Abstract
Sodalis glossinidius is a maternally transmitted secondary endosymbiont residing intracellularly in tissues of the tsetse flies, Glossina spp. In this study, we have used Tn5 mutagenesis and a negative selection procedure to derive a S. glossinidius mutant that is incapable of invading insect cells in vitro and is aposymbiotic when microinjected into tsetse. This mutant strain harbors Tn5 integrated into a chromosomal gene sharing high sequence identity with a type III secretion system invasion gene (invC) previously identified in Salmonella enterica. With the use of degenerate PCR, we have amplified a further six Sodalis inv/spa genes sharing high sequence identity with type III secretion system genes encoded by Salmonella pathogenicity island 1. Phylogenetic reconstructions based on the inv/spa genes of Sodalis and other members of the family Enterobacteriaceae have consistently identified a well-supported clade containing Sodalis and the enteric pathogens Shigella and Salmonella. These results suggest that Sodalis may have evolved from an ancestor with a parasitic intracellular lifestyle, possibly a latter-day entomopathogen. These observations lend credence to a hypothesis suggesting that vertically transmitted mutualistic endosymbionts evolve from horizontally transmitted parasites through a parasitism–mutualism continuum.
Although parasitism and mutualism may have radically different implications for host fitness, endosymbiotic bacteria participating in these relationships are known to share many similarities, including an intracellular habitat (1). The exploitation of an intracellular habitat is thought to have been one of the most important events in bacterial evolution, permitting significant environmental niche expansion and defining the arrival of intracellular pathogens and mutualistic endosymbionts (2, 3). Although there is a good understanding of the mechanisms contributing to bacterial pathogenesis, very little is known about interactions between bacterial endosymbionts and their host cells. Theoretical studies assume that there may be a tradeoff between the effectiveness of horizontal and vertical modes of transmission (4, 5). It has been predicted that mutualists evolve from parasites through an evolutionary continuum in which parasite virulence is attenuated and transmission strategy switches from horizontal to vertical (6). According to this theory, we might expect to find that pathogens and mutualistic endosymbionts harbor similar virulence determinants and utilize the same machinery to facilitate invasion and survival in host cells. In the present study, we explore these issues by investigating genes that coordinate insect cell invasion in Sodalis glossinidius, an intracellular secondary endosymbiont of the tsetse fly (Glossina spp.).
Three distinct endosymbiotic bacteria have been identified previously in the tissues of tsetse (7). Whereas one of these bacteria is known to be a parasitic Wolbachia, the remaining two are thought to be mutualists and have been classified as the primary and secondary endosymbionts of tsetse (named Wigglesworthia glossinidia and S. glossinidius, respectively) (8, 9). Sodalis is a bacterium found exclusively in tsetse flies residing both inter- and intracellularly in a number of different host tissues, including midgut, fat body, and hemolymph (9, 10). The symbiotic role of Sodalis remains unclear, because it has proved difficult to selectively eliminate either Sodalis or Wigglesworthia from tsetse without inducing sterility in the host. Phylogenetic reconstructions based on the 16S rDNA locus reveal that Sodalis is a member of the family Enterobacteriaceae, which is closely related to other intracellular secondary bacterial endosymbionts found in other insects such as the flour weevil Sitophilus zeamais and the aphid Acrythosiphon pisum (11–13). We are particularly interested in Sodalis as a study model because it is known that the association between this bacterium and tsetse has only recently been established. This association is evident from symbiont–host coevolution studies demonstrating the absence of phylogenetic congruence in the evolution of Sodalis and tsetse (11). Sodalis provides an excellent model for the study of host–symbiont interactions because of the availability of an in vitro Sodalis–insect cell coculture system (14). In addition, Sodalis is the only maternally transmitted insect endosymbiont to have been isolated and maintained in pure culture (9). In this study, we demonstrate the use of Tn5 transposon mutagenesis as a tool for generating random Sodalis mutants. With the use of an in vitro negative selection procedure, we have identified Sodalis mutants deficient in their ability to attach to and invade insect cells both in vitro and in vivo. Characterization of a noninvasive Tn5 Sodalis mutant has revealed that Sodalis relies on components of a type III secretion system to facilitate entry into insect cells.
Materials and Methods
Bacterial Strains, Cell Lines, and Culture Conditions.
Throughout this study, we used S. glossinidius type strain M1, a pure bacterial culture isolated from the hemolymph of laboratory colonized tsetse (9). S. glossinidius strain M1 was maintained by coculture in Aedes albopictus C6/36 cells (15) at 25°C in liquid Mitsuhashi–Maramorosch (MM) medium (15) supplemented with 20% (vol/vol) heat-inactivated FCS (ICN). Uninfected insect cell cultures were passaged every 10 days with a 1:10 split into fresh medium and were examined by Gimenez staining (16) and light microscopy. For cloning, S. glossinidius strain M1 was cultivated on MM agar plates composed of MM medium (without FCS) solidified by autoclaving after the addition of 1% (wt/vol) bacto-agar (Difco). MM agar plates were cultivated under microaerophilic conditions in sealed gas jars flushed with 10 vol of 5% O2/95% CO2.
Transposon Mutagenesis.
Electrocompetent S. glossinidius was prepared on ice from 100 ml of a 5-day-old log-phase culture of strain M1 (OD 600 nm = 0.3) by successively pelleting (6,000 × g, 10 min, 4°C) and resuspending bacteria in 25 ml, 2 ml, and finally 0.2 ml of sterile 10% (vol/vol) glycerol on ice. For electroporation, 50 μl of electrocompetent strain M1 was mixed on ice in a 0.1-cm-path cuvette with 20 ng of pUTkm1 DNA, and a single pulse was applied (1.9 kV, 25 μF, 200 Ω). After electroporation, bacteria were transferred to a flask containing 5 ml of sterile MM medium and allowed to recover by overnight incubation at 25°C. To select for bacteria with Tn5 transpositions, kanamycin was added to the flask containing recovering cells (final concentration: 20 μg/ml). After a further 3-day incubation at 25°C, kanamycin-resistant S. glossinidius was mixed with a suspension of insect cells (A. albopictus clone C6/36) in fresh MM at multiplicity of infection of ≈10 to allow invasive bacteria to adhere to and invade insect cells. After 24 h at 25°C, the cell suspension was vortexed gently to release attached bacterial cells. Insect cells (containing invasive S. glossinidius) were pelleted by low-speed centrifugation (1,500 × g, 5 min, 25°C). Nonadherent or noninvasive bacteria were collected by aspiration of the supernatant, providing enrichment for bacteria unable to invade insect cells. This enrichment procedure was repeated a further three times under identical conditions to ensure selection of nonadherent and noninvasive bacteria. Bacteria recovered from the supernatant of the final enrichment assay were cloned by plating on MM agar containing 20 μg/ml kanamycin. After 5 days of growth at 25°C under microaerophilic conditions (5% O2/95% CO2), 30 individual bacterial colonies (designated clones D1–D30) were isolated and inoculated separately into 5-ml 3-day-old cultures of A. albopictus C6/36 cells. To map the integration site for miniTn5 in the S. glossinidius invC mutant (clone D18), genomic DNA was isolated by an established procedure (17), and 10 μg of DNA was digested to completion in three separate reactions with restriction enzymes that do not cut miniTn5 (ClaI, SacII, XhoI). DNA from each digest was electrophoresed and fragments greater than 2.5 kb were excised from agarose gels, purified by electroelution, and ligated to an appropriately digested and dephosphorylated pBluescript SK+ (Stratagene). After cloning, recombinants carrying a miniTn5 with flanking S. glossinidius DNA were identified by survivor selection on LB agar supplemented with 20 μg/ml kanamycin.
Amplification and Nucleotide Sequencing of inv/spa Genes.
We designed redundant PCR primers to amplify invA, invB, invC, spaM, spaP, spaQ, and spaR, based on clustal alignments of the published Inv/Spa amino acid sequences of Salmonella enterica, Shigella flexneri, and Yersinia pestis. To amplify DNA segments by the PCR, 50–100 ng of template DNA was mixed in a 50-μl reaction with 50 pmol of each primer and 2.5 units of Pfu DNA polymerase (Stratagene). The PCR reactions consisted of an initial denaturation step (5 min at 95°C) followed by 30 cycles of denaturation (1 min at 95°C), annealing (1 min at 50°C), and extension (1 min/kb at 72°C). PCR products were purified with a Qiaquick PCR purification kit (Qiagen, Chatsworth, CA), and DNA sequences were determined in both directions by the chain termination method. Additional internal primers for use in PCR and sequencing were designed as sequence information became available.
Experimental Infection of Tsetse Flies with S. glossinidius Clone D18.
S. glossinidius D18 and S. glossinidius T1 (type strain M1 harboring the plasmid replicon pKT231) (18) were inoculated separately into 3-day-old mated female tsetse (Glossina morsitans morsitans) by a single intrathoracic microinjection of 1 μl of 0.85% saline containing 106 bacterial cells. The first puparium deposited by each of these flies was collected and maintained in the laboratory until emergence, when 1 μl of hemolymph was removed from each fly for PCR assay. To detect S. glossinidius clone D18 and clone T1 (pKT231), we used primer sets Tn5F/R and pKT231F/R (Table 1), which amplify an 850-bp fragment of miniTn5 and an 810-bp fragment of pKT231, respectively. PCR was conducted in standard 50-μl reactions containing 1 μl of hemolymph as a template along with 50 pmol of each primer and 2 units of Taq DNA polymerase (Promega). PCR reactions consisted of an initial denaturation step (5 min at 95°C) followed by 30 cycles of denaturation (1 min at 95°C), annealing (1 min at 52°C), and extension (1 min at 72°C).
Table 1.
PCR primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| Tn5F | ATGAGCCATATTCAACGGGAAAC |
| Tn5R | CCAGTGTTACAACCAATTAAC |
| pKT231F | TGGCTACCCATAAGCCT |
| pKT231R | CTCTTGCGCTGCCTCTCC |
| In1 (invC) | GGAAGCGCCGGCCAGGAG |
| In2 (spaM) | GCGGACACAGCCGTTGCC |
| In3 (invC) | ATGMGNGCNWSNYTNYT |
| In4 (invC) | TCACGCATCAAACGCATGC |
| In5 (invA) | GCXGARGTXGCXGCXMGXTT |
| In6 (invA) | ARXARXGCXGGDATYTG |
| In7 (spaP) | AAYGCXYTXGGXYTXCARCA |
| In8 (spaP) | AYCATCATCATXCCXARXGC |
| In9 (spaQ) | ARXGTYTGYTCYTGXARYTG |
| In10 (spaP) | GCXYTXGGXATGATGATG |
| In11 (invA) | TTYATGGGXWSXTTYTAYAT |
| In12 (invA) | GGCATGGCGTCCAGGGAGAACCGCG |
| In13 (invA) | CACGGTATGGAGCTTTCCGAGGCGC |
| In14 (invA) | TTGGTCTCCTGAATGCCG |
| In15 (spaQ) | GCGACGCTGGTGGGATTGCTGGTGG |
| In16 (spaS) | GTXGGYTTYTCXGTYTT |
| In17 (invA) | TTYGGXATHCARGARACXAA |
| In18 (invC) | GGCATTGCATCGTTCCAGTGAGGCG |
DNA Hybridization.
Genomic DNA was isolated from S. glossinidius strain M1 by a standard lysis and extraction procedure (17), and 5-μg aliquots of DNA were digested to completion with combinations of BamHI/XhoI and EcoRI/ClaI and separated by electrophoresis through a 0.7% agarose gel. Sodalis extrachromosomal DNA was isolated and separated by pulsed-field gel electrophoresis according to established methods (19). After electrophoresis, DNA was capillary transferred onto nylon Hybond-N+ membranes (Amersham Pharmacia) with the use of an alkaline transfer buffer (0.4 N NaOH). The 500-bp Sodalis invC PCR product was generated by PCR with primers In1 and In4 (Table 1) and labeled with [α-32P]dCTP with the Prime-It II random primer labeling kit (Stratagene). Blots were probed with the labeled PCR product and exposed to a phosphor screen overnight for visualization with a Molecular Imager FX System PhosphorImager (Bio-Rad).
Phylogenetic Analyses.
Nucleotide sequences for invA, invC, spaQ, and spaR were obtained from GenBank and aligned with the homologous sequences obtained for Sodalis in this study. Amino acid alignments were performed initially with clustal and checked manually. Phylogenetic analyses were performed on both nucleotide (using maximum likelihood methods in paup*) (20) and amino acid alignments (by using protein distance methods in phylip) (21). Bootstrap analyses were performed in phylip.
Nucleotide Sequence Accession Numbers.
The nucleotide sequence accession numbers for the inv/spa sequences used in this study are as follows: Bordetella bronchiseptica, AF172245 and AF049488; Chlamydia pneumoniae, AE001652, AE001617, and AE001663; Erwinia amylovora, L25828; Pseudomonas syringae, AF043444, L11582 and U07346; Ralstonia solanacearum, AJ245811; Rhizobium sp. NGR234, AE000107 and AE000108; S. enterica, M90846 and X73525; S. flexneri, M91664 and D13663; Xanthomonas campestris, M99176, U33548, and AF056246; Yersinia pestis, AF074612. The invA homologue of B. bronchiseptica was obtained from the Sanger Center Sequencing Project (base pairs 1057–3156 in contig 2016).
Results
Characterization of S. glossinidius Entry into A. albopictus C6/36 Cells.
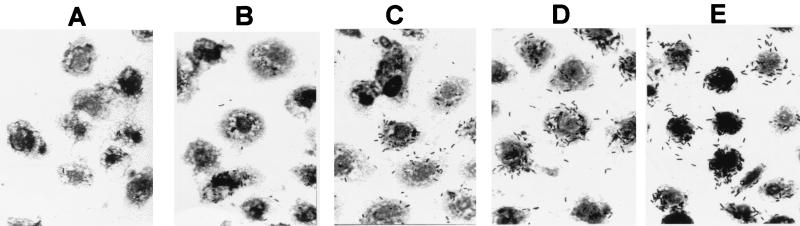
To determine the basic mechanism of invasion for Sodalis, we infected a monolayer culture of A. albopictus C6/36 cells with S. glossinidius type strain M1 at a low multiplicity of infection (≈1). Over a time course of 96 h, we removed samples of insect cells for Gimenez staining and microscopic examination. Intracellular Sodalis was first observed in host cell cytoplasm 8 h after the initial infection of the monolayer (Fig. 1A). After 24 h, we observed an increased density of S. glossinidius in infected cells, indicating that bacteria were dividing within the cytoplasm of insect cells (Fig. 1B). As the infection progressed over the next 72 h (Fig. 1 C–E), the density of bacterial infection within cells increased further until 96 h after infection, when insect cells began to undergo lysis. By 96 h after infection, all of the insect cells examined had become infected, indicating that S. glossinidius is capable of crossinfecting neighboring cells in the monolayer.
Figure 1.
Time course of infection. A. albopictus C6/36 cells were infected with wild-type Sodalis glossinidius strain M1 and examined at intervals by Gimenez staining and light microscopy. (A–E) 8, 24, 48, 72, and 96 h after infection, respectively.
Transposon Mutagenesis and in Vitro Negative Selection of a S. glossinidius Invasion Mutant.
We used a miniTn5 suicide vector pUTkm1 (22) to generate mutant Sodalis harboring random miniTn5 insertions. Mutagenized Sodalis was combined with cultured insect cells (A. albopictus cell line C6/36) at a low multiplicity of infection (≈10) to permit attachment and invasion of virulent bacteria. After allowing 24 h for bacterial invasion, noninvasive Sodalis was collected by separation from insect cells harboring invasive bacteria. This negative selection procedure was repeated three times to maximize the likelihood of obtaining S. glossinidius mutants incapable of invading insect cells. Thirty individual mutant Sodalis clones were recovered after negative selection, and these clones were inoculated into individual cultures of A. albopictus C6/36 cells to determine their invasive potential. Microscopy revealed that 19 of these clones had escaped the selection procedure and were still invasive, and a further 10 clones were deficient in their ability to attach to insect cells. A single clone, designated clone D18, was efficient in attaching to A. albopictus C6/36 cells but was unable to invade cultured insect cells over a 96-h period.
Tn5 Is Integrated into an invC Homologue in the Noninvasive S. glossinidius Mutant Clone D18.
To identify the integration site of miniTn5, we cloned DNA from S. glossinidius D18 after restriction with enzymes that do not cut miniTn5. Recombinant clones with captured miniTn5 were observed only when SacII-digested DNA was cloned; 10 randomly isolated SacII clones all contained a 3.3-kb SacII fragment. Southern blotting of SacII-digested genomic DNA with labeled 3.3-kb SacII fragment as a probe revealed only a single integration of miniTn5 in all recombinant clones (data not shown). Sequencing of the 3.3-kb SacII fragment revealed that integration of miniTn5 had occurred in a region of Sodalis DNA sharing high sequence identity with an invasion gene (invC) previously identified in S. enterica. In S. enterica, invC is known to be an essential component of a type III secretion system, located within the centisome 63 Salmonella pathogenicity island 1 (23, 24). Specifically, invC is known to encode a virulence ATPase sharing high amino acid sequence identity with the flagellar FliI protein and the F0F1-related ATPases (25).
Experimental Infection of Tsetse Flies with S. glossinidius Clone D18.
In a previous study, genetically modified Sodalis has been successfully reestablished in tsetse through intrathoracic microinjection (26). We have used an identical procedure to introduce S. glossinidius clone D18 and clone T1 into female tsetse to determine whether the invC mutant (D18) could persist in the insect and be transferred to interuterine progeny. Despite being able to detect clone D18 by PCR in hemolymph of adult females after microinjection (data not shown), we were unable to detect clone D18 in any of the progeny insects deposited by 30 female flies, each injected with clone D18. To ensure that our microinjection strategy was satisfactory, we used an identical strategy to microinject 12 female tsetse with 106 S. glossinidius clone T1 harboring the broad host range plasmid pKT231. Plasmid pKT231 was detected by PCR in the hemolymph of eight of the first offspring from each of the 12 microinjected female flies, indicating a success rate of >60% for the microinjection procedure (data not shown). These results show that loss of the invC gene and the invasive phenotype in S. glossinidius renders this bacterium aposymbiotic and unable to achieve vertical transmission to progeny.
PCR Amplification of invC and Other Components of the S. glossinidius Type III Secretion System.
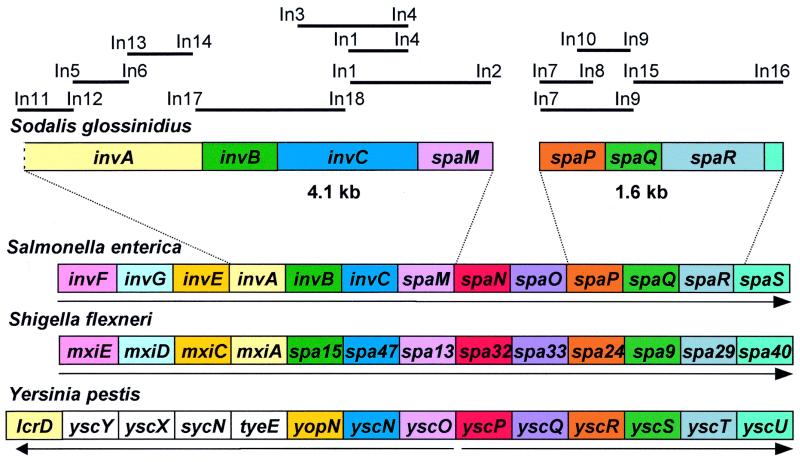
On the basis of the high level of sequence identity observed between the invC sequences of Salmonella and Sodalis, we designed PCR primers (Table 1) to amplify a full-length invC along with other selected homologues of genes found within Salmonella pathogenicity island 1. Degenerate PCR primers were designed according to conserved regions found in an amino acid sequence alignment of Inv/Spa proteins from selected enteric pathogens. With the use of combinations of degenerate PCR primers we amplified and cloned fragments of Sodalis DNA that were homologous to the published sequences of S. enterica invA, invB, invC, spaM, spaP, spaQ, and spaR found in the GenBank data library. Assuming that the organization of inv/spa genes would be similar for Sodalis, Salmonella spp., and other related enteric pathogens, we designed specific PCR primers to amplify flanking and intergenic sequences. With the use of combinations of degenerate and specific PCR primers we identified two contiguous Sodalis DNA sequences of 1.6 kb and 4.2 kb, illustrated in Fig. 2. ORF searches revealed that the 1.6-kb fragment harbored two intact putative ORFs (spaQ and spaR) alongside two putative partial ORFs (spaP and spaS), and the 4.2-kb fragment harbored two intact putative ORFs (invB and invC) and one partial putative ORF (invA). Three candidate spaM ORFs (sharing the same stop codon) were identified within the 4.2-kb fragment, coding putative polypeptides with 154-, 151-, or 114-aa residues. Because the homologues of spaM from S. enterica and S. flexneri are known to have 147- and 112-aa residues, respectively, and there is little conservation in SpaM amino acid sequences in different bacterial species, we were unable to determine the correct Sodalis spaM ORF. Overlapping ORFs were observed for the genes spaP and spaQ (four nucleotides), and noncoding sequences were observed in the intergenic regions between invA and invB (29 nucleotides), invB and invC (11 nucleotides), and spaQ and spaR (4 nucleotides).
Figure 2.
Organization of the inv/spa genes of Sodalis glossinidius and selected enteric pathogens. Homologous genes are decorated with the same color. Arrows indicate the direction of transcription of the inv/spa genes. The Yersinia pestis genes without color have no defined homologues in the other bacterial species. Solid lines above the gene organization represent the PCR products amplified in this study with the primers listed in Table 1. Gene designations are those published by Hueck (31).
DNA Hybridization.
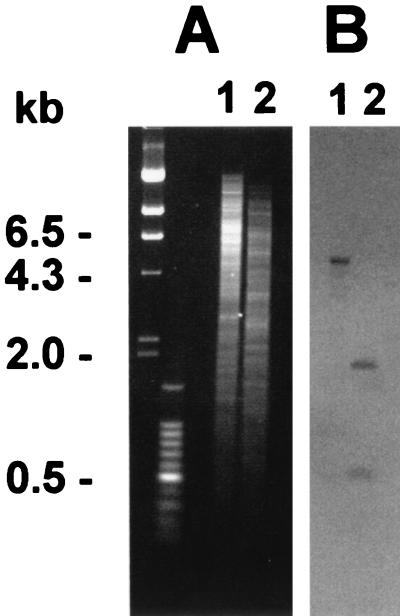
Southern blotting and hybridization revealed that the invC probe hybridized to a 6-kb BamHI/XhoI fragment and 450-bp and 1.9-kb EcoRI/ClaI fragments (Fig. 3). Sequence analysis confirmed the presence of the EcoRI and ClaI sites, giving rise to a 450-bp fragment within the invC ORF. Because Sodalis is known to harbor large extrachromosomal elements that can be resolved only by pulsed-field gel electrophoresis (19), Southern blots of pulsed-field gel electrophoresis-separated extrachromosomal DNA were screened separately with the use of the invC probe and a traI (DNA helicase) probe known to hybridize to Sodalis plasmid DNA (27). Whereas the traI probe hybridized as expected with Sodalis extrachromosomal DNA (data not shown), no hybridization was detected when an identical blot was screened with the invC probe, indicating that the Sodalis inv/spa genes have a chromosomal location.
Figure 3.
Southern hybridization of S. glossinidius strain M1 genomic DNA with a Sodalis invC probe generated by PCR. (A) Genomic DNA digested with BamHI/XhoI (lane 1) and EcoRI/ClaI (lane 2) separated on a 0.7% agarose gel. (B) Southern blot of A probed with the 500-bp Sodalis invC PCR product. Molecular size markers are indicated (Left) .
Phylogenetic Analysis of the Sodalis inv/spa Genes.
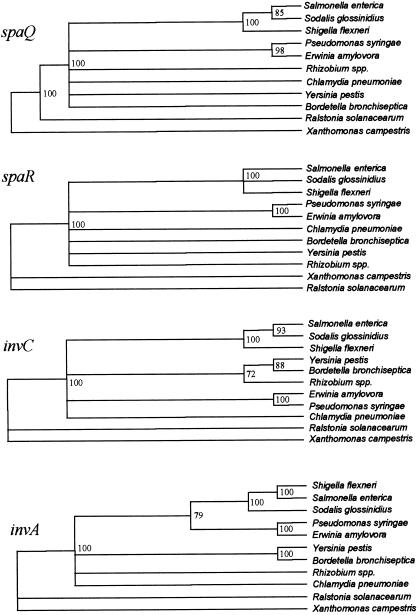
To avoid potential complications arising from differential GC content in the inv/spa genes (Table 2), we compared amino acid sequences of Inv/Spa proteins from Sodalis and selected members of the family Enterobacteriaceae. All four gene trees (Fig. 4) consistently identified a clade containing Salmonella, Sodalis, and Shigella supported by 100% of bootstrap resamples. Alignments of genes encoding spaQ (110 aa) and invC (469 aa) showed Shigella to outgroup a Salmonella/Sodalis clade, invA (765 aa) showed Sodalis to outgroup a Shigella/Salmonella clade, and spaR (305 aa) did not resolve the internal structure of this clade. Many of the deeper relationships between these genes could not be consistently resolved. Very similar results were obtained from maximum likelihood analyses of the corresponding nucleotide sequences (not shown). Results were robust with respect to details of the alignment, remaining similar after removal of Indels.
Table 2.
GC content (%) of inv/spa homologs
| invA | invB | invC | spaQ | spaR | mean | genome | |
|---|---|---|---|---|---|---|---|
| Sodalis glossinidius | 46.5 | 46.1 | 57.6 | 51.0 | 53.5 | 50.9 | 53.5 |
| Salmonella enterica | 45.5 | 43.5 | 54.3 | 46.7 | 49.4 | 48.4 | 52.0 |
| Shigella flexneri | 34.2 | 33.3 | 39.2 | 34.5 | 32.7 | 35.2 | 51.0 |
Figure 4.
Phylogenetic relationships between Inv/Spa proteins from selected enteric pathogens as revealed by phylogenetic reconstruction from protein distance matrices calculated with protdist in the phylip package. Only clades supported by over 70% of bootstrap resamples are shown as resolved. Sodalis Inv/Spa proteins consistently group with Salmonella and Shigella according to all four gene trees.
Discussion
Insect endosymbionts have been classified into two distinct groups according to host tissue distribution, with “primary (P) endosymbionts” residing in specialized cells (mycetocytes) and “secondary (S) endosymbionts” residing in multiple cell types. Molecular coevolution studies show that this distinction reflects the age of the association between symbiont and host because only P-endosymbionts have congruent host–symbiont phylogenies indicating that their associations are ancient (28). Tsetse flies harbor both P-endosymbionts (W. glossinidia) located in mycetocytes and S-endosymbionts (S. glossinidius) that are found to infect a wide range of tsetse tissues, including mycetocyte sheath cells. Comparisons of host 18S rDNA and symbiont 16S rDNA sequences in different tsetse species reveal that only the P-endosymbiont (W. glossinidia) has evolved concordantly with the insect host, having been in symbiosis with tsetse for at least 40 million years (29). The relative ages of the associations between tsetse and their P- and S-endosymbionts are illustrated by a comparison of 16S rDNA pairwise distances for the P- and S-endosymbionts of the two most distantly related tsetse species. These data reveal that whereas Wigglesworthia strains in the two most distantly related hosts have 82/1,180 base differences in 16S rDNA, Sodalis strains in the same two species have only 4/1,026 base differences (11). Although these data indicate that the Wigglesworthia symbiosis is clearly much older than the Sodalis symbiosis, the differences observed in Sodalis strains from distantly related tsetse may be indicative of a lack of recent horizontal transmission events for Sodalis. In addition, the observation of distinct extrachromosomal DNA profiles in Sodalis strains isolated from different tsetse species provides further evidence to suggest that there has been little or no recent horizontal transfer of Sodalis between different host species (19, 27). Although it is impossible to rule out a horizontal component in the transmission of Sodalis, the available evidence indicates that the maternal (vertical) strategy is the predominant route of transmission for Sodalis. For intracellular bacteria adopting a symbiotic lifestyle involving vertical transmission, a process of reductive evolution is known to result in the loss of genes that are essential in the free-living state but are not required for a symbiotic lifestyle (30). Although there is no direct evidence for genomic reduction in Sodalis, phenotypic tests have demonstrated that Sodalis has a very inactive biochemical profile in comparison with related (free-living) members of the family Enterobacteriaceae (9). These test results indicate that many of the genes that are characteristically maintained by members of the family Enterobacteriaceae have been lost or silenced in Sodalis, suggesting a reductive adaptation to the symbiotic lifestyle.
In the present study, we have demonstrated that Sodalis relies on components of a type III secretion system to facilitate entry into insect cells in vitro. Type III secretion systems have been identified in a number of pathogens (31) as well as in symbiotic Rhizobium spp. (32). Here we report a description of such a system in an endosymbiont with a predominantly vertical transmission strategy. Many cognate virulence gene clusters (including Salmonella pathogenicity island 1) are located on horizontally transferable elements or as chromosomal integrons in the form of so-called pathogenicity islands (33). The horizontal acquisition of these elements is known to have been critical to the evolution of many important bacterial pathogens (34, 35). Although homologues of the Salmonella pathogenicity island 1 inv/spa genes have been identified in a number of enteric pathogens, the inv/spa complex is not thought to be ancestral to the family Enterobacteriaceae (36). Because the inv/spa complex was present in the last common ancestor of the contemporary Salmonellae, it was suggested that the inv/spa genes may have been acquired by Salmonella and Shigella independently through separate horizontal transfer events (37). In the present study, phylogenetic reconstructions of the inv/spa gene trees consistently identify Salmonella, Shigella, and Sodalis as members of a distinct clade, suggesting a common ancestry for the inv/spa genes of these three bacterial genera. Whereas the inv/spa genes of Shigella are known to be plasmid borne and have a low GC content relative to the remainder of the Shigella genome, the inv/spa genes of Salmonella and Sodalis are chromosomal and have a GC content equal to those of their respective genomes (Table 2). The location and GC content of the Shigella invasion genes are indicative of recent horizontal acquisition (38), and for this reason it seems unlikely that Shigella has served as a donor for the inv/spa complex of either Salmonella or Sodalis. Because the inv/spa genes of Sodalis and Salmonella are substantially more GC rich than the Shigella sequences, it seems unlikely that either of these bacteria served as the donor for the horizontal transfer of inv/spa to Shigella.
Having a lifestyle preventing contact with bacteria living outside the host, endosymbionts with vertical transmission strategies are known to have a greatly reduced opportunity for de novo gene acquisition (30). Consequently, in comparison to pathogens and horizontally transmitted symbionts such as Rhizobium spp., a vertically transmitted endosymbiont such as Sodalis would have a greatly reduced opportunity to acquire a type III secretion system through horizontal transfer. Therefore it seems likely that Sodalis acquired the inv/spa genes before the establishment of a symbiotic relationship with tsetse. In this study, we were unable to infect tsetse with a noninvasive Sodalis mutant, indicating that the inv/spa genes may be critical to the symbiotic lifestyle of this bacterium, providing further evidence that these genes may be ancestral to the symbiosis. As an evolving endosymbiont having only recently established vertical transmission in tsetse, it appears that Sodalis is utilizing virulence determinants that are phylogenetically most closely related to those identified previously in the enteric pathogens S. flexneri and S. enterica. Because endosymbionts closely related to Sodalis have been identified in taxonomically unrelated host insects (10, 11), it is plausible that before the establishment of an endosymbiotic lifestyle in tsetse, the ancestor of Sodalis was an insect pathogen infecting a diverse range of insect taxa.
Acknowledgments
We gratefully acknowledge Roeland van Ham (Instituto Nacional de Tecnica Aeroespacial, Madrid, Spain), who provided a critique of this manuscript. This document is an output from a project funded by the Department for International Development for the benefit of developing countries (C.D.). The views expressed are not necessarily those of the Department for International Development. D.T.H. and S.C.W. are supported by the Wellcome Trust.
Abbreviation
- MM
Mitsuhashi–Maramorosch medium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF306649 and AF306650).
See commentary on page 1338.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021450998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021450998
References
- 1.Hentschel U, Steinert M, Hacker J. Trends Microbiol. 2000;8:226–231. doi: 10.1016/s0966-842x(00)01758-3. [DOI] [PubMed] [Google Scholar]
- 2.Steinert M, Hentschel U, Hacker J. Naturwissenschaften. 2000;87:1–11. doi: 10.1007/s001140050001. [DOI] [PubMed] [Google Scholar]
- 3.Corsaro D, Venditti D, Padula M, Valassina M. Crit Rev Microbiol. 1999;25:39–79. doi: 10.1080/10408419991299167. [DOI] [PubMed] [Google Scholar]
- 4.Frank S A. Proc R Soc Lond B. 1996;263:339–344. doi: 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- 5.Frank S A. Evolution (Lawrence, KS) 1993;47:1721–1732. doi: 10.1111/j.1558-5646.1993.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 6.Ewald P. Ann NY Acad Sci. 1987;503:295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x. [DOI] [PubMed] [Google Scholar]
- 7.Beard C B, O'Neill S L, Mason P W, Mandelco L, Woese C R, Tesh R B, Richards F F, Aksoy S. Insect Mol Biol. 1993;1:123–131. doi: 10.1111/j.1365-2583.1993.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 8.Aksoy S. Int J Syst Bacteriol. 1995;45:848–851. doi: 10.1099/00207713-45-4-848. [DOI] [PubMed] [Google Scholar]
- 9.Dale C, Maudlin I. Int J Syst Bacteriol. 1999;49:267–275. doi: 10.1099/00207713-49-1-267. [DOI] [PubMed] [Google Scholar]
- 10.Aksoy S, Chen X, Hypsa V. Insect Mol Biol. 1997;6:183–190. doi: 10.1111/j.1365-2583.1997.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 11.Aksoy S, Pourhosseini A, Chow A. Insect Mol Biol. 1995;4:15–22. doi: 10.1111/j.1365-2583.1995.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 12.Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P. J Mol Evol. 1998;47:52–61. doi: 10.1007/pl00006362. [DOI] [PubMed] [Google Scholar]
- 13.Fukatsu T, Nikoh N, Kawai R, Koga R. Appl Environ Microbiol. 2000;66:2748–2758. doi: 10.1128/aem.66.7.2748-2758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welburn S C, Maudlin I, Ellis D S. Ann Trop Med Parasitol. 1987;81:331–335. doi: 10.1080/00034983.1987.11812127. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi A. J Gen Virol. 1978;40:531. doi: 10.1099/0022-1317-40-3-531. [DOI] [PubMed] [Google Scholar]
- 16.Gimenez D. Stain Technol. 1964;39:135–140. doi: 10.3109/10520296409061219. [DOI] [PubMed] [Google Scholar]
- 17.Welburn S C, Gibson W A. Parasitology. 1989;98:81–84. doi: 10.1017/s0031182000059710. [DOI] [PubMed] [Google Scholar]
- 18.Bagdasarian M, Lurz R, Ruckert B, Franklin F C, Bagdasarian M M, Frey J, Timmis K N. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 19.Welburn S C, Dale C. In: The Molecular Biology of Insect Disease Vectors. Crampton J M, Beard C B, Louis C, editors. London: Chapman & Hall; 1997. pp. 547–554. [Google Scholar]
- 20.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2000. [Google Scholar]
- 21.Felsentein J. phylip (Phylogeny Inference Package) Seattle: Univ. of Washington; 1993. , Version 3.573. [Google Scholar]
- 22.De Lorenzo V, Herrero M, Jakubzik U, Timmis K N. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galán J E. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 24.Kubori T, Sukhan A, Aizawa S-I, Galan J E. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. . (First Published August 15, 2000; 10.1073/pnas.170128997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eichelberg K, Ginocchio C, Galán J E. J Bacteriol. 1994;176:4501–4510. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Q, Aksoy S. Insect Mol Biol. 1999;8:125–132. doi: 10.1046/j.1365-2583.1999.810125.x. [DOI] [PubMed] [Google Scholar]
- 27.Dale C. Ph.D. thesis. Liverpool, U.K.: Univ. of Liverpool; 1997. [Google Scholar]
- 28.Moran N A, Baumann P. Curr Opin Microbiol. 2000;3:270–275. doi: 10.1016/s1369-5274(00)00088-6. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Li S, Aksoy S. J Mol Evol. 1999;48:49–58. doi: 10.1007/pl00006444. [DOI] [PubMed] [Google Scholar]
- 30.Wernegreen J J, Ochman H, Jones I B, Moran N A. J Bacteriol. 2000;182:3867–3869. doi: 10.1128/jb.182.13.3867-3869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viprey V, Del Greco A, Golinowski W, Broughton W J, Perret X. Mol Microbiol. 1998;28:1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee C A. Infect Agents Dis. 1996;5:1–7. [PubMed] [Google Scholar]
- 34.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 35.Groisman E A, Ochman H. Cell. 1996;29:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 36.Boyd E F, Li J, Ochman H, Selander R K. J Bacteriol. 1997;179:1985–1991. doi: 10.1128/jb.179.6.1985-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hale T L. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]