Abstract
The regulation of gene expression is an important determinant of organismal phenotype and evolution. However, the widespread recognition of this fact occurred long after the synthesis of evolution and genetics. Here, we give a brief sketch of thoughts regarding gene regulation in the history of evolution and genetics. We then review the development of genome-wide studies of gene regulatory variation in the context of the location and mode of action of the causative genetic changes. In particular, we review mapping of the genetic basis of expression variation through expression quantitative trait locus studies and measuring the cis/trans component of expression variation in allele-specific expression studies. We conclude by proposing a systematic integration of ideas that combines global mapping studies, cis/trans tests and modern population genetics methodologies, in order to directly estimate the forces acting on regulatory variation within and between species.
Keywords: gene regulation, expression evolution, allele-specific expression, expression QTL, cis-regulation, trans-regulation
1. Introduction
Deciphering the causes and consequences of phenotypic variation is at the heart of understanding evolution and natural selection. Although biologists today acknowledge the importance of gene regulation in the evolution of form as spectacularly evidenced through the study of development (Carroll 2008), the relationship between genetics and development was at one time a yawning gulf. The rise of the authority of Mendelian genetics ushered in by T.H. Morgan led to a view where genes were treated as trait-causing elements linearly arrayed on chromosomes (Sapp 1987). Many workers, especially embryologists, recognized that this explanation for phenotypes had to be incomplete, as the exquisite differentiation of tissues in the genetically identical cells of developing embryos attested. This perspective derived in large part from the inability to reconcile two observations: (i) that genes in some important sense influenced traits and (ii) that genetically identical cells in different tissues produced vastly different cellular phenotypes. This ‘paradox of nuclear equivalence’ (Sapp 1987; Comfort 1999) led many embryologists to conclude that the cytoplasm (as opposed to the chromosome-containing nucleus) housed the crucial elements leading to embryological differentiation. This emphasis on cytoplasmic factors led to a conflict between embryology and the burgeoning field of genetics, which focused on the nucleus. Morgan (1926) addressed this issue directly:
The confusion that is met with sometimes in the literature has resulted from a failure to keep apart the phenomenon of heredity, which deals with the transmission of the heredity units, and the phenomena of embryonic development that take place almost exclusively by changes in the cytoplasm.
While we now recognize that such conflicts are unnecessary for our understanding of genetics and embryology, this was not appreciated at the time. As a result, embryology, a discipline that is profoundly related to gene expression regulation, was divested of much of its authority in its apparent conflict with the ascendant discipline of genetics, quite possibly delaying work on gene regulation. Morgan's role in this conflict is interesting in its own right, as he was trained as an embryologist and originally subscribed to the notion that regulating factors from the cytoplasm played an important role in the development of embryological phenotypes, a perspective he defended vigorously in the 1890s (Gilbert 1987). His perspective was influenced strongly by his relationship with E.B. Wilson and his work on Drosophila, eventually leading to Morgan's position as the foremost proponent of the chromosomal theory of inheritance (Gilbert 1987; Sapp 1987).
This apparent dichotomy between genetics and development was a source of tension among biologists who knew that a synthesis between these concepts and evolution would have to come at some point. It was in tackling this problem that regulation of gene expression was first adduced to explain development empirically (Comfort 2001b). One way of resolving this apparent paradox was positing that the action of genes was in some way controlled by other elements such that the action of genes could be altered in time or space. Indeed, Haldane had argued for the importance of the ‘time of action of genes’ as early as 1932 (Haldane 1932), and Wright reviewed many genetically based head abnormalities in guinea pigs, in part arguing that many hereditary factors were clearly controlled at particular stages of development and/or regulated general features of growth at particular stages of development, as early as 1934 (Wright 1934). McClintock's work in maize genetics on the Ac/Ds (activator/dissociator) system and Spm (suppressor mutator) systems adduced genetic evidence in favour of just such a set of concepts (McClintock 1956, 1961; Comfort 1999, 2001a,b). While McClintock's contribution is now usually viewed as synonymous with dissecting the phenomenon of transposable elements, it has been cogently argued both by herself and by historians of science that she originally viewed her own work as the first steps in a solution to the gene regulation problem (McClintock 1956, 1961; Comfort 1999, 2001b). Interestingly, McClintock's investigation of ‘controlling elements’ in maize demonstrated the most significant hallmarks of modern gene regulatory control, including analogues to cis-regulatory elements and trans-activating factors that could be either tightly linked, or unlinked altogether (McClintock 1961), though this was not widely recognized by her peers at the time (Comfort 1999, 2001b).
The first example of a gene regulatory system to gain wide acceptance was the lac operon in Escherichia coli proposed by Jacob & Monod, for which the Nobel Prize in Physiology or Medicine was awarded in 1965. The simplicity and clarity of the operon model left little doubt that Jacob & Monod had elucidated an important mechanism of gene regulation (Jacob & Monod 1961). By the mid-1960s, the concept of gene expression regulation, controlled by the coordination of factors that could be either linked or unlinked to the target gene, had gained currency throughout the world of genetics.
While these groundbreaking insights into controlling the action of genes were clearly important to the organismal phenotype, interest in evolutionary genetics was soon to be monopolized by studies of variation in peptide sequences spurred by electrophoretic techniques first applied to populations in the mid-1960s (Harris 1966; Hubby & Lewontin 1966; Lewontin & Hubby 1966). Meanwhile, work on the intersection of evolution and gene expression lay largely dormant until the 1970s. At around the same time, Susumu Ohno was considering the origin of new genes with novel functions via gene duplication (Ohno 1970). Arguing that mutational load imposes a finite limit on the number of genes an organism can maintain, he concluded that genetic functional innovation could not be entirely explained by his preferred mechanism, gene duplication. Consequently, he invoked regulatory innovation in combination with gene duplication to explain the origin of new functions in genomes, asserting that ‘the creation of additional regulatory systems contributed more to big leaps in evolution than did the creation of new structural genes’ (Ohno 1972). Just a few years later, King & Wilson (1975) wrote what was to become an especially influential paper, arguing that comparisons between human and chimpanzee peptide sequences showed too few differences to account for their observed morphological differences, though they actually provided no guidance as to what level of divergence would have satisfied this intuition. In any event, King and Wilson speculated that this apparent discrepancy could be resolved if the morphological differences were largely determined by differences in gene regulation. This study has been widely cited as one of the earliest clear arguments in favour of the relative importance of regulatory evolution compared with protein evolution and remains a touchstone for studies of gene regulation in the context of evolution of development. However, because of technical difficulties as well as a preoccupation with studying the evolution of protein coding sequence (Nevo et al. 1984; Lewontin 1991), the systematic study of evolution of gene regulation did not start in earnest until recent years. Despite this delay, the idea that regulatory evolution, and in particular cis-regulatory evolution, is crucial to major evolutionary innovation has gained considerable currency (Carroll 2008), though well-reasoned counter-arguments to the importance of regulatory evolution have been offered (Hoekstra & Coyne 2007).
2. Types of gene regulation
Following Jacob & Monod's seminal work, a great deal was learned about the general properties of mutations that modify the way a gene product is regulated. The canonical example is in fact the E. coli lac operon elucidated by Jacob & Monod, a virtual microcosm illustrating the basic elements of gene regulation that concern many modern studies, even in eukaryotes. We now take this opportunity for a brief digression into explaining the classic lac operon, for it concisely illustrates all of the essential elements, and serves as a familiar point of reference for organizing and clarifying subsequent discussion (Jacob & Monod 1961; Lodish et al. 1999).
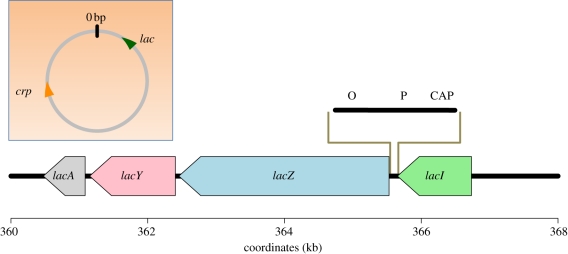
Briefly, the system involves a set of three non-regulatory genes that are expressed together as a polycistronic mRNA. The function of these three genes is to enable the bacterial cell to metabolize lactose in the absence of glucose. These enzymes of the lac operon (composed of the genes lacZ, lacY and lacA) are expressed at high levels when two conditions are met: (i) glucose is absent and (ii) lactose is present. This activation is coordinated through two pairs of elements, both of which operate in broadly analogous ways. Each pair is similar in that it is composed of a stretch of DNA upstream of the lac operon and a protein that binds to it (figure 1).
Figure 1.
The regulation of the lac operon. Each coloured arrow represents the coding region of a peptide, with the direction of the arrow pointing from 5′ to 3′ in the direction of transcription. The three genes lacZ, lacY and lacA are three genes whose messages are all part of a single polycistronic messenger RNA. Their protein products are β-galactosidase, β-galactosidase permease and β-galactosidase transacetylase, respectively. The lacI gene codes the lac repressor. The enlarged approximately 130 bp segment between lacZ and lacI represents the cis-regulatory DNA upstream of the lac polycistron. O represents the operator, P, the promoter and CAP is the CAP protein binding site. The coordinates represent positions in the E. coli genome in kilobases. Inset: The E. coli circular chromosome showing that the crp gene encoding the CAP protein lies approximately 1.5 Mb distant from the lac operon.
One pair acts through repression of gene expression. Under ‘typical’ laboratory conditions, E. coli is cultured in an environment containing abundant glucose and lacking lactose. Under these conditions, a protein encoded by lacI binds to a DNA sequence, called the operator, just upstream of the genes coding for the lac enzymes. When the protein encoded by lacI binds to the operator, it interferes with RNA polymerase binding to the promoter upstream of the lac genes, thereby repressing transcription. The stretch of DNA encoding lacI is fewer than 150 bp upstream of the lac cistron and the operator lies between the lac cistron and lacI (figure 1). When lactose is present, lactose molecules bind to the repressor encoded by lacI, which induces a conformational change, releasing the repressor from binding to the DNA, which was previously blocking RNA polymerase. Thus, the lacI/operator pair is an example of a diffusible factor that binds to a DNA motif adjacent to the lac operon. Notably, both elements are encoded in sequence very closely linked to the lac operon.
Another type of regulation is carried out by the gene crp that produces a product called the CAP protein. The CAP protein is a regulatory protein that binds to another DNA sequence between the lacI and the lac operon sequences near the lac promoter (figure 1). This second regulatory pair differs from the lacI/operator in two crucial aspects. Firstly, the crp gene is more than 1.5 Mb distant from the lac operon, which is quite distant for the E. coli genome. Secondly, CAP acts as an activator instead of a repressor in the presence of cyclic AMP; cAMP is indicative of a low level of glucose. Thus, when lactose is present, the repressor lacI is inactive and when glucose is absent, the activator is active.
It is also worth mentioning that the CAP protein is a trans-regulatory factor that influences the expression of many genes, including operons encoding genes responsible for catabolizing galactose and arabinose. In this sense, the CAP protein is a promiscuous regulator in that a change in crp may result in expression changes in multiple downstream targets. The repressor encoded by lacI, on the other hand, is specific for regulating the lac operon.
In this lac operon example, we distinguish between two dichotomous ways of classifying gene regulatory systems (table 1). The first case is in how the regulatory element acts. If the regulatory element is a diffusible element like lacI or the CAP protein, it is by definition a trans-acting factor. On the other hand, if it is a linked DNA element like the operator, the promoter or the CAP binding sequence whose action is non-diffusible and allele-specific, we call it a cis-acting element. In the second classification system, instead of identifying the regulatory element's mode of action, we classify the element according to its position with respect to the gene(s) it regulates. If it maps very close to a gene it regulates, we call it ‘local’. Otherwise, we call it ‘distant’. Both the lacI and the lac operator are local regulators of the lac operon, though one is trans-acting and the other cis-activating. On the other hand, the crp gene is distant. The distinction between local and distant is somewhat arbitrary if a regulatory element maps to the same chromosome as the gene it regulates. Usually, it is convenient to classify a regulatory variant as distant when it maps outside of the region expected to contain the regulatory sequence, though this distinction is by necessity operational and depends in part on the resolution of the method used to map the variation as well as the organism in which the mapping takes place. In eukaryotic genomes, an enhancer is a distantly linked DNA sequence (i.e. one type of cis-element) that can regulate gene expression even though it is distant. Thus, although a trans-activating factor tends to be distant, it can be local, and although cis-activating elements tend to be local, they can be distant.
Table 1.
Categorizing the mode of action and the location of regulatory elements with examples from the E. coli system.
| cis | trans | |
|---|---|---|
| local | operators, promoters | neighbouring transcription factors |
| lac operator | lacI | |
| distant | enhancers | distant transcription factors |
| Shh ZPA enhancera | Crp |
aIn the case of distant/cis regulators, there are no good examples in the E. coli system, so we report one example from mammals in the sonic hedgehog zone polarizing activity enhancer (Shh ZPA enhancer) which is a cis factor that regulates sonic hedgehog from up to 1 Mb away (Lettice et al. 2002; Jeong et al. 2006).
For the remainder of this review, we shall focus on the genetic architecture of gene expression variation. We regret that space constraints prevent us from considering other recent work pertaining to evolution of gene regulation such as evolution of cis elements at the sequence level (reviewed in Wray et al. 2003; Wittkopp 2006; Wray 2007) or evolutionary models that consider the mutational variance of expression phenotypes (reviewed in Gilad et al. 2006; Fay & Wittkopp 2008).
3. Strategies for determining the architecture of expression level variation
The lac operon serves as an ideal cartoon framework on which to hang the concepts of modern high-throughput evolutionary genomics studies of regulation of gene expression levels (Jacob & Monod 1961). Assaying gene regulation treats variation in levels of the mRNA of a focal gene as the phenotype, where the focal gene is analogous to the lac operon. Variation in either the cis-regulatory elements (closely linked DNA elements, e.g. analogues of the promoter and operator) or the trans-regulatory factors (diffusible factors that can influence expression of a gene whether or not it is adjacent to it, e.g. the lacI gene and the crp gene) can lead to change in the level of expression of the focal gene.
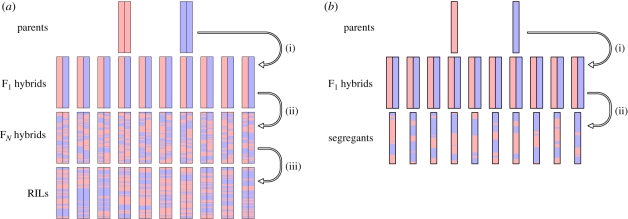
Currently, two strategies are most commonly used for assaying expression levels for the purpose of uncovering the nature of their genetic bases. The first method, expression quantitative trait locus (eQTL) mapping, takes advantage of methodology pioneered for mapping morphological quantitative traits. Typically, two or more inbred strains are used to construct a large panel of recombinants (Gilad et al. 2008). Such recombinant strains possess genomes composed of segments originating from the parental strains that are randomly shuffled by one or more generations of recombination to form mosaic chromosomes (figure 2a). In order to obtain strains homozygous for all (or nearly all) loci, the recombinants are isogenized, either through sibling mating for many generations, or through genetic methods suitable for the genetic system of interest (e.g. tetrad dissection in Saccharomyces cerevisiae or by using balancer chromosomes in Drosophila melanogaster).
Figure 2.
(a) Recombinant inbred line (RIL) creation for QTL mapping. In this strategy, the founding strains are two unrelated isogenic strains. (i) In the first generation, a cross between the two strains is made to obtain F1 hybrids. (ii) The F1 hybrids are crossed through random mating for N generations to produce FN hybrids. (iii) The FN hybrids are subjected to a method of isogenization, whereby all, or nearly all, of the genome is made homozygous. This is typically done using inbreeding, though chromosomes can also be completely isogenized using genetic techniques such as crosses to a balancer stock (in Drosophila). (b) Yeast segregant line creation for QTL mapping. A single generation of recombination can be easily obtained in S. cerevisiae with a single cross (Winzeler et al. 1998). (i) Two haploid yeast strains of opposite mating types are crossed to one another to obtain diploid F1 hybrids. (ii) The F1 hybrids are then made to undergo sporulation, leading to haploids that have undergone recombination during meiosis. Since these segregants are now haploid, there is no need for inbreeding or other isogenization, as the segregants cannot become heterozygous until mated again.
In order to determine which segments of a recombinant genome derive from which source strains, a set of genetic markers is required. These markers must be at a resolution suited to the expected size of the segments; the smaller the segments, the greater the density of markers required. While any marker is suitable, genome-wide studies almost universally employ single nucleotide polymorphisms (SNPs) to genotype the recombinant lines. Each recombinant strain is also subjected to measurement of the phenotype of interest. For gene regulation, the transcript level is treated as a quantitative trait. QTL mapping is particularly convenient, as it only requires measuring the expression level of recombinant inbred lines (RILs), a requirement that is easily met by widely available microarray methodologies (Ranz & Machado 2006). The first genome-scale QTL studies used microarrays to assay both the SNPs and the expression level of such RILs across the whole genome simultaneously (Brem et al. 2002). The genotypic and phenotypic information are then integrated using a suitable QTL algorithm to produce a map for each expression phenotype.
QTLs that are distantly separated from the genes they regulate are most likely to be diffusible trans-regulating factors. Any expression variation mapped to the crp gene is one such trans variant drawn from the lac operon example. On the other hand, QTLs that map adjacent to the genes they regulate are good candidates for being cis variants. The operator and promoter sequences are the relevant examples from the lac operon. Occasionally, regulatory variants map to a region near the gene they regulate, but are actually influencing a diffusible trans factor instead of a cis element, as would be the case for a regulatory mutant affecting the lacI gene. However, without an extremely dense marker panel combined with RILs shuffled very finely, a strict QTL framework lacks the resolution to distinguish trans factors immediately adjacent to the genes they regulate from cis elements that map equally closely. Thus, QTL studies would have a very difficult time distinguishing variation in lacI from variation in the operator it binds to. We prefer to use the local/distant dichotomy adopted by Rockman & Kruglyak (2006) in place of the cis/trans dichotomy in the cases that exhibit such ambiguity (table 1).
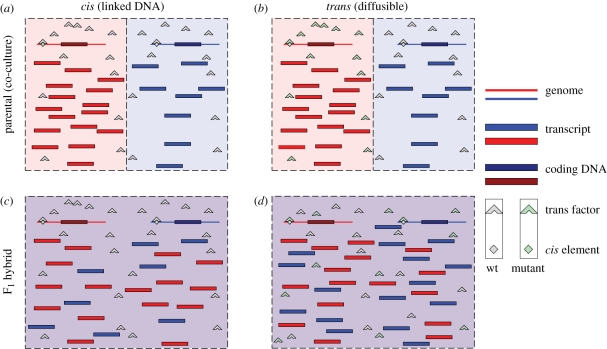
A complementary strategy for discovering genetic variation in transcriptional regulation employs comparisons between two parental strains and their F1 hybrids (figure 3), and is closely related to the classic cis/trans test that inspired it. However, instead of merely comparing the expression of hybrids and parents, the allelic expression for each is examined. F1 hybrids can reveal cis-regulatory variation owing to the fact that trans variation should be equalized by the two alleles sharing the same set of diffusible elements like transcription factors. If the expression level is not equal in the F1 progeny, this implies that no diffusible element can equalize the expression levels, leading to the conclusion that the difference is entirely or at least in part encoded in cis. In such comparisons, the key insight requires comparisons of allele-specific expression (ASE) patterns between the parents and offspring. The methodological requirement for measuring ASE can be met through pyrosequencing for moderate numbers of genes (Wittkopp et al. 2004, 2008; Sung et al. 2009). For expression studies on a more global scale, methods like microarrays (Tirosh et al. 2009; Zhang & Borevitz 2009) or high-throughput short-read sequencing (Nagalakshmi et al. 2008; Emerson et al. 2010) are required.
Figure 3.
Illustration of the detection of cis/trans effects by comparing parents and F1 hybrids. The dashed borders indicate cell boundaries. The colours red and blue represent two different genotypes. The mutant regulatory elements are both mutations that are present only in the red parental strain and confer a twofold increase in the transcript level. The top row represents the expectation for expression at a locus in the parental comparisons for (a) cis mutations and (b) trans mutations. The second row indicates the expected expression patterns of (c) cis mutations and (d) trans mutations in the offspring genomes.
In ASE experiments, variation is detected in a comparison of expression levels between two alleles in homozygous or hemizygous parental strains (figure 3, top row). Then the difference in expression between alleles is assayed again in the heterozygote (F1). If the original pattern of variation is the same between the two comparisons (figure 3, first column), the expression variation cannot be attributed to the action of a diffusible regulatory factor, as such a factor would be readily shared between loci in a hybrid individual. Thus, such examples are identified as cis mutations, and are likely to be DNA regulatory sequences. On the other hand, if regulatory variation between the parents is not observed in the hybrids (figure 3, second column), this suggests strongly that the variation in the expression level is the result of a diffusible trans-regulatory factor. Any situation other than these two extreme cases indicates the presence of both cis and trans variation.
Under this experimental strategy, in contrast to eQTL strategies, the locations of the elements responsible for the expression variation cannot be mapped at all, thus making it unsuitable for classifying regulatory mutations as local or distant. However, unlike in eQTL mapping, this strategy can easily distinguish between a cis-regulatory element and a trans element closely linked to the gene it regulates. Furthermore, this strategy requires measurement of many fewer strains, as only each parent and their single F1 hybrid need to be assayed.
4. Expression qtl mapping: measuring local and distant expression level variation
The first study of eQTLs on a truly genomic scale was conducted in yeast (Brem et al. 2002), though QTL studies of anonymous protein abundance had been carried out on a moderate number of proteins in maize using two-dimensional gel electrophoresis as early as 1994 (Damerval et al. 1994). Brem et al. established that not only was expression variation within species widespread, but also expression patterns were frequently multi-genic. In the yeast system, characterizing a large number of recombinant chromosomes is relatively easy (figure 2b) (Winzeler et al. 1998). Many expression phenotypes could be mapped to multiple genomic regions. Moreover, it was found that many expression phenotypes mapped to a small number of distant-acting regions. Another study working in the same system examined the variation in distant-acting factors more deeply (Yvert et al. 2003). In this study, they found that most variation was due to distant-acting variation and that a total of 1265 expression phenotypes displayed linkages that mapped to 13 regions, and that most of the linkages found in the previous study (Brem et al. 2002) were recapitulated in this one. Thus, many genes are regulated by distant-acting ‘hotspots’. To compare this result with the lac operon cartoon above, the dominant mode of trans-regulatory variation was found to be analogous to how variation would influence genes regulated by the CAP protein. A relatively small number of variants were responsible for influencing the expression level of many downstream genes.
In further support of the idea that a few distant-acting factors control the expression of many genes, Yvert et al. (2003) applied hierarchical clustering to the dataset and determined that many of these genes were co-regulated. Surprisingly, regions determined to contain distant-acting eQTL hotspots were not enriched for transcription factors, indicating that variation in transcription factors may be removed by natural selection. Whether there is a commonality to such hotspot regions has yet to be determined. Additional studies of the yeast eQTLs indicated that not only were variable expression phenotypes usually multi-genic, but they also were involved in complex genetic interactions (Brem & Kruglyak 2005; Brem et al. 2005). The widespread presence of such epistatic interactions poses challenges for identifying QTLs, though progress is being made in solving these problems (Brem et al. 2005; Storey et al. 2005). Similar results were obtained in Arabidopsis thaliana, where it was shown that only approximately one-third of eQTLs were local and that many of the distant eQTLs mapped to hotspots (West et al. 2007).
Refinements to eQTL analyses allow more specific inferences to be made concerning the mode of action of regulatory variants. By incorporating ASE assay techniques (reviewed below), it is possible to both map the regulatory variants and determine whether they are acting in cis configuration or in trans configuration, independent of where the eQTL maps to (Doss et al. 2005; Ronald et al. 2005a,b).
5. Allele-specific assays for measuring cis and trans expression variation
While eQTL studies provide genome coordinates for regulatory variants, alone it cannot distinguish between cis- and trans-regulatory variants. However, this information can be gleaned by comparing the ASE levels of parental strains with that of their F1 hybrids, though at the cost of requiring allele-specific assays. As reviewed above, F1 hybrids can reveal cis-regulatory variation owing to the fact that trans variation should be equalized by the two alleles sharing the same set of diffusible elements like transcription factors (figure 3). Early ASE studies took advantage of this property, demonstrating across multiple individuals that alleles of the same gene exhibited differential expression in heterozygotes for both humans (Yan et al. 2002; Bray et al. 2003; Lo et al. 2003; Serre et al. 2008) and mice (Cowles et al. 2002). Two of these studies sampled multiple tissues (Cowles et al. 2002; Lo et al. 2003) and, indeed, Cowles et al. (2002) discovered tissue-specific variation between alleles, indicating that the action of cis-acting regulatory elements could mediate tissue-specific phenotypic variation.
While the hybrid studies are rather useful for detecting cis variation, information about trans variation can only be gained by comparing the ASE between the homozygous parental strains and their F1 progeny. This approach has been exploited to great effect in model systems where constructing inbred parental lines and their F1 hybrid is easily done. This approach has been applied both between individuals from within the same species in maize (Stupar & Springer 2006; Springer & Stupar 2007a,b; Stupar et al. 2007, 2008; Guo et al. 2008), yeast (Sung et al. 2009; Emerson et al. 2010), Drosophila simulans (Main et al. 2009) and A. thaliana (Zhang & Borevitz 2009) and between individuals from closely related species in Drosophila (Ranz et al. 2004; Wittkopp et al. 2004; Landry et al. 2005) and Saccharomyces (Tirosh et al. 2009).
The work in the maize system has indicated that variation between different strains is largely dominated by cis-acting variation (Stupar & Springer 2006). Interestingly, this new genome-wide ASE technique was also applied to the old problem of explaining hybrid vigour or heterosis, a phenomenon for which maize is well known. Two competing hypotheses have long been in contention: overdominance and dominance (Crow 1998; Charlesworth & Willis 2009). In attempting to investigate why heterosis is so common in maize, answers have been sought through examining the dominance of variation found through ASE studies, although current work is inconclusive and can only provide speculations on the precise relationship between the dominance of ASE patterns and hybrid vigour (Springer & Stupar 2007b; Stupar et al. 2007, 2008). The intraspecific studies in yeast and Arabidopsis differ from the maize studies in that they either find a substantially larger number of genes shown to be acting in trans fashion than in cis (Sung et al. 2009; Emerson et al. 2010) or similar numbers of both, with a slightly larger number of trans interactions (Zhang & Borevitz 2009). The study in D. simulans showed that cis ASE variation predominated over trans variation, though this was observed only in a sample of five genes (Main et al. 2009).
The studies of interspecific ASE conducted in Drosophila take advantage of the ability to cross D. melanogaster to its sibling species D. simulans. Both studies indicated that expression differences were very common between species (Ranz et al. 2003; Wittkopp et al. 2004). Wittkopp et al. (2004) further suggested that cis variation was much more frequent than trans variation. Though Ranz et al. (2004) did not measure ASE in hybrids, they did compare the expression level of F1 hybrid strains to their inbred parental strains and found that transgressive segregation of expression variation (i.e. the expression level of the heterozygote is outside the range defined by the parental expression level) was common, a phenomenon that is quite common for many phenotypes in interspecific hybrids (Rieseberg et al. 2003). In a yeast cross between Saccharomyces cerevisiae and Saccharomyces paradoxus, it was also found that species differences were predominantly cis variants. Interestingly, it was found for both flies and yeast that when a given gene exhibits variation in both cis- and trans-regulation, the mutations frequently show variation in opposite directions. This apparent excess of compensatory variation would seem to indicate that the gene expression level is under stabilizing selection (Landry et al. 2005; Tirosh et al. 2009).
Finally, two studies in flies and yeast (Wittkopp et al. 2008; Emerson et al. 2010) have compared the results of within- and between-species variation in ASE (i.e. polymorphism and divergence, respectively) to evaluate the contribution of natural selection to species differences. The logic behind comparing within-species variation with between-species variation is predicated on predictions derived from the neutral theory of molecular evolution (Kreitman & Aguade 1986; McDonald & Kreitman 1991). Under the hypothesis of no selection (i.e. neutrality), the number of substitutions between two species for a particular category of mutation is proportional to the time since speciation and the mutation rate: SDiv ∝ tμ. For polymorphism, the number of sites segregating within the population is proportional to the effective population size of the population, a quantity related to the sample size, and the mutation rate: SPoly ∝ Ne ksampμ. As a result, the relative amount of variation between polymorphism and divergence in every given neutral category of mutation should be the same quantity, which is independent of mutation rate: SDiv / SPoly ∝ t/Neksamp. In comparing different categories of mutation (e.g. comparing cis variation to trans variation), we expect that if both categories are neutrally evolving, then they should exhibit similar polymorphism/divergence ratios.
For the two studies mentioned above, it was determined that cis differences dominated between species to a larger extent than would be predicted given the levels of expression polymorphism, or in other words, the divergence/polymorphism ratio for cis mutations was much larger than that for trans mutations, implicating the role of natural selection in governing between-species expression differences. In the Emerson et al. study, which compared intraspecific ASE data to interspecific ASE data generated by Tirosh et al. (2009), it was further demonstrated that trans variation was strongly correlated between polymorphism and divergence, indicating that it was more likely to follow a neutral model of evolution, thus implicating natural selection in shaping the cis divergence, which showed no such correlation.
6. Prospects: investigating the action of natural selection on expression variation
Progress in high-throughput sequencing technology has offered the ability to rapidly genotype entire genomes with less effort and cost, making obtaining dense panels of markers much more feasible than even just a few years ago. Similarly, entire transcriptomes can be measured simultaneously with improving methodology in microarrays as well as high-throughput sequencing technology like RNA-seq (Nagalakshmi et al. 2008). Given these tools, we can now conceive of study designs tailor-made to answer detailed questions about how natural expression variation is shaped by evolutionary forces, including natural selection, genetic drift and demographic processes. Such studies will soon be limited primarily only by the effort and expense required to construct and maintain the biological materials.
To date, global investigations of natural selection on expression have been studied from a perspective that treats expression phenotypes in ways analogous to how morphological traits are treated (Gilad et al. 2006; Fay & Wittkopp 2008). While such studies have had remarkable success in identifying violations of neutral predictions, there is no clear direction to bring such studies into population genetics frameworks that treat genotype frequencies instead of phenotypic variance. While single-gene investigations of expression polymorphism have met with success in examining the selective forces shaping regulatory variation (reviewed in Rockman & Kruglyak 2006), these studies are not global in nature. Recent work (Wittkopp et al. 2008; Emerson et al. 2010) has made inroads into ranking the relative importance of natural selection on cis and trans variation by taking advantage of comparisons between polymorphism and divergence. Emerson et al. (2010) have treated significant expression variants as mutations in order to place their data into a framework more amenable to site-based models prevalent for analysing nucleotide population data. While such studies are capable of evaluating the relative importance of cis and trans in the evolution of gene expression, they do not offer a clear way to measure the selection intensity directly. Such direct measurement of the magnitude of natural selection would be an important advance in understanding phenotypic evolution.
Fortunately, recent work in population genetics has made it possible to estimate the selection coefficients of classes of natural variation by examining the statistical properties of population samples. Such studies have made great strides in quantifying one of the most important aspects of evolutionary genetics: the distribution of fitness effects. By sampling genetic variation from natural populations, recent models are capable of parametrizing evolutionary models using predictions that diffusion theory makes about the histogram of allele frequencies, or the site frequency spectrum (SFS) (Sawyer & Hartl 1992; Bustamante et al. 2002; Williamson et al. 2005; Boyko et al. 2008). These models have met with great success in estimating both the demographic parameters as well as the selection parameters of population samples. One of the most attractive aspects of such frameworks is that they do not rely on models of sequence or phenotypic evolution for their inferences. They require only the identification of mutations and the measurement of their frequencies.
One way of treating expression variation in this framework is to apply QTL mapping strategies to population genetic samples of individuals in model organisms. As mentioned above, eQTL strategies have been applied to small numbers of individuals (usually two) and have had great success in identifying regulatory variants. Thus far, it has not been feasible to combine eQTL perspectives with population genetics perspectives. Recently, the mouse and fly communities (Churchill et al. 2004; Macdonald & Long 2007) have been working towards design of RILs constructed from a large number of founder individuals. Such a design is quite amenable to population genetic analysis, provided that the demography of individuals making up the founders can be modelled using current population genetics methodologies.
Once a large RIL panel has been constructed and genotyped, it can be subjected to measurement of phenotypes, mirroring previous eQTL studies. Since the founder lines of most model systems (especially yeast and fruitflies) can be sequenced relatively easily, it is possible to correlate QTLs with specific sequence changes. As long as the founder lines are chosen with an eye towards a tractable demographic sample, any mutations discovered in such a sample can be subjected to population genetic analysis. Recent methods provide for estimation of both natural selection parameters and demographic parameters from collections of genome-wide data (Williamson et al. 2005; Boyko et al. 2008), provided the histogram of allele frequencies (or SFS) can be collected from the data.
Once QTLs have been identified, their SFS can be subjected to population genetic parameter estimation using various methods such as the Poisson random field (Williamson et al. 2005; Boyko et al. 2008) or transition matrix methods (Keightley & Eyre-Walker 2007), enabling a direct investigation of the distribution of fitness effects influencing natural variation in gene regulation. However, given the imperfect nature of QTL studies, it is likely that both ascertainment bias and error will influence the SFS observed from such studies. Indeed, it is well recognized that inferences from population genetics analyses suffer when ascertainment bias is present (Clark et al. 2005). However, if the influence of ascertainment bias and error can be estimated, their influence can be incorporated into estimation of selection parameters (Emerson et al. 2008).
With such advances, we can envision bringing expression variation into the frameworks that are being explored for SNPs at present. Since the demographic models can be fitted by silent SNPs, these models can be very powerful examinations of the action of natural selection on natural expression.
Acknowledgements
We thank Brian Charlesworth and two anonymous reviewers for their helpful comments and suggestions. We also thank Michael McDonald, Gene Ng and Bernhard Schaefke for discussion. This work was supported by Academia Sinica, Taiwan, and by NIH grants GM30998 and GM081724.
Footnotes
One contribution of 18 to a Discussion Meeting Issue ‘Genetics and the causes of evolution: 150 years of progress since Darwin’.
References
- Boyko A. R., et al. 2008Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4, e1000083 (doi:10.1371/journal.pgen.1000083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. J., Buckland P. R., Owen M. J., O'Donovan M. C.2003Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 113, 149–153 (doi:10.1007/s00439-003-0956-y) [DOI] [PubMed] [Google Scholar]
- Brem R., Kruglyak L.2005The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Natl Acad. Sci. USA 102, 1572–1577 (doi:10.1073/pnas.0408709102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Yvert G., Clinton R., Kruglyak L.2002Genetic dissection of transcriptional regulation in budding yeast. Science 296, 752–755 (doi:10.1126/science.1069516) [DOI] [PubMed] [Google Scholar]
- Brem R., Storey J., Whittle J., Kruglyak L.2005Genetic interactions between polymorphisms that affect gene expression in yeast. Nature 436, 701–703 (doi:10.1038/nature03865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. D., Nielsen R., Sawyer S. A., Olsen K. M., Purugganan M. D., Hartl D. L.2002The cost of inbreeding in Arabidopsis. Nature 416, 531–534 (doi:10.1038/416531a) [DOI] [PubMed] [Google Scholar]
- Carroll S. B.2008Evo–devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36 (doi:10.1016/j.cell.2008.06.030) [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Willis J. H.2009The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796 (doi:10.1038/nrg2664) [DOI] [PubMed] [Google Scholar]
- Churchill G., et al. 2004The collaborative cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36, 1133–1137 (doi:10.1038/ng1104-1133) [DOI] [PubMed] [Google Scholar]
- Clark A. G., Hubisz M. J., Bustamante C. D., Williamson S. H., Nielsen R.2005Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 15, 1496–1502 (doi:10.1101/gr.4107905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comfort N. C.1999‘The real point is control’: the reception of Barbara McClintock's controlling elements. J. Hist. Biol. 32, 133–162 (doi:10.1023/A:1004468625863) [DOI] [PubMed] [Google Scholar]
- Comfort N. C.2001aFrom controlling elements to transposons: Barbara McClintock and the Nobel prize. Trends Biochem. Sci. 26, 454–457 [DOI] [PubMed] [Google Scholar]
- Comfort N. C.2001bThe tangled field: Barbara McClintock's search for the patterns of genetic control. Cambridge, MA: Harvard University Press [Google Scholar]
- Cowles C., Hirschhorn J., Altshuler D., Lander E.2002Detection of regulatory variation in mouse genes. Nat. Genet. 32, 432–437 (doi:10.1038/ng992) [DOI] [PubMed] [Google Scholar]
- Crow J. F.199890 years ago: the beginning of hybrid maize. Genetics 148, 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C., Maurice A., Josse J. M., Devienne D.1994Quantitative trait loci underlying gene-product variation—a novel perspective for analyzing regulation of genome expression. Genetics 137, 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss S., Schadt E., Drake T., Lusis A.2005Cis-acting expression quantitative trait loci in mice. Genome Res. 15, 681–691 (doi:10.1101/gr.3216905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J. J., Cardoso-Moreira M., Borevitz J. O., Long M.2008Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320, 1629–1631 (doi:10.1126/science.1158078) [DOI] [PubMed] [Google Scholar]
- Emerson J. J., Hsieh L.-C., Sung H.-M., Wang T.-Y., Huang C.-J., Lu H. H., Lu M.-Y., Wu S.-H., Li W.-H.2010Natural selection on cis and trans regulation in yeasts. Genome Res. 20, 826–836 (doi:10.1101/gr.101576.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wittkopp P. J.2008Evaluating the role of natural selection in the evolution of gene regulation. Heredity 100, 191–199 (doi:10.1038/sj.hdy.6801000) [DOI] [PubMed] [Google Scholar]
- Gilad Y., Oshlack A., Rifkin S. A.2006Natural selection on gene expression. Trends Genet. 22, 456–461 (doi:10.1016/j.tig.2006.06.002) [DOI] [PubMed] [Google Scholar]
- Gilad Y., Rifkin S. A., Pritchard J. K.2008Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 24, 408–415 (doi:10.1016/j.tig.2008.06.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S. F.1987In friendly disagreement—Wilson, Morgan, and the embryological origins of the gene theory. Am. Zool. 27, 797–806 [Google Scholar]
- Guo M., Yang S., Rupe M., Hu B., Bickel D. R., Arthur L., Smith O.2008Genome-wide allele-specific expression analysis using massively parallel signature sequencing (MPSSTM) reveals cis- and trans-effects on gene expression in maize hybrid meristem tissue. Plant Mol. Biol. 66, 551–563 (doi:10.1007/s11103-008-9290-z) [DOI] [PubMed] [Google Scholar]
- Haldane J.1932The time of action of genes, and its bearing on some evolutionary problems. Am. Nat. 66, 5–24 [Google Scholar]
- Harris H.1966Enzyme polymorphisms in man. Proc. R. Soc. Lond. B 164, 298–310 (doi:10.1098/rspb.1966.0032) [DOI] [PubMed] [Google Scholar]
- Hoekstra H. E., Coyne J. A.2007The locus of evolution: evo devo and the genetics of adaptation. Evol. Int. J. Organic Evol. 61, 995–1016 (doi:10.1111/j.1558-5646.2007.00105.x) [DOI] [PubMed] [Google Scholar]
- Hubby J. L., Lewontin R. C.1966A molecular approach to study of genic heterozygosity in natural populations. I. Number of alleles at different loci in Drosophila pseudoobscura. Genetics 54, 577–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Monod J.1961Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 [DOI] [PubMed] [Google Scholar]
- Jeong Y., El-Jaick K., Roessler E., Muenke M., Epstein D. J.2006A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 133, 761–772 (doi:10.1242/dev.02239) [DOI] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A.2007Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177, 2251–2261 (doi:10.1534/genetics.107.080663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. C., Wilson A. C.1975Evolution at 2 levels in humans and chimpanzees. Science 188, 107–116 (doi:10.1126/science.1090005) [DOI] [PubMed] [Google Scholar]
- Kreitman M. E., Aguade M.1986Excess polymorphism at the adh locus in Drosophila melanogaster. Genetics 114, 93–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry C. R., Wittkopp P. J., Taubes C. H., Ranz J. M., Clark A. G., Hartl D. L.2005Compensatory cis–trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171, 1813–1822 (doi:10.1534/genetics.105.047449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice L., et al. 2002Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl Acad. Sci. USA 99, 7548–7553 (doi:10.1073/pnas.112212199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C.199125 years ago in genetics—electrophoresis in the development of evolutionary genetics—milestone or millstone. Genetics 128, 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C., Hubby J. L.1966A molecular approach to study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54, 595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H., Wang Z., Hu Y., Yang H., Gere S., Buetow K., Lee M.2003Allelic variation in gene expression is common in the human genome. Genome Res. 13, 1855–1862 (doi:10.1101/gr.1006603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H., Berk A., Zipursky L., Matsudaira P., Baltimore D., Darnell J.1999Molecular cell biology. New York, NY: W. H. Freeman [Google Scholar]
- Macdonald S. J., Long A. D.2007Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176, 1261–1281 (doi:10.1534/genetics.106.069641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main B. J., Bickel R. D., McIntyre L. M., Graze R. M., Calabrese P. P., Nuzhdin S. V.2009Allele-specific expression assays using Solexa. BMC Genomics 10, (doi:10.1186/1471-2164-10-422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B.1956Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 21, 197–216 (doi:10.1101/SQB.1956.021.01.017) [DOI] [PubMed] [Google Scholar]
- McClintock B.1961Some parallels between gene control systems in maize and in bacteria. Am. Nat. 95, 265–277 [Google Scholar]
- McDonald J. H., Kreitman M.1991Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 (doi:10.1038/351652a0) [DOI] [PubMed] [Google Scholar]
- Morgan T. H.1926Genetics and the physiology of development. Am. Nat. 60, 489–515 [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M.2008The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (doi:10.1126/science.1158441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E., Beiles A., Ben-Shlomo R.1984The evolutionary significance of genetic diversity: ecological, demographic and life history correlates. Lect. Notes Biomath. 53, 13–213 [Google Scholar]
- Ohno S.1970Evolution by gene duplication. Berlin, Germany: Springer-Verlag [Google Scholar]
- Ohno S.1972An argument for genetic simplicity of man and other mammals. J. Hum. Evol. 1, 651–662 (doi:10.1016/0047-2484(72)90011-5) [Google Scholar]
- Ranz J. M., Machado C. A.2006Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol. Evol. 21, 29–37 (doi:10.1016/j.tree.2005.09.002) [DOI] [PubMed] [Google Scholar]
- Ranz J. M., Castillo-Davis C. I., Meiklejohn C. D., Hartl D. L.2003Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300, 1742–1745 (doi:10.1126/science.1085881) [DOI] [PubMed] [Google Scholar]
- Ranz J. M., Namgyal K., Gibson G., Hartl D. L.2004Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 14, 373–379 (doi:10.1101/gr.2019804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Widmer A., Arntz A. M., Burke J. M.2003The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Phil. Trans. R. Soc. Lond. B 358, 1141–1147 (doi:10.1098/rstb.2003.1283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L.2006Genetics of global gene expression. Nat. Rev. Genet. 7, 862–872 (doi:10.1038/nrg1964) [DOI] [PubMed] [Google Scholar]
- Ronald J., Akey J., Whittle J., Smith E., Yvert G., Kruglyak L.2005aSimultaneous genotyping, gene-expression measurement, and detection of allele-specific expression with oligonucleotide arrays. Genome Res. 15, 284–291 (doi:10.1101/gr.2850605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J., Brem R., Whittle J., Kruglyak L.2005bLocal regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 1, 213–222 (doi:10.1371/journal.pgen.0010025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp J.1987Beyond the gene: cytoplasmic inheritance and the struggle for authority in genetics. Oxford, UK: Oxford University Press [Google Scholar]
- Sawyer S. A., Hartl D. L.1992Population genetics of polymorphism and divergence. Genetics 132, 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre D., et al. 2008Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic Cis-acting mechanisms regulating gene expression. PLoS Biol. 4, e1000006 (doi:10.1371/journal.pgen.1000006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N. M., Stupar R. M.2007aAllele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19, 2391–2402 (doi:10.1105/tpc.107.052258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N. M., Stupar R. M.2007bAllelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res. 17, 264–275 (doi:10.1101/gr.5347007) [DOI] [PubMed] [Google Scholar]
- Storey J., Akey J., Kruglyak L.2005Multiple locus linkage analysis of genomewide expression in yeast. PLoS Biol. 3, 1380–1390 (doi:10.1371/journal.pbio.0030267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar R. M., Springer N. M.2006Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F-1 hybrid. Genetics 173, 2199–2210 (doi:10.1534/genetics.106.060699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar R. M., Hermanson P. J., Springer N. M.2007Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol. 145, 411–425 (doi:10.1104/pp.107.101428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar R. M., Gardiner J. M., Oldre A. G., Haun W. J., Chandler V. L., Springer N. M.2008Gene expression analyses in maize inbreds and hybrids with varying levels of heterosis. BMC Plant Biol. 8, 33 (doi:10.1186/1471-2229-8-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H.-M., et al. 2009Roles of trans and cis variation in yeast intraspecies evolution of gene expression. Mol. Biol. Evol. 26, 2533–2538 (doi:10.1093/molbev/msp171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Reikhav S., Levy A. A., Barkai N.2009A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324, 659–662 (doi:10.1126/science.1169766) [DOI] [PubMed] [Google Scholar]
- West M. A. L., Kim K., Kliebenstein D. J., van Leeuwen H., Michelmore R. W., Doerge R. W., Clair D. A. S.2007Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175, 1441–1450 (doi:10.1534/genetics.106.064972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S. H., Hernandez R., Fledel-Alon A., Zhu L., Nielsen R., Bustamante C. D.2005Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl Acad. Sci. USA 102, 7882–7887 (doi:10.1073/pnas.0502300102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., et al. 1998Direct allelic variation scanning of the yeast genome. Science 281, 1194–1197 (doi:10.1126/science.281.5380.1194) [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J.2006Evolution of cis-regulatory sequence and function in Diptera. Heredity 97, 139–147 (doi:10.1038/sj.hdy.6800869) [DOI] [PubMed] [Google Scholar]
- Wittkopp P., Haerum B. K., Clark A. G.2004Evolutionary changes in cis and trans gene regulation. Nature 430, 85–88 (doi:10.1038/nature02698) [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G.2008Regulatory changes underlying expression differences within and between Drosophila species. Nat. Genet. 40, 346–350 (doi:10.1038/ng.77) [DOI] [PubMed] [Google Scholar]
- Wray G. A.2007The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216 (doi:10.1038/nrg2063) [DOI] [PubMed] [Google Scholar]
- Wray G. A., Hahn M. W., Abouheif E., Balhoff J. P., Pizer M., Rockman M. V., Romano L. A.2003The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20, 1377–1419 (doi:10.1093/molbev/msg140) [DOI] [PubMed] [Google Scholar]
- Wright S.1934Genetics of abnormal growth in the guinea pig. Cold Spring Harb. Symp. Quant. Biol. 2, 137–147 (doi:10.1101/SQB.1934.002.01.019) [Google Scholar]
- Yan H., Yuan W., Velculescu V., Vogelstein B., Kinzler K.2002Allelic variation in human gene expression. Science 297, 1143 (doi:10.1126/science.1072545) [DOI] [PubMed] [Google Scholar]
- Yvert G., Brem R., Whittle J., Akey J., Foss E., Smith E., Mackelprang R., Kruglyak L.2003Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35, 57–64 (doi:10.1038/ng1222) [DOI] [PubMed] [Google Scholar]
- Zhang X., Borevitz J. O.2009Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics 182, 943–954 (doi:10.1534/genetics.109.103499) [DOI] [PMC free article] [PubMed] [Google Scholar]