Abstract
Human genetic variation is likely to be responsible for a substantial fraction of the variability in complex traits including drug response. Single nucleotide polymorphisms (SNPs) have been implicated in drug response using genome-wide association studies as well as candidate-gene approaches. A more comprehensive catalogue of human genetic variation should complement the current large-scale genotypic dataset from the International HapMap Project, which focuses on common genetic variants. The 1000 Genomes Project (KGP) is an international research effort that aims to provide the most comprehensive map of human genetic variation using next-generation sequencing platforms. Due to the lack of convenient tools, however, it is a challenge for the pharmacogenetic research community to take advantage of these data. We present here a new database of some pharmacogenes of particular interest to pharmacogenetic researchers. Our database provides a convenient portal for immediate utilization of the newly released KGP data in pharmacogenetic studies.
Keywords: pharmacogenetics, pharmacogene, single nucleotide polymorphism, next generation sequencing, database
Introduction
During the past decade, the concept of personalized medicine has gained increasing popularity among industry and healthcare practitioners. In a new era where personalized medicine becomes routine, doctors would be able to manage a patient’s healthcare based on the individual patient’s specific characteristics including gender, age as well as genetic make-up. Personalized medicine would especially benefit oncology patients, because of serious toxicities (e.g., nephrotoxicity [1]) associated with chemotherapy in the clinic. Similar to common diseases, drug response and toxicity are also likely complex traits, which may be determined by multiple genes and non-genetic factors including drug dosage. In addition to studies focusing on well-characterized candidate genes, some recent genome-wide studies using human lymphoblastoid cell lines (LCLs) also demonstrated that genetic variants such as single nucleotide polymorphisms (SNPs) account for a substantial fraction of the variation in response to anticancer agents of different mechanisms [2].
To successfully identify genetic variants associated with drug response, it is necessary to have a comprehensive catalogue of human genetic variation. The availability of extensive genotypic data (> 3 million SNPs) from the International HapMap Project [3,4] has begun to allow investigators to use the HapMap LCLs as a model for pharmacogenomic discovery [2]. However, the HapMap Project focused largely on cataloguing common genetic variation such as SNPs with a minor allele frequency (MAF) greater than 0.05 in a population [3,4], although some rare variation is, of course, included in the HapMap. Considering the fact that there are more than 10 million SNPs in the human genomes, there are likely to be many more rare (and some additional common) genetic variants not covered in the current HapMap dataset. Furthermore, recent pharmacogenomic studies suggest that common variants covered in the HapMap Project can explain only a fraction (less than 50%) of the variation in drug response [2]. Although other types of genetic variations (e.g., copy number variants, CNVs) and non-genetic factors could be responsible for the remaining variation, the unknown, untyped common or rare variants could help explain the drug response variation. Several large-scale deep resequencing projects such as the SeattleSNPs Project (http://pga.gs.washington.edu) are working to comprehensively catalogue genetic variations in certain candidate genes [5]. In contrast, the 1000 Genomes Project (http://www.1000genomes.org) (KGP) has an ambitious goal to establish a detailed catalogue of human genetic variation in at least 1000 human genomes from world-wide populations [6] using next-generation sequencing technologies [7]. The specified aims of this project are to identify > 95% of the variants with allele frequencies > 1% in parts of the human genome that can be sequenced, as well as to identify > 95% of the variants with allele frequencies > 0.1 - 0.5% in exons [8]. Prospectively, once integrated with other public resources (e.g., the HapMap resource [9]), the KGP data have the potential to greatly benefit pharmacogenetic or pharmacogenomic research using these samples. To take advantage of these new data, we present here a database that is designed for convenient access to the KGP data on some pharmacogenetic candidate genes, the Very Important Pharmacogenes (VIPs) as maintained by the PharmGKB [10] (Pharmacogenetics and Pharmacogenomics Knowledge Base, http://www.PharmGKB.org). Though the KGP provides a web-based browser (http://browser.1000genomes.org) for viewing the four genomes in the high coverage pilot, our database can be used to access 57 additional genomes (61 in total) overlapped with the HapMap CEU (Caucasians from Utah, USA) and YRI (Yoruba people from Ibadan, Nigeria) samples in the low coverage pilot. We also show an example to demonstrate the utility of our database in pharmacogenetic studies.
Database Description
Very Important Pharmacogenes
The VIP project of PharmGKB [10] provides annotated information about genes and variants of particular relevance for pharmacogenetics and pharmacogenomics. The current VIP (accessed on March 2, 2009, http://www.pharmgkb.org/search/annotatedGene/) list is comprised of 39 genes including those encoding transporters (e.g., ABCB1) and CYP450s (e.g., CYP2A6). Detailed pharmacogenetic and/or pharmacogenomic annotations based on the literature are available for these genes.
The KGP Data
The 1000 Genomes Project was launched in January, 2008. The first set of SNP calls representing the preliminary analysis of four genomes of HapMap LCLs (3 samples from a CEU parents-child trio and 1 YRI sample) were recently released (Dec., 2008) as the high coverage (> 20×) pilot. The SNP calls on the CEU trio (father: NA12891; mother: NA12892; child: NA12878) were based on the Illumina platform (mostly paired end 35bp reads). The SNP calls on the YRI sample (NA19240) were detected using the Applied Biosystems SOLiD (Sequencing by Oligo Ligation and Detection) sequencing platform. More recently, sequence data and SNP calls on 603 genomes (March, 2009) in the low coverage pilot were also released. We downloaded KGP data of the currently available 36 CEU samples and 25 YRI samples including FASTQ files (nucleotides and quality assessments), SNP calls, and Binary Simple Alignment / Map files (BAM) from ftp://ftp-trace.ncbi.nih.gov/1000genomes/, as well as FASTA files for the human genome reference assembly from ftp://ftp.ensembl.org/pub/current_fasta/homo_sapiens/dna/.
Database Implementation
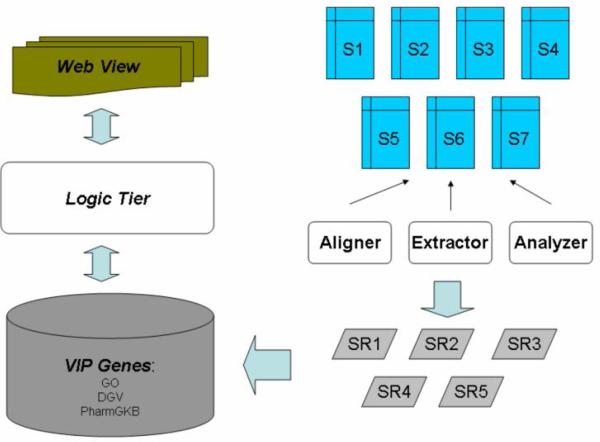
Figure 1 shows the technical architecture of the infrastructure. The Si’s are storage devices for FASTA, BAM, and FASTQ files containing nucleotides and single-byte encoded quality scores. The SRi’s are results data including extracted sequence data, short reads, and (KGP’s and our own) alignments. We wrote our own Extractor (for the FASTQ files) and Analyzer (for summarization), and invoke the tool (SAMtools, http://samtools.sourceforge.net/samtools-c.shtml) for the (binary) Sequence Alignment / Map format (for multiple alignments) used by the Sanger Institute. Summary analysis data, as well as VIP annotations from Gene Ontology (GO) [11], PharmGKB, and the Database of Genomic Variants (DGV) [12], are stored in a relational data store (MySQL). A logic tier handles all executions of queries and separates the execution rules from the other layers, including a web presentation layer.
Fig. 1.
The technical architecture of the infrastructure.
Si’s are storage devices. SRi’s are results data derived from the application of the Aligner, Extractor, and the Analyzer modules. Summary analysis data, as well as VIP annotations from Gene Ontology (GO), PharmGKB, and the Database of Genomic Variants (DGV), are stored in a relational data store. A logic tier handles all executions of queries.
Database Features
The database (http://genemed1.bsd.uchicago.edu/pharmacodb/thougen/) is organized as gene-centric that allows queries for individual genes. The current version covers the following features: 1) Query for gene sequences based on the human genome reference assembly (NCBI build 36); 2) Query for alignment using the reference assembly, the four KGP genomes (high coverage); 3) Query for novel SNPs as well as known HapMap SNPs (genotypes and MAF); 4) Query for PharmGKB annotations as well as other information such as GO terms.
Application of the Database
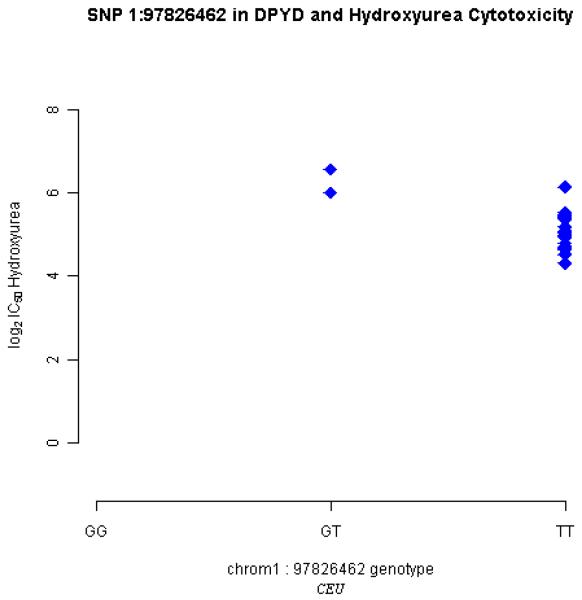
DPYD (dihydropyrimidine dehydrogenase), a pyrimidine catabolic enzyme, could be implicated in the toxicity of hydroxyurea like other DNA antimetabolites [13,14]. We observed significant correlation between the expression of this gene and LCL sensitivity to hydroxyurea (unpublished data). Yet, no associations were identified using the known HapMap SNPs (MAF>5%, HapMap Release 23a). We extracted genotypic data on 96 novel tagging SNPs (r2>0.8, MAF≥10%) in the 36 CEU samples from our database and performed linear regression with drug response data (log2 transformed IC50, concentration required for 50% of cellular growth inhibition) on hydroxyurea (unpublished data). Figure 2 shows an example in which one new association relationship (nominal P=0.001, Padjusted<0.10 after Bonferroni correction) was identified. This example demonstrates the potential utility of our database in pharmacogenetic studies even at the early stage of the KGP, though functional validation may be necessary to evaluate this particular finding.
Fig. 2.
A novel SNP of DPYD associated with the cytotoxicity of hydroxyurea.
SNP Chr1_97826462 (P=0.001, MAF=10%) was associated with hudroxyurea response in the CEU samples. CEU: Caucasians from Utah, USA; IC50: concentration required for 50% of cellular growth inhibition. Standard SNP IDs are not yet available for the novel SNPs. They are currently designated by their genomic positions (NCBI build 36).
Discussion
We developed a new database of some well-characterized pharmacogenes from the PharmGKB [10], which serves the community by developing, implementing, and disseminating a public genotype-phenotype resource focused on pharmacogenetics and pharmacogenomics. To leverage the available resources, the design of our pharmacogene database was compatible to the PharmGKB, therefore, allowing our database to be integrated into the PharmGKB in the future.
Our database of pharmacogenes was designed particularly to meet the challenges faced by researchers in the pharmacogenetic research community to evaluate and utilize the newly released KGP sequence data. For example, our database can be used to extract novel KGP genetic variants of candidate genes to be analyzed in pharmacogenetic studies (Fig. 2). Compared with the default browser provided by the KGP, our database has several advantages. First, our database is focused on the pharmacogenes that are of particular interest to the pharmacogenetic research community. Convenient links to resources such as the PharmGKB website, DGV [12] and GO [11] allow researchers to access important and relevant information on these genes. Second, our database incorporates all available CEU and YRI samples in the KGP, therefore, can be integrated with the HapMap resource [9]. In contrast, the current KGP browser can only be used to view the four genomes in the high coverage pilot. Third, our database will be updated regularly to reflect any new release of the KGP data and updates in the VIP list. Fourth, the current VIP-focused database can be expanded to include whole genome data in the future.
Technically, the current KGP data were generated using the Illumina Genome Analyzer and the Applied Biosystems SOLiD platforms. These platforms differ significantly in terms of cost, sequencing chemistry, amplification approach and performance (e.g., length of reads) [7]. The accuracy of their sequencing reads and associated quality, however, are not yet comprehensively evaluated. Research on technical quality might help the users evaluate the reliability of these data, though some minimal quality assessment has been included in the current release. On the other hand, other novel sequencing technologies such as single-molecule sequencing [15] are becoming available to the research community. Finally, another limitation of the KGP data is that they represent EBV (Epstein Barr Virus)-transformed LCLs. The effect of EBV transformation and cell line culture/passage on the DNA sequences of these samples has not been comprehensively evaluated.
Acknowledgements
This work was funded through the Pharmacogenetics of Anticancer Agents Research Group (http://www.pharmacogenetics.org) by the NIH/NIGMS grant U01GM61393, data deposits are supported by U01GM61374 (http://www.pharmgkb.org) and NIH/NCI Breast SPORE P50 CA125183. The authors are grateful to Drs. John Cunningham and Bala Poonkuzhali for sharing the hydroxyurea phenotype data.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Piccart MJ, Lamb H, Vermorken JB. Current and future potential roles of the platinum drugs in the treatment of ovarian cancer. Ann Oncol. 2001;12:1195–1203. doi: 10.1023/a:1012259625746. [DOI] [PubMed] [Google Scholar]
- 2.Zhang W, Huang RS, Dolan ME. Cell-based models for discovery of pharmacogenomic markers of anticancer agent toxicity. Trends in Cancer Research. 2009;4:1–13. [PMC free article] [PubMed] [Google Scholar]
- 3.The International HapMap Consorium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 4.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Dolan ME. Beyond the HapMap genotypic data: prospects of deep resequencing projects. Curr Bioinformatics. 2008;3:178–182. doi: 10.2174/157489308785909232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuehn BM. 1000 Genomes Project promises closer look at variation in human genome. Jama. 2008;300:2715. doi: 10.1001/jama.2008.823. [DOI] [PubMed] [Google Scholar]
- 7.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 8.The 1000 Genomes Project Meeting Report: A workshop to plan a deep catalog of human genetic variation; 2007; http://www.1000genomes.org. [Google Scholar]
- 9.Zhang W, Ratain MJ, Dolan ME. The HapMap Resource is Providing New Insights into Ourselves and its Application to Pharmacogenomics. Bioinform Biol Insights. 2008;2:15–23. doi: 10.4137/bbi.s455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 11.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 13.Salgado J, Zabalegui N, Gil C, Monreal I, Rodriguez J, Garcia-Foncillas J. Polymorphisms in the thymidylate synthase and dihydropyrimidine dehydrogenase genes predict response and toxicity to capecitabine-raltitrexed in colorectal cancer. Oncol Rep. 2007;17:325–328. [PubMed] [Google Scholar]
- 14.Huang RS, Ratain MJ. Pharmacogenetics and pharmacogenomics of anticancer agents. CA Cancer J Clin. 2009;59:42–55. doi: 10.3322/caac.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]