Abstract
Genomic DNA sequences are an irreplaceable source for reconstructing the vanished past of living organisms. Based on updated sequence data, this paper summarizes our studies on species divergence time, ancient population size and functional loss of genes in the primate lineage leading to modern humans (Homo sapiens sapiens). The inter- and intraspecific comparisons of DNA sequences suggest that the human lineage experienced a rather severe bottleneck in the Middle Pleistocene, throughout which period the subdivided African population played a predominant role in shaping the genetic architecture of modern humans. Also, published and newly identified human-specific pseudogenes (HSPs) are enumerated in order to infer their significance for human evolution. Of the 121 candidate genes obtained, authentic HSPs turn out to comprise only 25 olfactory receptor genes, four T cell receptor genes and nine other genes. The fixation of HSPs has been too rare over the past 6–7 Myr to account for species differences between humans and chimpanzees.
Keywords: primates, modern humans, ancestral polymorphism, pseudogenes
1. Introduction
The last two decades have witnessed explosive advances in molecular evolutionary studies that are based on a large amount of DNA sequence information. Darwin's dream of reconstructing the tree of life has come true and much light has been thrown on the origin of man and its history (Darwin 1859, 1871).
Using all the available DNA sequences as of 2009, we review our genetics studies on primates with special reference to the origin and demographic history of modern humans (Homo sapiens sapiens). Section 2 addresses the species divergence time and ancient population size of six primate species. Two main conclusions are drawn regarding the rather ancient divergences of major primate taxa and rather large ancestral population sizes. Section 3 is concerned with the origin of modern humans. To distinguish between two alternative hypotheses for their origin, we re-examine DNA polymorphism data on 37 loci in the three major ethnic groups. At individual loci, we determine the most ancient type of genes in a sample, the time to the most recent common ancestor (TMRCA), and the place or group in which the most ancient type of genes occurs most frequently (PMRCA).
Section 4 enumerates human-specific pseudogenes (HSPs), in order to understand their role in human evolution and relationships to palaeo-environments. Unexpectedly, authentic HSPs are more limited than presently claimed, thereby bringing into question the functional loss of genes as a major driving force in human evolution. Finally, a short perspective is given on human evolutionary genetics.
2. Primate divergence and demography
Except for the extreme conditions that may be found with endangered species, any bisexual diploid species is almost always genetically polymorphic. The larger the effective population size (Ne), the more ancient the origin of the polymorphism. DNA sequences at a locus chosen from a population are necessarily derived from the most recent common ancestor (MRCA), in the absence of recombination. Owing to randomness in the reproduction process, the time (τ) at which a randomly selected pair of alleles at a locus can be traced back to the MRCA is a random variable. Under selective neutrality (Kimura 1968), the probability distribution of τ is exponential with the average value of 2Ne generations (Kingman 1982). If this species splits into two populations, both must initially inherit more or less the same set of polymorphisms that were present in the ancestral species. As time elapses, the descendant populations gradually differentiate from each other and evolve into new reproductively isolated species. In t years or t/g generations (with a generation time of g years) after the populations split from each other, the extent of the inherited ancestral polymorphism at a given locus decreases and, eventually, only one ancestral gene lineage remains in each descendant species. Of course, this does not necessarily mean that these descendant species are genetically monomorphic, since new mutations continuously accumulate and cause differentiation from the ancestral gene lineage.
For orthologous gene pairs at different loci sampled from two extant species with a divergence time t, we can observe a set of the number (k) of nucleotide differences per site that have accumulated at each locus since the MRCA τg + t years ago. The value of k differs from locus to locus and is governed by the probability laws for the coalescence time τ and the stochastic nature of accumulating nucleotide substitutions in a gene lineage. For a given set of DNA sequence data for a pair of species, we have developed a maximum-likelihood (ML) method to infer t and ancestral Ne (Takahata et al. 1995). In this method, t and Ne are scaled by the nucleotide substitution rate (μ) per year per site such that y = 2tμ stands for the net nucleotide difference between the two extant species and x = 4Negμ stands for the nucleotide diversity in the ancestral species.
Since the ML method was originally based on several simplified assumptions, Yang (1997) extended to the case where the rate of nucleotide substitutions may differ among loci. Yang (2002) and Rannala & Yang (2003) further developed the Markov chain Monte Carlo (MCMC) method for the more general case where more than two extant species are included in a sample and the number of DNA sequences may differ among loci. While the current MCMC method cannot be applied to synonymous sites, it permits us to use other types of DNA sequence data at multiple loci from multiple extant species simultaneously.
The MCMC method was previously applied to 53 intergenic sequence data from four primate species (Chen & Li 2001). The method yielded smaller estimates of Ne (Rannala & Yang 2003) than the ML for synonymous sites (Takahata & Satta 1997; Takahata 2001). The small MCMC estimates may be attributable to the nature of the data because the ML method also gave rather small estimates of ancestral Ne for the same data (Satta et al. 2004). Nonetheless, it is instructive to note the strong dependence of MCMC estimates on the prior distribution. The posterior mean tends to be confined to local areas near a given prior mean if the prior standard deviation (s.d.) is assumed to be small. In the opposite case of a large prior s.d., the posterior mean tends to differ greatly from the prior mean, whereas the posterior s.d. becomes correspondingly large. We tested if the previous result in Rannala & Yang (2003) is robust to the prior distribution. Our tentative conclusion for the MCMC method is that we must assume that the prior s.d. is no smaller than the prior mean.
With this in mind, we used both the ML and MCMC methods to re-examine autosomal DNA sequence data available for six primate species (electronic supplementary material, figure S1). The data include 53 intergenic sequences (Chen & Li 2001) together with additional orthologous sequences from Old and New World monkeys, 17 intron sequences (O'hUigin et al. 2002) and 13 intergenic sequences newly retrieved from databases (electronic supplementary material, table S1). In total, we used 83 loci and to our knowledge, this is the largest dataset to be analysed with the inclusion of six primate species.
There are two exceptionally large datasets—the 58 Mb BAC end sequences (BES) for humans and chimpanzees (Fujiyama et al. 2002) and 18 Mb shotgun sequences for humans, chimpanzees, gorillas and macaques (Patterson et al. 2006). We exclude these datasets from the present analysis, mainly because they were thoroughly examined in Satta et al. (2004) and Burgess & Yang (2008), respectively, and because the number of primate species studied was four at most.
The ML or MCMC method yielded estimates of y (or y/2) and x for six primate species (table 1). It is clear that the ML estimates are more similar to the MCMC estimates for the second set of broader priors than the first set. To convert y and x to t and Ne, we must know the nuisance parameters μ and g. It has long been argued that the nucleotide substitution rate has slowed down in hominoids and that the generation time as a life-history trait has been greatly lengthened in human evolution (Bogin 2009). If we assume that the human and the orangutan diverged 14 Myr ago, μ becomes a little smaller than 10−9. With this estimate, both methods roughly estimate the 30 Myr separation time between hominoids and Old World monkeys and the 45 Myr separation time between hominoids and New World monkeys (cf. Kumar & Hedges 1998; Takahata 2001). However, for the human and chimpanzee divergence to be at least 6 Myr ago, μ in this hominoid dataset must be as small as 0.7 × 10−9, which is consistent with the rate slow-down hypothesis.
Table 1.
The ML and MCMC estimates (%) of y/2 = tμ and x = 4Negμ based on 83 loci sampled from humans (H), chimpanzees (C), gorillas (G), orangutans (O), Old World monkeys (M) and New World monkeys (N). In the MCMC estimates, all species specified by subscripts are used, whereas in the ML estimates, H and the most distantly related species specified by the subscripts are used. See electronic supplementary material, table S1 and figure S1 for detail.
| MCMC1a |
MCMC2a |
||||
|---|---|---|---|---|---|
| ML | prior-1 | posterior-1 | prior-2 | posterior-2 | |
| xHC | 0.35 | 0.10 ± 0.10 | 0.27 ± 0.11 | 1.00 ± 1.00 | 0.43 ± 0.19 |
| xHCG | 0.39 | 0.10 ± 0.10 | 0.38 ± 0.06 | 1.00 ± 1.00 | 0.39 ± 0.06 |
| xHCGO | 0.52 | 0.10 ± 0.10 | 0.24 ± 0.12 | 1.00 ± 1.00 | 0.36 ± 0.10 |
| xHCGOM | 1.03 | 0.10 ± 0.10 | 0.55 ± 0.12 | 1.00 ± 1.00 | 0.74 ± 0.16 |
| xHCGOMN | 2.73 | 0.10 ± 0.10 | 1.54 ± 0.24 | 1.00 ± 1.00 | 2.39 ± 0.40 |
| y/2HC | 0.41 | 0.50 ± 0.11 | 0.45 ± 0.03 | 0.50 ± 0.11 | 0.42 ± 0.04 |
| y/2HCG | 0.53 | 0.66 ± 0.15 | 0.55 ± 0.03 | 0.66 ± 0.15 | 0.55 ± 0.03 |
| y/2HCGO | 1.23 | 1.40 ± 0.37 | 1.40 ± 0.06 | 1.40 ± 0.37 | 1.35 ± 0.05 |
| y/2HCGOM | 2.42 | 3.00 ± 0.60 | 2.65 ± 0.08 | 3.00 ± 0.60 | 2.57 ± 0.09 |
| y/2HCGOMN | 4.00 | 5.00 ± 0.80 | 4.59 ± 0.15 | 5.00 ± 0.80 | 4.35 ± 0.16 |
aTwo sets of prior mean and standard errors are examined.
On the other hand, the estimate of x for the hominoid ancestor is about 0.4 per cent, which is five times larger than the extent of the DNA polymorphism in the extant human population (e.g. Li & Sadler 1991; Yu et al. 2002; Nachman et al. 2004; Zhao et al. 2006). In addition, the generation time in the hominoid ancestor is likely to have been 10 years, suggesting that Ne = 105 rather than 104, as in the extant human population (Takahata 1993; Takahata et al. 1995).
We are concerned about the possibility that our large estimates of x in the case of large y values (table 1) may result from computational problems. By computer simulation with 100 loci, we found that both ML and MCMC methods can recover the assumed values reasonably well even in the case where x is as small as 0.04 per cent and y is as large as 20 per cent. Thus, the large Ne in the early primate ancestor does not appear to be a computational artefact.
3. Modern human demography
After splitting from the chimpanzee lineage 6–7 Myr ago, the human lineage has undergone significant changes in morphology, physiology and behaviour (Leakey 1994). Before the emergence of the genus Homo, a number of hominid speciation events occurred in Africa in the Pliocene. Something unusual took place about 2 Myr ago, around which time Homo erectus migrated from Africa to Southeast Asia. The second Out-of-Africa event took place much later, involving modern humans that had spread over the world by 20 000 years ago. The origin of modern humans has long been debated, particularly with respect to the possibility of interbreeding between the expanding modern humans and the original inhabitants (Cann et al. 1987; Takahata 1993; Wolpoff et al. 2000; Takahata et al. 2001; Klein & Takahata 2002; Satta & Takahata 2002; Templeton 2002).
In our dataset, the present human population is subdivided into three major groups, consisting of Africans (Af), Europeans (Eu) and Asians (As). The Hispanic population sample, genotyped in the National Institute of Environmental Health Sciences (NIEHS), is treated separately, although it can be regarded as an admixture group between Europeans and descendants of Asians (Amerinds). The pattern and extent of DNA polymorphisms differ from one group to another for historical reasons.
Previously, Takahata et al. (2001) examined 10 X-chromosomal loci, five autosomal loci, mitochondrial DNA (mtDNA) and one Y-chromosomal locus (YAP). The TMRCA ranges from about 0.2 Myr for haploid mtDNA and YAP to 1.6 Myr for both X-chromosomal and autosomal loci, whereas the PMRCA is mostly assigned to Africans. Incidentally, TMRCA or the time scale of DNA polymorphism in living human populations encompasses that of the entire history of the genus Homo. DNA polymorphism thus reflects the demographic history of Homo. In particular, PMRCA contains information about relative population sizes or different population structures for the three major groups and the lengths of their histories. If one group has dominated in these respects, it is likely that the PMRCAs for individual loci are unevenly distributed among the groups. However, the sample size or the length of DNA sequences was not sufficiently large at some loci. Subsequently, more DNA polymorphism data have been accumulated, yielding more reliable estimates.
Here, based on the maximum-parsimony method for estimating the MRCA sequence in a human sample with one chimpanzee orthologue, we re-examine the TMRCAs and the PMRCAs at 37 loci with each having a worldwide sample of greater than or equal to 60 chromosomes (table 2). Of these loci, 18 are previously reported and the remaining 19 come from randomly retrieved NIEHS genotype data from which haploid sequences are inferred. The estimated TMRCAs for autosomal and X-linked loci range from 0.3 Myr at PLCG1 to 3.1 Myr at APOE. The average TMRCA at the 31 autosomal loci alone becomes 1.24 Myr, if humans and chimpanzees diverged 6 Myr ago. The extant polymorphisms at most loci in the human population were thus generated in the Pleistocene period. Some exceptions are EDN, CMAH, ASAH1, CD209, APOE and RRM2P4 loci, at which the TMRCA is greater than 2 Myr. Since there are no such loci among the 19 loci derived from NIEHS single nucleotide polymorphism data, there might be some bias towards reporting highly polymorphic loci in the literature. In any event, such a high proportion of six out of 31 autosomal loci (19%) with a TMRCA greater than 2 Myr may indicate a significant demographic change in the human population during the Pleistocene.
Table 2.
TMRCA and PMRCA at 37 genomic regions. The results of the first 10 loci are taken from Takahata et al. (2001) and Satta & Takahata (2004) and those of the next eight loci are taken from Hayakawa et al. (2006), Zhao et al. (2006), Kim & Satta (2008), Patin et al. (2006), Barreiro et al. (2005), Fullerton et al. (2000), Cox et al. (2008) and Yu et al. (2002).
| regions | chromosome | length (bp) | sample size | TMRCA (Myr)a | PMRCAb |
|---|---|---|---|---|---|
| HFE | 6 | 11 214 | 60 | 1.08 | Af |
| HBB | 11 | 2998 | 264 | 1.63 | Af |
| ECP | 14 | 1203 | 108 | 0.51 | Af |
| EDN | 14 | 1214 | 134 | 3.03 | Af |
| MC1R | 16 | 954 | 242 | 0.85 | Af |
| HBA | 16 | 350 | 276 | 1.43 | Af |
| ZFX | X | 1215 | 335 | 1.21 | Af |
| Xq13.3 | X | 10 163 | 69 | 0.67 | Af |
| MAOA | X | 4260 | 146 | 1.43 | Af |
| mtDNA | mt | 610 | 189 | 0.20 | Af |
| CMAH | 6 | 7302 | 132 | 2.90 | Eu |
| 6p22 | 6 | 12 113 | 122 | 0.60 | Af |
| ASAH1 | 8 | 4358 | 60 | 2.40 | Af |
| NAT1 | 8 | 2605 | 160 | 2.01 | As |
| CD209 | 19 | 5500 | 254 | 2.80 | Af |
| APOE | 19 | 5491 | 192 | 3.11 | Af |
| RRM2P4 | X | 5667 | 253 | 2.33 | Af |
| DACH2 | X | 10 346 | 62 | 1.20 | Af |
| ENO1 | 1 | 6165 | 174 | 0.33 | Af |
| MAD2L2 | 1 | 5018 | 172 | 0.59 | Af |
| ODC1 | 2 | 8003 | 174 | 1.00 | Af |
| ATOX1 | 5 | 7546 | 168 | 0.08 | Af |
| MAPK9 | 5 | 6780 | 176 | 0.70 | Af |
| RAD1 | 5 | 7684 | 174 | 1.19 | Af |
| SEPP1 | 5 | 10 108 | 174 | 0.43 | Af |
| VNN3 | 6 | 7684 | 156 | 0.93 | Af |
| MSH5 | 6 | 4745 | 148 | 0.48 | Af |
| PEO1 | 10 | 8598 | 172 | 0.59 | Eu |
| PRDX3 | 10 | 11 316 | 140 | 0.78 | Af |
| CSK | 15 | 8586 | 166 | 0.69 | Hs |
| DUT | 15 | 11 453 | 164 | 1.00 | Af |
| TGFB1I1 | 16 | 6719 | 164 | 0.67 | Af |
| EPX | 17 | 7549 | 166 | 0.47 | Af |
| PLCG1 | 20 | 11 039 | 170 | 0.32 | Af |
| SPO11 | 20 | 11 724 | 150 | 1.01 | Af |
| GABPA | 21 | 5851 | 172 | 1.09 | Af |
| TBX1 | 22 | 4488 | 178 | 0.77 | Af |
aEstimates are either taken from the original papers or made based on the assumption of the 6 Myr divergence time between humans and chimpanzees.
b‘Af’, ‘Eu’, ‘Hs’, and ‘As’ stand for Africans, Europeans, Hispanics, and Asians, respectively. For instance, ‘Af’ indicates that the ancestral haplotype is most frequent in Africans.
In fact, since the average TMRCA is roughly equal to 4Neg years under neutrality, Ne becomes 1.55 × 104 from the observed average TMRCA = 1.24 Myr and g = 20. There are also other statistics for estimating Ne from polymorphism data. One is the number (s) of segregating sites per site (Watterson 1975). With the average s value being 0.11 per cent in our sample, we can estimate Ne as 1.40 × 104 from Watterson's formula and the assumption of μ = 10−9 per site per year. Thus, Ne becomes about 1.5 × 104 in both estimates. If μ is as small as 0.7 × 10−9 as mentioned earlier, the Ne values become correspondingly large. These estimates of Ne are at least 1.5 times greater than the previous estimate of 104 (Takahata 1993) but smaller than 105 for the common ancestral population of humans and chimpanzees as mentioned in §2.
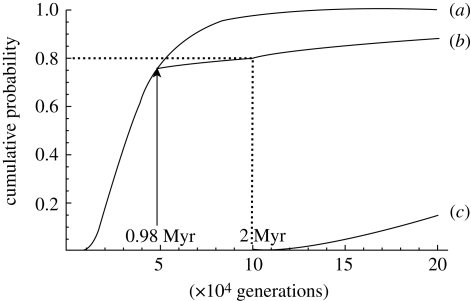
One of us suggested a one-order reduction in population size during the Pleistocene or a Pleistocene bottleneck in human evolution (Takahata 1993). Actually, under a demographic model of a constant Ne = 104, the probability that 60 genes sampled for a locus coalesce to the MRCA within the past 2 Myr or 105 generations is as high as 0.98 (Takahata & Nei 1985). On the other hand, if Ne = 105, the same probability becomes as small as 0.004. For simplicity, we assume a sudden Pleistocene bottleneck model with Ne = 105 before tb years and Ne = 104 after tb years. We then determine the most likely value of tb, for TMRCA greater than 2 Myr to occur among 19 per cent of the loci. The tb value thus estimated is 0.98 Myr (figure 1) and suggests that the bottleneck occurred during the Middle Pleistocene. The subsequent population expansion in the Upper Pleistocene and Holocene is too recent to alter the conclusion in any significant way.
Figure 1.
The probability of TMRCA smaller than t for a sample of 60 DNA sequences at a locus; see eqn (7) in Takahata & Nei (1985) and note that the exponent in eqn (7b) should contain a minus sign. The curves (a), (b) and (c) represent the case of Ne = 104 throughout, the case of Ne = 104 for t < tb and 105 for t ≥ tb where tb = 0.98 Myr, and the case of Ne = 105 throughout, respectively. The generation time is assumed to be 20 years.
The PMRCA analysis indicates that, in 33 of the 37 cases (89%), Africans possess the most ancient type of genes, whereas non-Africans generally possess derived types of genes. Africans have thus maintained about eight times more distinct gene lineages than non-Africans. This PMRCA or lineage asymmetry may be attributed to an extremely large effective size or a more subdivided population structure of Africans relative to non-Africans. However, since it is unrealistic to assume that the effective size of the entire non-Africans was as small as 103, the African subdivision hypothesis is more likely. In this scenario, a necessary condition is the existence of some African subpopulations that have not directly exchanged migrants with non-Africans (Satta & Takahata 2002, 2004) and that could retain ancestral types of genes. It appears that no comparable subpopulation structure has existed in Eurasia, even though H. erectus occupied the area and inherited correspondingly ancient types of genes.
Modern human descendants migrating out of Africa might have encountered and interbred with former H. erectus inhabitants. Our PMRCA analysis suggests that genes that were maintained in Africa and that spread over Eurasia have by and large swamped those genes that were inherited by descendants of H. erectus. There is little or no strong genetic signal for multi-regional origins of modern humans (Wolpoff et al. 2000).
4. Functional loss of genes
Olson (1999) argued that functional loss of genes can frequently occur by means of numerous molecular causes and proposed the less-is-more hypothesis. The hypothesis is based on the observation that a large fraction of genetic functions of a genome are dispensable and on the speculation that selection may permit emergence of a less complete genome. Likewise, one of us (Takahata 1999) emphasized that dispensability of genes should be taken as evidence for relationships between the gene function and the physical and biological environments. One good example of such a non-functional gene is the gulonolactone oxidase (GLO) gene in primates, whose diet contains sufficient amounts of vitamin C. Given this improved diet, functional loss of the gene is less costly or even more beneficial than biosynthesis of vitamin C from γ-gulonolactone (Linster & Van Schaftingen 2007).
Genes often die, but whether or not such dead genes or pseudogenes can be fixed in a population in the context of the selectively relevant environment is a completely different matter. Conversely, it is possible to understand the biological implications of functional loss of genes in relation to palaeo-environments under which the pseudogenes arose and were evolutionarily accepted.
Examining the human and chimpanzee genomes in silico, Wang et al. (2006), Hahn & Lee (2006) and Hahn et al. (2007), the International Human Genome Sequencing Consortium (IHGSC 2001) and others (e.g. Torrents et al. 2003; Go & Niimura 2008) collectively enumerated more than 120 ‘HSPs’. Of these, 14 pseudogenes are polymorphic and the remaining ones have supposedly been fixed in the human population (table 3 and electronic supplementary material, table S2). However, since only the human and chimpanzee genomes were examined in most in silico studies, HSPs simply imply that they are disrupted by mutations in the human, but not in the chimpanzee. It is possible that some of these pseudogenes are also non-functional in other primates. In addition, these HSPs may include processed pseudogenes, truly functional genes that are misclassified as pseudogenes or pseudogenes without functional orthologues in non-human primates. We exclude all of these as HSP candidates.
Table 3.
Examination of human specific pseudogenes (HSPs). (Criteria: a, the presence of closely related paralogues with sequence divergences of less than 10%; b, the presence of pseudogenes in non-human Catarrhini; c, processed pseudogenes; d, misclassified as pseudogenes; e, these pseudogenes are actually absent in the genome of either humans or non-human Catarrhini.)
| criteria2 |
|||||||
|---|---|---|---|---|---|---|---|
| fixed candidates | no.1 | a | b | c | d | e | no. of HSPs |
| T-cell receptor genes | 4 | 0 | 0 | 0 | 0 | 0 | 4 |
| olfactory receptor genes | 53 | 10 | 5 | 0 | 1 | 12 | 25 |
| taste receptor genes | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| other genes | 48 | 9 | 18 | 5 | 6 | 7 | 93 |
| subtotal | 107 | 20 | 24 | 5 | 7 | 19 | 38 |
| polymorphic candidates | 14 | 4 | 1 | 2 | 0 | 1 | 8 |
| total | 121 | 24 | 25 | 7 | 7 | 21 | 46 |
1The number of HSP candidates thus far identified.
2The five criteria (a to e) for the exclusion as HSPs are not mutually exclusive and there are six genes that are excluded by two different criteria.
3The nine HSPs are CMAH (Hayakawa et al. 2006), GLRA4 (IHGSC 2001), MBL1 (Wang et al. 2006), MYH16 (Stedman et al. 2004), ZNF850 (Wang et al. 2006), S100A15 (Hahn et al. 2007), SIGLEC13 (Angata et al. 2004), TDH (Edgar 2002), and KRT41 (Winter et al. 2001). See electronic supplementary material, table S2 for detail.
Perhaps more importantly, many HSPs identified thus far belong to multi-gene families. If there exist any closely related copies (paralogues) of a given pseudogene in the human genome, the functional loss of a copy is likely to be selectively neutral and to have nothing to do with the environment. To exclude this case too, we set an operational cut-off value of nucleotide substitutions kc between a candidate pseudogene and a functional paralogue. Namely, wherever there exists a closely related functional paralogue with kc ≤ 0.1 in the human genome, we exclude such a trivial pseudogene from the HSPs considered in our study.
The application of the above criteria to 107 fixed pseudogenes has left only 25 olfactory receptor (OR) pseudogenes and 13 other pseudogenes (table 3). The latter group of pseudogenes comprises four T cell receptors (TCR), CMAH, GLRA4, MBL1, MHY16, SIGLEC-13, TDH, KRT41 and two other less characterized genes. An immediate consequence is that the number of fixed HSPs is much smaller than previously claimed. This substantial reduction results, in part, from the inaccurate/incomplete genome database in non-human primates or the presence of closely related duplicated genes in the human genome or both and, in part, from the absence of orthologues in non-human primate genomes.
From the observation that the total 38 pseudogenes have been fixed in the human population, the overall fixation rate is 5–6 per genome per millon years or 2.2 × 10−10 per locus per year if the human genome contains 25 000 loci. We note, however, that the fixation rate differs considerably from one gene family to another. Large multi-gene families such as OR and TCR appear to have evolved with high rates. On the other hand, even apparently unique genes such as CMAH and TDH have also lost their functions. We tried to date the functional loss of seven unique genes to the exclusion of ZNF850P with a highly repetitive motif as well as SIGLEC13 that is completely deleted from the human genome (Angata et al. 2004). Of particular interest are the functional losses of GLRA4 and TDH, which occurred in this order, since both are involved in glycine metabolism or glycine transmittance and glycine acts as a neuro-transmitter in the mammalian central nervous system.
Because the number of authentic HSPs is discouragingly small, the interspecies differences between humans and chimpanzees cannot be entirely attributed to the functional loss of genes. In this respect, we have compared gene expression profiles in the skin of humans and chimpanzees and found that there are about 180 gene loci at each of which the human skin expresses greater than 100 times more transcripts than the chimpanzee skin or vice versa (data not shown). Although our experiment with microarray analyses are not exhaustive for other tissues and organs, we are inclined to agree with the supposition of Zuckerkandl & Pauling (1965), who proposed, ‘many phenotypic differences may be the result of changes in the patterns of timing and rate of activity of structural genes rather than of changes in functional properties of the polypeptides as a result of changes in amino-acid sequence.’ Functional loss of genes is certainly one extreme case of regulatory changes, but some other changes at the expression level appear to have played more important roles in human evolution.
5. Perspectives
When we initiated our studies reviewed in this article, only a limited number of pertinent DNA sequences were available. This situation has changed dramatically during the last two decades, followed by various innovations in theoretical and computational methods. Furthermore, genome-wide comparisons in large samples within and among species will soon offer new insights into significant evolutionary problems. One hundred and fifty years ago, Darwin (1859) eloquently concluded in The Origin:
Thus, from the war of nature, from famine and death, the most exalted object which we are capable of conceiving, namely, the production of the higher animals directly follows. There is grandeur in this view of life, with its several powers, having been originally breathed by the Creator into a few forms or into one; and that, whilst this planet has gone cycling on according to the fixed gravity, from so simple a beginning endless forms most beautiful and most wonderful have been and are being evolved. (Darwin 1859, p. 459)
To us, this ending is echoed in Brenner's (1991) remark that ‘because we have no direct access to the processes of evolution and can only study its contemporary products and relics of the past, it is here that the creative imagination plays an important role in the scientific endeavour.’ However, at the deepest level of the contemporary products, we have abundant informational relics at hand that surely would substantiate Darwin's thesis.
Acknowledgements
We thank B. Charlesworth for his helpful comments on the manuscript and the Royal Society for hospitality.
Footnotes
Present address: Institute of Molecular Evolutionary Genetics and Department of Biology, Pennsylvania State University, 311 Mueller Laboratory, University Park, PA 16802, USA.
Present address: Institute for Amphibian Biology, Graduate School of Science, Hiroshima University, Higashihiroshima 739-8526, Japan.
One contribution of 18 to a Discussion Meeting Issue ‘Genetics and the causes of evolution: 150 years of progress since Darwin’.
The first two authors contributed equally to the study.
References
- Angata T., Margulies E., Green E., Varki A.2004Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. Proc. Natl Acad. Sci. USA 101, 13 251–13 256 (doi:10.1073/pnas.0404833101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro L. B., Patin E., Neyrolles O., Cann H. M., Gicquel B., Quintana-Murci L.2005The heritage of pathogen pressures and ancient demography in the human innate-immunity CD209/CD209L region. Am. J. Hum. Genet. 77, 869–886 (doi:10.1086/497613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogin B.2009Childhood, adolescence, and longevity: a multilevel model of the evolution of reserve capacity in human life history. Am. J. Hum. Biol. 21, 567–577 (doi:10.1002/ajhb.20895) [DOI] [PubMed] [Google Scholar]
- Brenner S.1991Summary and concluding remarks. In Evolution of life (eds Osawa S., Honjo S. T.), pp. 453–456 Tokyo, Japan: Springer [Google Scholar]
- Burgess R., Yang Z.2008Estimation of hominoid ancestral population sizes under Bayesian coalescent models incorporating mutation rate variation and sequence errors. Mol. Biol. Evol. 25, 1979–1994 (doi:10.1093/molbev/msn148) [DOI] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C.1987Mitochondrial DNA and human evolution. Nature 325, 31–36 (doi:10.1038/325031a0) [DOI] [PubMed] [Google Scholar]
- Chen F.-C., Li H.2001Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68, 444–456 (doi:10.1086/318206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. P., Mendez F. L., Karafet T. M., Pilkington M. M., Kingan S. B., Destro-Bisol G., Strassmann B. I., Hammer M. F.2008Testing for archaic hominin admixture on the X chromosome: model likelihoods for the modern human RRM2P4 region from summaries of genealogical topology under the structured coalescent. Genetics 178, 427–437 (doi:10.1534/genetics.107.080432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C.1859The origin of species by means of natural selection. London, UK: John Murray [Google Scholar]
- Darwin C.1871The descent of man, and selection in relation to sex. London, UK: John Murray [Google Scholar]
- Edgar A.2002The human L-threonine 3-dehydrogenase gene is an expressed pseudogene. BMC Genet. 3, 18 (doi:10.1186/1471-2156-3-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama A., et al. 2002Construction and analysis of a human–chimpanzee comparative clone map. Science 295, 131–134 (doi:10.1126/science.1065199) [DOI] [PubMed] [Google Scholar]
- Fullerton S. M., et al. 2000Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am. J. Hum. Genet 67, 881–900 (doi:10.1086/303070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y., Niimura Y.2008Similar numbers but different repertoires of olfactory receptor genes in humans and chimpanzees. Mol. Biol. Evol. 25, 1897–1907 (doi:10.1093/molbev/msn135) [DOI] [PubMed] [Google Scholar]
- Hahn Y., Lee B.2006Human-specific nonsense mutations identified by genome sequence comparisons. Hum. Genet. 119, 169–178 (doi:10.1007/s00439-005-0125-6) [DOI] [PubMed] [Google Scholar]
- Hahn Y., Jeong S., Lee B.2007Inactivation of MOXD2 and S100A15A by exon deletion during human evolution. Mol. Biol. Evol. 24, 2203–2212 (doi:10.1093/molbev/msm146) [DOI] [PubMed] [Google Scholar]
- Hayakawa T., Aki I., Varki A., Satta Y., Takahata N.2006Fixation of the human-specific CMP-N-acetylneuraminic acid hydroxylase pseudogene and implications of haplotype diversity for human evolution. Genetics 172, 1139–1146 (doi:10.1534/genetics.105.046995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- IHGSC (International Human Genome Sequencing Consortium) 2001Initial sequencing and analysis of the human genome. Nature 409, 860–921 (doi:10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- Kim H. L., Satta Y.2008Population genetic analysis of the N-acylsphingosine amidohydrolase gene associated with mental activity in humans. Genetics 178, 1505–1515 (doi:10.1534/genetics.107.083691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M.1968Evolutionary rate at the molecular level. Nature 217, 624–626 (doi:10.1038/217624a0) [DOI] [PubMed] [Google Scholar]
- Kingman J. F. C.1982On the genealogy in large populations. J. Appl. Prob. 19A, 27–43 [Google Scholar]
- Klein J., Takahata N.2002Where do we come from? The molecular evidence for human descent. Berlin, Germany: Springer [Google Scholar]
- Kumar S., Hedges S. B.1998A molecular timescale for vertebrate evolution. Nature 392, 917–919 (doi:10.1038/31927) [DOI] [PubMed] [Google Scholar]
- Leakey R.1994The origin of humankind. New York, NY: BasicBooks [Google Scholar]
- Li W.-H., Sadler L. A.1991Low nucleotide diversity in man. Genetics 129, 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster C. L., Van Schaftingen E.2007Vitamin C: biosynthesis, recycling and degradation in mammals. FEBS J. 274, 1–22 [DOI] [PubMed] [Google Scholar]
- Nachman M. W., D'Agostino S. L., Tillquist C. R., Mobasher Z., Hammer M. F.2004Nucleotide variation at Msn and Alas2, two genes flanking the centromere of the X chromosome in humans. Genetics 167, 423–437 (doi:10.1534/genetics.167.1.423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'hUigin C., Satta Y., Takahata N., Klein J.2002Contribution of homoplasy and of ancestral polymorphism to the evolution of genes in anthropoid primates. Mol. Biol. Evol. 19, 1501–1513 [DOI] [PubMed] [Google Scholar]
- Olson M. V.1999When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64, 18–23 (doi:10.1086/302219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patin E., et al. 2006Deciphering the ancient and complex evolutionary history of human arylamine N-acetyltransferase genes. Am. J. Hum. Genet. 78, 423–436 (doi:10.1086/500614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Richter D. J., Gnerre S., Lander E. S., Reich D.2006Genetic evidence for complex speciation of humans and chimpanzees. Nature 441, 1103–1108 (doi:10.1038/nature04789) [DOI] [PubMed] [Google Scholar]
- Rannala B., Yang Z.2003Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164, 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satta Y., Takahata N.2002Out of Africa with regional interbreeding? Modern human origins. BioEssays 24, 871–875 (doi:10.1002/bies.10166) [DOI] [PubMed] [Google Scholar]
- Satta Y., Takahata N.2004The distribution of the ancestral haplotype in finite stepping-stone models with population expansion. Mol. Ecol. 13, 877–886 (doi:10.1046/j.1365-294X.2003.02069.x) [DOI] [PubMed] [Google Scholar]
- Satta Y., Hickerson M., Watanabe H., O'hUigin C., Klein J.2004Ancestral population sizes and species divergence times in the primate lineage on the basis of intron and BAC end sequences. J. Mol. Evol. 59, 478–487 (doi:10.1007/s00239-004-2639-2) [DOI] [PubMed] [Google Scholar]
- Stedman H. H., et al. 2004Myosin gene mutation correlates with anatomical changes in the human lineage. Nature 428, 415–418 (doi:10.1038/nature02358) [DOI] [PubMed] [Google Scholar]
- Takahata N.1993Allelic genealogy and human evolution. Mol. Biol. Evol. 10, 2–22 [DOI] [PubMed] [Google Scholar]
- Takahata N.1999Genetic understanding of mutually sustaining biodiversities (in Japanese). AERA Mook (Asahi Shimbun) 54, 166–175 [Google Scholar]
- Takahata N.2001Molecular phylogeny and demographic history of humans. In Humanity from African naissance to coming millennia (eds Tobias P. V., Raath M. A., Moggi-Cecchi J., Doyle G. A.), pp. 299–305 Johannesburg, South Africa: Firenze University Press [Google Scholar]
- Takahata N., Nei M.1985Gene genealogy and variance of interpopulational nucleotide differences. Genetics 110, 325–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Satta Y.1997Evolution of the primate lineage leading to modern humans: phylogenetic and demographic inferences from DNA sequences. Proc. Natl Acad. Sci. USA 94, 4811–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Satta Y., Klein J.1995Divergence time and population size in the lineage leading to modern humans. Theory Popul. Biol. 48, 198–221 [DOI] [PubMed] [Google Scholar]
- Takahata N., Lee S.-H., Satta Y.2001Testing multiregionality of modern human origins. Mol. Biol. Evol. 18, 172–183 [DOI] [PubMed] [Google Scholar]
- Templeton A.2002Out of Africa again and again. Nature 416, 45–51 (doi:10.1038/416045a) [DOI] [PubMed] [Google Scholar]
- Torrents D., Suyama M., Zdobnov E., Bork P.2003A genome-wide survey of human pseudogenes. Genome Res. 13, 2559–2567 (doi:10.1101/gr.1455503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Grus W. E., Zhang J.2006Gene losses during human origins. PLoS Biol. 4, 366–377 (doi:10.1371/journal.pbio.0040052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A.1975On the number of segregating sites genetical models without recombination. Theory Popul. Biol. 7, 256–276 [DOI] [PubMed] [Google Scholar]
- Winter H., Langbein L., Krawczak M., Cooper D. N., Jave-Suarez L. F., Rogers M. A., Praetzel S., Heidt P. J., Schweizer J.2001Human type I hair keratin pseudogene phihHaA has functional orthologs in the chimpanzee and gorilla: evidence for recent inactivation of the human gene after the Pan-Homo divergence. Hum. Genet. 108, 37–42 (doi:10.1007/s004390000439) [DOI] [PubMed] [Google Scholar]
- Wolpoff M. H., Hawks J., Caspari R.2000Multiregional, not multiple origins. Am. J. Phys. Anthropol. 112, 129–136 [DOI] [PubMed] [Google Scholar]
- Yang Z.1997On the estimation of ancestral population sizes of modern humans. Genet. Res. 69, 111–116 [DOI] [PubMed] [Google Scholar]
- Yang Z.2002Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics 162, 1811–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Fu Y.-X., Li H.2002DNA polymorphism in a worldwide sample of human X chromosomes. Mol. Biol. Evol. 19, 2131–2141 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Yu N., Fu Y.-X., Li H.2006Nucleotide variation and haplotype diversity in a 10-kb noncoding region in three continental human populations. Genetics 174, 399–409 (doi:10.1534/genetics.106.060301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L.1965Evolutionary divergence and convergence in proteins. In Evolving genes and proteins (eds Bryson V., Vogel H.), pp. 97–166 New York, NY: Academic Press [Google Scholar]