Abstract
Two pectate lyases were identified from Paenibacillus amylolyticus 27C64; both enzymes demonstrated activity on methylated pectin in addition to polygalacturonic acid. PelA is in a subclass of the pectate lyase family III. PelB shows some features of pectate lyase family I but is highly divergent.
Pectinases have many industrial applications, including uses in food and textile production (9, 12). Additionally, pectinases are important for the degradation of biomass, where pectin can comprise a significant portion of plant structure (5, 6). The degradation of pectin requires methylesterases and depolymerases. Pectin methylesterases are responsible for the hydrolysis of methylester linkages from the polygalacturonic acid (PGA) backbone (24), while pectin depolymerases act upon the polygalacturonate backbone and belong to one of two families, polygalacturonases or lyases. Polygalacturonases hydrolytically cleave the polygalacturonate chain, while lyases cleave by β-elimination, giving a Δ4,5-unsaturated product (10, 19). There are two types of lyases: pectate lyases (PLs), which cleave unesterified polygalacturonate, and pectin lyases, which cleave methylesterified pectin.
Paenibacillus amylolyticus strain 27C64, isolated from the larval hindgut of the aquatic crane fly, Tipula abdominalis, possesses a wide range of lignocellulose-degrading enzymes. This study describes two pectate lyases from P. amylolyticus that display unusual activity by combining traits of pectate and pectin lyases (2, 7, 21, 22).
Identification of pelA and pelB.
A library containing 2- to 5-kb chromosomal fragments of P. amylolyticus strain 27C64 was constructed in Escherichia coli DH5α. Two pectinase-positive clones were identified after screening approximately 6,000 clones on polygalacturonase medium (23) and subcloned for characterization (Table 1). The first clone carried a 2-kb insert with an open reading frame (ORF) of 669 bp (pelA, GenBank accession no. GU289919); a putative ribosomal binding site and promoter was located upstream of the ATG start codon. The deduced protein sequence of the ORF is 222 amino acids and contains an N-terminal region with features of a Bacillus signal peptide, the most likely cleavage site being between amino acids 26 and 27 (17).
TABLE 1.
Cloning strains and plasmids used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristics | Reference or source |
|---|---|---|
| Bacterial strain Escherichia coli DH5α | F−endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG φ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | 16 |
| Plasmids | ||
| pUC19 | lacZα+; Apr | Invitrogen (Carlsbad, CA) |
| pEDH13C2 | pUC19 derivative; P. amylolyticus DNA fragment with pelA | This study |
| pEDH27 | pUC19 derivative; pelA+ | This study |
| pUC19-19F6 | pUC19 derivative; P. amylolyticus DNA fragment with pelB | This study |
| pWEB1 | pUC19 derivative; pelB+ | This study |
| Oligonucleotides | ||
| PLAscF (for pelA) | 5′-GTACAGGGCCCGGATCCTTGACATGATAGAAGCACTCTACTATATTCTAGTGCTTCTACGGTTCTGTGGGACAA-3′ | This study |
| PLAscR (for pelA) | 5′-CGATCAAGCTTGGGCCCGAGCGGCCGCCTCGAGTCCACATGGTTTGGAGCATTTCG-3′ | This study |
| ScpelBF (for pelB) | 5′-GCAGTGAGCTCTTGACATGATAGAAGCACTCTACTATATTCTAGTTATACTTATCGGGAGGAATCG-3′ | This study |
| ScpelBR (for pelB) | 5′-CATGGGATCCCGGAGCGCTTAACTTAGTAACTC-3′ | This study |
The second pectinase-positive clone had a 1.5-kb insert with a single ORF of 1,176 bp (pelB, GenBank accession no. GU289920). Located upstream of pelB was a putative ribosomal binding site and promoter element. The deduced protein sequence of the ORF is 302 amino acids and contains an N-terminal region with features of a Bacillus signal peptide, the most likely cleavage site being between amino acids 30 and 31 (17).
Identification and characterization of PelA.
By protein-protein BLAST (blastp) with the NCBI database (1), PelA exhibited homology to pectate lyases within family III (PL3). PelA was 95% identical to PelA from Paenibacillus barcinonensis (21), 78% identical to Bacillus sp. KSM-P15 pectate lyase (7), 55% identical to Bacillus subtilis PelC (22), 54% identical to Bacillus licheniformis YvpA, and 53% identical to Bacillus sp. P-2850 pectate lyase (see Table S1 in the supplemental material for GenBank accession numbers). Three of four signature blocks of conserved residues for PL3 enzymes (20) are found in PelA, but as in its homologous enzymes, the fourth block of residues is not conserved; it is replaced by another domain not found in other pectate lyases (22) (see Fig. S1). PelA appears to belong to a subgroup of family PL3 enzymes from saprophytic bacteria (22) which includes P. barcinonensis PelA, Bacillus sp. KSM-P15 PL, B. subtilis PelC, B. licheniformis YvpA, and Bacillus sp. P-2850 PL.
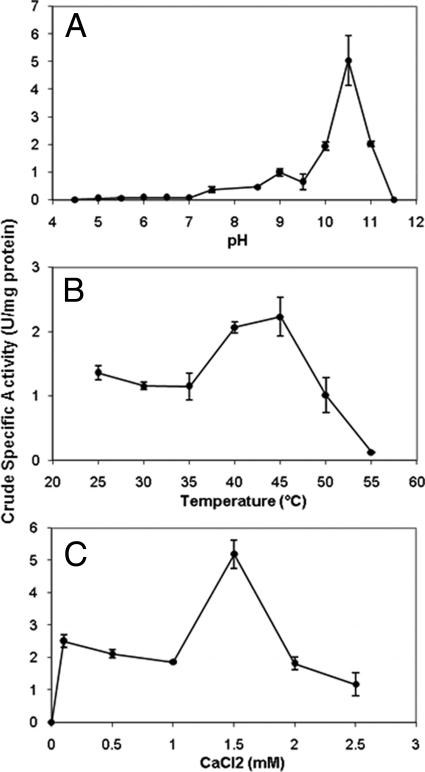
PelA exhibited pectate lyase activity on polygalacturonic acid (PGA) but did not show xylanase or cellulase activity with model substrates and liberation of reducing sugars. Pectate lyase assays were performed as described previously (3, 21) with E. coli DH5α clone cell extracts prepared by sonication, and pH, temperature, and CaCl2 optima were determined (Fig. 1). E. coli DH5α(pUC19) extracts had no detectable pectate lyase activity, and PelA was closely related to characterized proteins; therefore, further studies with purified enzyme were not performed. CaCl2 was necessary for activity, as it is for all known pectate lyases (11).
FIG. 1.
P. amylolyticus strain 27C64 PelA optima for pH (A), temperature (B), and CaCl2 (C). The pH optimum was determined at 40°C with 1 mM CaCl2, the temperature optimum was determined at pH 10.5 in a range of 25 to 55°C, and the CaCl2 concentration optimum was determined at pH 10.5 and 45°C in a range of 0 to 2.5 mM. One unit of enzyme activity was defined as the amount of enzyme that produces 1 μmol Δ4,5-unsaturated product from PGA per minute under assay conditions; specific activity is reported as U/mg protein.
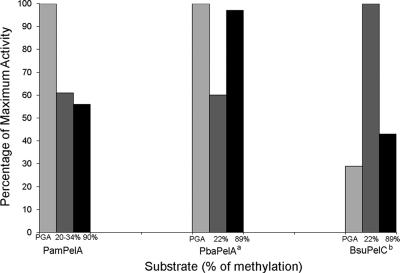
Activity of PelA on citrus pectin was also investigated (Fig. 2), and assays with 20 to 34% and 90% methylesterified citrus pectin demonstrated activity at 61% and 56% of the maximum activity on PGA, respectively (Fig. 2). The high activity of PelA on both PGA and pectins with low and high levels of methylation, although also observed for PelA from P. barcinonensis and PelC from B. subtilis (21, 22), is rare among the pectate lyases described to date.
FIG. 2.
Activity on different pectic substrates for P. amylolyticus strain 27C64 pectate lyase A (PamPelA), P. barcinonensis pectate lyase A (PbaPelA), and B. subtilis pectate lyase C (BsuPelC). a, data from reference 21; b, data from reference 22. P. amylolyticus strain 27C64 PelA, P. barcinonensis PelA, and B. subtilis PelC substrate utilization ranges, with activity on PGA as well as pectin with any degree of methylation, are unique among the pectate lyases described to date.
Identification and characterization of PelB.
PelB showed low homology to family I pectate lyases (PL1) with 28% identity to Bacillus sp. YA-14 PelK (13), B. licheniformis Pel (18), and B. subtilis reference strain 168 Pel (15); 27% identity to Thermotoga maritima PelA (14); and 26% identity to B. subtilis BS-2 Pel and Bacillus amyloliquefaciens TB-2 Pel using protein-protein BLAST (blastp) with the NCBI database (1) (see Table S1 in the supplemental material for GenBank accession numbers). Compared with other pectate lyases, PelB contains all three of the conserved calcium binding sites (25), six of the seven conserved thermostable PL1 sites (Gly47, Val81, Ile83, Leu87, Arg157, and Val214), all of the three conserved catalytic sites, and 9 of 10 invariant residues found in all pectate lyases (see Fig. S2).
Three conserved sequence patterns are also typically contained in pectate lyases: vWiDH, VxxRxPxxRxGxxHxxxxN, and AxDIKGxxxxVTxS. PelB and its closest homologs contain the vWiDH and VxxRxPxxRxGxxHxxxxN regions, while the final conserved sequence, AxDIKGxxxxVTxS, is mostly conserved in PelB.
Four highly conserved consecutive Asn ladder positions which help to stabilize the β bend of the protein structure are found in all PL enzymes. PelB contains only half of the conserved Asn ladders, whereas all of the other enzymes being compared to PelB in this study contain all four of the conserved Asn ladders (8).
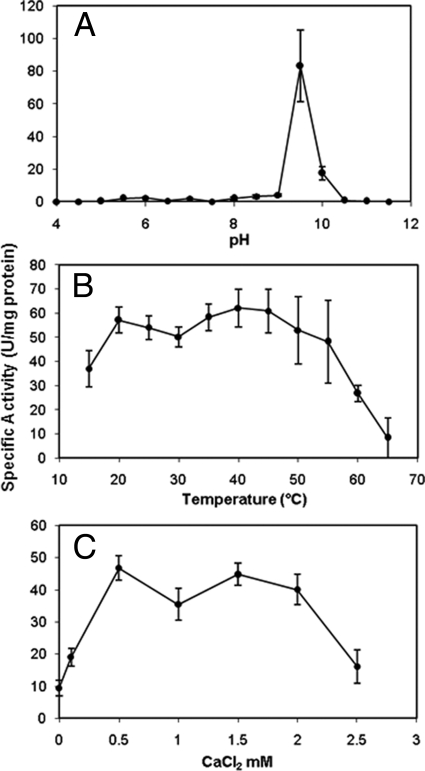
Pectate lyase assays for PelB were performed as with PelA but with E. coli DH5α clone concentrated supernatant. Denaturing SDS-PAGE separation and native zymogram analysis (4) of E. coli DH5α(pWEB1) supernatant with ImageJ analysis showed that PelB was 86% of the supernatant protein, and it was the only band with pectinase activity on zymograms. PelB pH, temperature, and CaCl2 optima were determined (Fig. 3). The enzyme preparation contained 0.115 mM calcium, but dependence on calcium was determined by studies with 0.5 M EDTA, in which no activity was detected (data not shown).
FIG. 3.
P. amylolyticus strain 27C64 PelB optima for pH (A), temperature (B), and CaCl2 (C). The pH optimum was determined at 40°C with 1 mM CaCl2, the temperature optimum was determined at pH 9.5 in a range of 15 to 65°C, and the CaCl2 concentration optimum was determined at pH 9.5 and 55°C in a range of 0 to 2.5 mM.
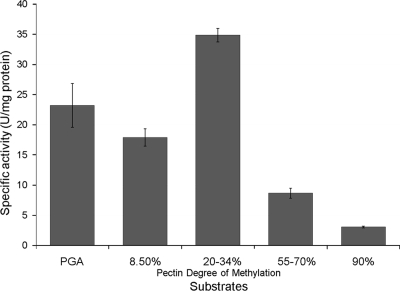
PelB was evaluated for its ability to cleave PGA and methylated pectin by using a range of pectic substrates (Fig. 4). PelB showed the highest activity on 20 to 34% methylated pectin but retained 67%, 51%, 25%, and 1% of its maximum activity on polygalacturonic acid and 8.5%, 55 to 70%, and 90% methylated pectin, respectively.
FIG. 4.
P. amylolyticus strain 27C64 pectate lyase B activity on different pectic substrates. PelB showed the highest activity on 20 to 34% methylated pectin but retained 67%, 51%, 25%, and 1% of its maximum activity on polygalacturonic acid and 8.5%, 55 to 70%, and 90% methylated pectin, respectively, providing evidence that PelB is active on PGA as well as highly methylated pectin.
P. amylolyticus 27C64 PelA and PelB are the first pectate lyases described in P. amylolyticus and show an unusual combination of pectate lyase and pectin lyase activity by degrading both polygalacturonic acid and highly methylated pectin, respectively. A subgroup of PL family III, which now includes PelA, demonstrates this broad substrate specificity; however, enzymes of closest homology or shared structure to PelB do not. While PelB has structural features in line with pectate lyase family I, it is missing some of the conserved amino acid regions and one of the three extracellular pectate lyase superfamily conserved amino acid regions (8, 25). Recently sequenced genomes and metagenomes suggest that enzymes with significant homology to PelB exist. The characterization of these enzymes will determine if they share the substrate specificity of PelB and if they form a new subclass or family of pectate lyases.
Nucleotide sequence accession numbers.
Sequences for pelA and pelB are available in GenBank under accession numbers GU289919 and GU289920, respectively.
Supplementary Material
Acknowledgments
We thank Harry Gilbert (University of Georgia, Athens, GA) and Bernard Henrissat (Architecture et Fonction des Macromolécules Biologiques, Marseille, France) for technical help with enzyme classification.
Footnotes
Published ahead of print on 9 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berensmeier, S., S. A. Singh, J. Meens, and K. Buchholz. 2004. Cloning of the pelA gene from Bacillus licheniformis 14A and biochemical characterization of recombinant, thermostable, high-alkaline pectate lyase. Appl. Biochem. Biotechnol. 64:560-567. [DOI] [PubMed] [Google Scholar]
- 3.Collmer, A., J. L. Ried, and M. S. Mount. 1988. Assay methods for pectic enzymes. Methods Enzymol. 161:329-335. [Google Scholar]
- 4.Coughlan, M. P. 1990. Microbial enzymes and biotechnology II. Elsevier Applied Science, New York, NY.
- 5.Doran, J. B., J. Cripe, M. Sutton, and B. Foster. 2000. Fermentations of pectin-rich biomass with recombinant bacteria to produce fuel ethanol. Appl. Biochem. Biotechnol. 86:141-152. [DOI] [PubMed] [Google Scholar]
- 6.Doran Peterson, J. B. 2003. Pectin-rich biorefinery for production of ethanol and commodity chemicals. Ethanol Producer Mag. 9:26-29. [Google Scholar]
- 7.Hatada, Y., K. Saito, K. Koike, T. Yoshimatsu, T. Ozawa, T. Kobayashi, and S. Ito. 2000. Deduced amino-acid sequence and possible catalytic residues of a novel pectate lyase from an alkaliphilic strain of Bacillus. Eur. J. Biochem. 267:2268-2275. [DOI] [PubMed] [Google Scholar]
- 8.Henrissat, B., S. E. Heffron, M. D. Yoder, S. E. Lietzke, and F. Jurnak. 1995. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 107:963-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoondal, G. S., R. P. Tiwari, R. Tewari, N. Dahiya, and Q. K. Beg. 2002. Microbial alkaline pectinases and their industrial applications: a review. Appl. Microbiol. Biotechnol. 59:409-418. [DOI] [PubMed] [Google Scholar]
- 10.Jayani, R. S., S. Saxena, and R. Gupta. 2005. Microbial pectinolytic enzymes: a review. Process Biochem. 40:2931-2944. [Google Scholar]
- 11.Jurnak, F., N. Kita, M. Garrett, S. E. Heffron, R. Scavetta, C. Boyd, and N. Keen. 1996. Functional implications of the three-dimensional structures of pectate lyases, p. 295-308. In J. Visser and A. G. J. Voragen (ed.), Pectin and pectinases, vol. 14. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 12.Kashyap, D. R., P. K. Vohra, S. Chopra, and R. Tewari. 2001. Applications of pectinases in the commercial sector: a review. Bioresour. Technol. 77:215-227. [DOI] [PubMed] [Google Scholar]
- 13.Kim, J. M., H. K. Park, D. Y. Yum, B. K. Hahm, D. H. Bai, and J. H. Yu. 1994. Nucleotide sequence of the pectate lyase gene from alkali-tolerant Bacillus sp. YA-14. Biosci. Biotechnol. Biochem. 58:947-949. [DOI] [PubMed] [Google Scholar]
- 14.Kluskens, L. D., G.-J. W. M. Van Alebeek, A. G. J. Voragen, W. M. De Vos, and J. Van Der Oost. 2003. Molecular and biochemical characterization of the thermoactive family 1 pectate lyase from the hyperthermophilic bacterium Thermotoga maritima. Biochem. J. 370:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunst, F., et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 16.Meselson, M., and R. Yuan. 1968. DNA restriction enzyme from E. coli. Nature 217:1110-1114. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, J., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 18.Rey, M. W., P. Ramaiya, B. A. Nelson, S. D. Brody-Karpin, E. J. Zaretsky, M. Tang, A. Lopez de Leon, H. Xiang, V. Gusti, I. G. Clausen, P. B. Olsen, M. D. Rasmussen, J. T. Anderson, P. L. Jorgensen, T. S. Larsen, A. Sorokin, A. Bolotin, A. Lapidus, N. Galleron, S. D. Ehrlich, and R. M. Berka. 2004. Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol. 5:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai, T., T. Sakamoto, J. Hallaert, and E. J. Vandamme. 1993. Pectin, pectinase, and protopectinase: production, properties, and applications. Adv. Appl. Microbiol. 39:231-294. [DOI] [PubMed] [Google Scholar]
- 20.Shevchik, V. E., J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1997. Pectate lyase PelI of Erwinia chrysanthemi 3937 belongs to a new family. J. Bacteriol. 179:7321-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soriano, M., A. Blanco, P. Diaz, and F. I. Javier Pastor. 2000. An unusual pectate lyase from a Bacillus sp. with high activity on pectin: cloning and characterization. Microbiology 146:89-95. [DOI] [PubMed] [Google Scholar]
- 22.Soriano, M., P. Diaz, and F. I. Javier Pastor. 2006. Pectate lyase C from Bacillus subtilis: a novel endo-cleaving enzyme with activity on highly methylated pectin. Microbiology 152:617-625. [DOI] [PubMed] [Google Scholar]
- 23.Starr, M. P., A. K. Chatterjee, P. B. Starr, and G. E. Buchanan. 1977. Enzymatic degradation of polygalacturonic acid by Yersinia and Klebsiella species in relation to clinical laboratory procedures. J. Clin. Microbiol. 6:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitaker, J. R. 1984. Pectic substances, pectic enzymes and haze formation in fruit juices. Enzyme Microb. Technol. 6:341-347. [Google Scholar]
- 25.Xiao, Z. H., H. Bergeron, S. Grosse, et al. 2007. Improving the thermostability and activity of a pectate lyase by single amino acid substitutions using a strategy based on Tm-guided sequence alignment. Appl. Environ. Microbiol. 10:1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.