulation of the intracellular free iron pool by Dpr provides oxygen tolerance to Streptococcus mutans. J. Bacteriol. 186:5997-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto, Y., M. Higuchi, L. B. Poole, and Y. Kamio. 2000. Identification of a new gene responsible for the oxygen tolerance in aerobic life of Streptococcus mutans. Biosci. Biotechnol. Biochem. 64:1106-1109. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto, Y., M. Higuchi, L. B. Poole, and Y. Kamio. 2000. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J. Bacteriol. 182:3740-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.