Abstract
Convergence—the independent evolution of the same trait by two or more taxa—has long been of interest to evolutionary biologists, but only recently has the molecular basis of phenotypic convergence been identified. Here, we highlight studies of rapid evolution of cryptic coloration in vertebrates to demonstrate that phenotypic convergence can occur at multiple levels: mutations, genes and gene function. We first show that different genes can be responsible for convergent phenotypes even among closely related populations, for example, in the pale beach mice inhabiting Florida's Gulf and Atlantic coasts. By contrast, the exact same mutation can create similar phenotypes in distantly related species such as mice and mammoths. Next, we show that different mutations in the same gene need not be functionally equivalent to produce similar phenotypes. For example, separate mutations produce divergent protein function but convergent pale coloration in two lizard species. Similarly, mutations that alter the expression of a gene in different ways can, nevertheless, result in similar phenotypes, as demonstrated by sister species of deer mice. Together these studies underscore the importance of identifying not only the genes, but also the precise mutations and their effects on protein function, that contribute to adaptation and highlight how convergence can occur at different genetic levels.
Keywords: adaptation, Agouti, colour, melanocortin-1 receptor, parallel evolution, Peromyscus
1. Introduction
Convergence—the repeated evolution of similar phenotypes serving the same ecological function in two or more taxa—has long been of interest to evolutionary biologists. On the one hand, phenotypic convergence among populations or species experiencing similar environmental pressures strongly suggests that these traits have evolved by natural selection (Harvey & Pagel 1991). On the other hand, convergence at the genetic level (i.e. the same genes and/or mutations are responsible for similar phenotypes) suggests that genetic constraints limit the available variation upon which natural selection can act, thereby influencing the course of evolutionary change (Wake 1991; Gould 2002). Convergent evolution, thus, informs us about the ultimate and proximate mechanisms generating diversity and can reveal the extent to which the evolutionary process is both repeatable and predictable (Gould 2002; Conway Morris 2003).
Over the last two decades, molecular phylogenetic analyses in a wide range of taxa have shown that natural selection frequently results in the evolution of convergent phenotypes both within (e.g. Brower 1994; Wang & Shaffer 2008) and between species (e.g. Losos et al. 1998; Blackledge & Gillespie 2004). More recently, however, increased power to identify genes responsible for adaptive traits in natural populations (reviewed in Ellegren & Sheldon 2008; Stinchcombe & Hoekstra 2008; Mackay et al. 2009; Slate et al. 2009) has permitted examination of the proximate mechanisms of convergent evolution as well (reviewed in Hoekstra & Coyne 2007; Stern & Orgogozo 2008, 2009; Gompel & Prud'homme 2009). There are multiple cases in which the same genes are responsible for adaptation in distantly related taxa and, conversely, multiple cases in which different genes produce similar phenotypes in closely related taxa (Arendt & Reznick 2008). Thus, evolutionary distance does not necessarily predict genetic mechanisms underlying convergent adaptations (for this reason, we do not distinguish between ‘convergent’ and ‘parallel’ evolution; Arendt & Reznick 2008). Because there are relatively few studies that have precisely characterized the molecular changes responsible for adaptive phenotypic change, it is unclear how often ‘genetic convergence’ (i.e. same gene) is mirrored at the mutational and functional levels.
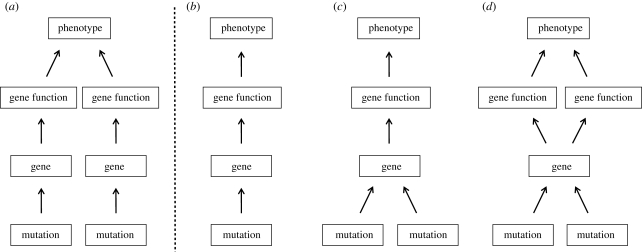
In terms of taxonomic breadth, genetic and developmental mechanism and ecological function, pigmentation is one of the best-studied morphological features in natural populations of animals (reviewed in Hoekstra 2006; Protas & Patel 2008; Hubbard et al. 2010). This wealth of knowledge about pigmentation allows us to study convergence in colour at multiple levels of biological organization—from phenotype to mutational mechanism. In this review, we describe recent pigmentation studies performed in natural populations that reveal how convergent phenotypes in different taxa can occur by the evolution of (i) different genes; (ii) the same gene and the same mutation; (iii) the same gene but different functionally distinct mutations; or (iv) the same gene but different functionally equivalent mutations (figure 1). In the discussion, we consider how identifying the precise mutations, genes and functional mechanisms responsible for convergent phenotypes will allow us to address fundamental questions about the genetic basis of adaptation and, more generally, the evolutionary process.
Figure 1.
Genetic convergence at multiple levels. Similar phenotypes, serving the same ecological function, can evolve by: (a) changes in different genes; (b) the same mutation in the same gene; (c) different mutations in the same gene that have similar functional consequences and (d) different mutations in the same gene that affect gene function or expression in different ways.
2. Pigmentation as a model system
Colour phenotype in most vertebrates is largely determined by two factors: the spatial distribution of pigments across the body and the type of pigments deposited along individual hairs, feathers or scales. Although the cellular and molecular events governing pigmentation patterning are poorly understood, the genetic and physiological bases of pigment synthesis and deposition have been widely studied in mammals (Jackson 1994; Jackson et al. 1994). The type of pigment produced is governed largely by the interaction of two genes: the melanocortin-1 receptor (Mc1r) and its antagonist, Agouti (Barsh 1996). The signalling activity of the transmembrane Mc1-receptor at the surface of pigment-producing cells (i.e. melanocytes) leads to the production of the dark (brown-black) pigment, eumelanin. The binding of its ligand, the secreted molecule Agouti, causes a switch to light (yellow-red) phaeomelanin production. Studies in Mus have shown that Agouti is likely to be involved in both pigment pattern and pigment-type switching by producing two transcriptional isoforms: the first is expressed in the ventral skin and is associated with dorsal–ventral differences in pigmentation (Bultman et al. 1994; Vrieling et al. 1994) and the second is expressed in a timed pulse during hair growth and is responsible for the banded hair pattern typical of most rodent hairs (Vrieling et al. 1994).
In addition to Mc1r and Agouti, a growing list of pigmentation genes identified in laboratory models provides a wealth of candidate genes to understand the mechanisms underlying the convergence of colour phenotypes at different levels of biological organization (Hoekstra 2006; Hofreiter & Schöneberg 2010; Hubbard et al. 2010). At the genetic level, mutations in genes involved in the same functional pathway may be more likely to produce comparable colour phenotypes (e.g. changes in the regulation of melanocyte migration generally cause differences in the distribution of pigments across the body, whereas changes in the enzymatic reactions governing melanin synthesis generally cause differences in the type and distribution of pigment along individual hairs). At the phenotypic level, different colour variants can serve the same ecological function: for example, both a reduction in the pigmented body regions and a switch to the lighter pigment type in hairs can provide camouflage in light-coloured habitats.
Here, we focus on the convergent evolution of colour phenotypes that confer the same ecological function: crypsis. Cryptic coloration minimizes detection by visually hunting predators. Arguably, the simplest form of crypsis is the correlation between dorsal pigmentation and substrate colour in terrestrial vertebrates (e.g. Dice & Blossom 1937). Because of the wide range of substrate colours—ranging from black lava flows to white sand dunes—that have formed independently in geographically dispersed regions, crypsis provides an ideal opportunity to examine phenotypic convergence.
3. Different genes produce similar phenotypes
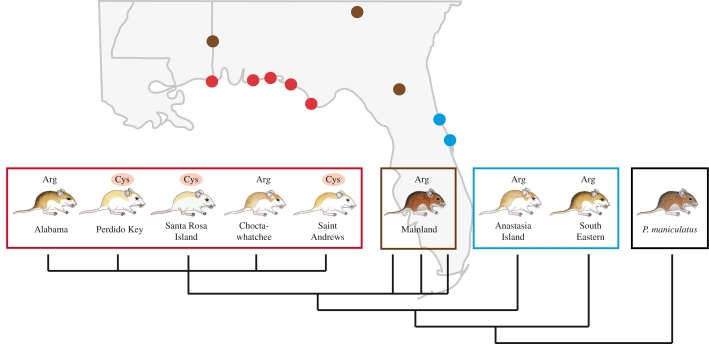
There are several known cases in which mutations in different genes can result in similar phenotypic effects. One striking example involves the dorsal coat colour of oldfield mice (Peromyscus polionotus), which closely match their local substrate and thereby reduce the attack rate of predators (Vignieri et al. 2010). These mice typically inhabit densely vegetated oldfields with dark loamy soils in the southeastern USA, where they have a dark brown dorsal coat and a grey ventrum. However, mice that have colonized the sandy coastal dunes of the Gulf and Atlantic coasts of Florida have a significantly lighter colour (figure 2; Howell 1920; Sumner 1929; Mullen & Hoekstra 2008). In general, these ‘beach mice’ (i) differ in the distribution of pigmentation, largely associated with an upward shift in their dorsal–ventral boundary, and (ii) in areas that are pigmented, have both a lower density of pigment and more light phaeomelanin than mainland mice (Steiner et al. 2007). Although all beach mice are lighter than mainland mice, different subspecies vary significantly in several pigmentation traits, and remarkably, Atlantic Coast subspecies are more similar in colour pattern to Gulf Coast subspecies than their neighbouring subspecies (Steiner et al. 2009). An intraspecific phylogeny of Gulf and Atlantic Coast subspecies, however, shows that beach mice are not monophyletic (figure 2), and the hypothesis that there was a single origin of the light ‘beach mouse’ phenotype can be rejected statistically (Steiner et al. 2009). Thus, the lighter overall colour of Gulf and Atlantic beach mice probably represents the independent evolution of camouflage. In this system, then, one can ask whether the same genes were targets of adaptive change multiple times or whether different genes were modified to produce similar phenotypic results.
Figure 2.
Light coloration in beach mice has evolved independently through changes in different genes. Map of the southeastern USA, in which circles represent collecting locales for P. polionotus: three mainland subspecies (brown), five Gulf Coast (red) and the two Atlantic Coast (blue) beach mouse subspecies. Atlantic and Gulf Coast beach mice have independently evolved lighter coloration, compared with their dark ancestor (cartoons). A mutation (Arg65Cys) in Mc1r contributes to light coat colour in Gulf Coast subspecies, but not in Atlantic Coast subspecies. The most common amino acid at site 65 in each subspecies is shown: ancestral (Arg) and derived (Cys, highlighted in light red). The schematized tree represents the phylogenetic relationships of P. polionotus subspecies (P. maniculatus is shown as an outgroup; adapted from Steiner et al. 2009).
As a first step in determining the genetic basis of colour variation in these mice, Hoekstra and co-workers generated a genetic intercross between a mainland subspecies (P. polionotus subgriseus) and the Santa Rosa Island beach mouse subspecies (P. p. leucocephalus) from Florida's Gulf Coast. Using a quantitative trait loci (QTL) approach, three genomic regions, which together explain most of the colour variation between the two subspecies, were identified (Steiner et al. 2007). These regions each contain a candidate pigment gene, Mc1r (Hoekstra et al. 2006), Agouti (Steiner et al. 2007) and Corin (Jacobs-Palmer et al. submitted), which encodes a serine protease acting upstream of Agouti signalling (Enshell-Seijffers et al. 2008). Changes in both Agouti and Corin mRNA expression level are statistically associated with colour differences, but the causal mutations in these loci have yet to be identified. Although there are no measurable differences in Mc1r expression levels between beach and mainland mice (Steiner et al. 2007), a single amino acid substitution in the first intracellular region of the Mc1r protein (Arg65Cys) is strongly associated with colour differences. Cell-based functional assays confirm that the Mc1r Arg65Cys mutation reduces ligand binding, causing a decrease in receptor signalling, and consequently a reduction in the production of dark eumelanin (figure 3b; Hoekstra et al. 2006). This single mutation explains approximately 30 per cent (depending on the specific trait) of the colour difference between mainland and beach mice in this cross. In nature, 65Cys is fixed and derived in Santa Rosa Island beach mice and absent from mainland populations (Hoekstra et al. 2006; Mullen et al. 2009).
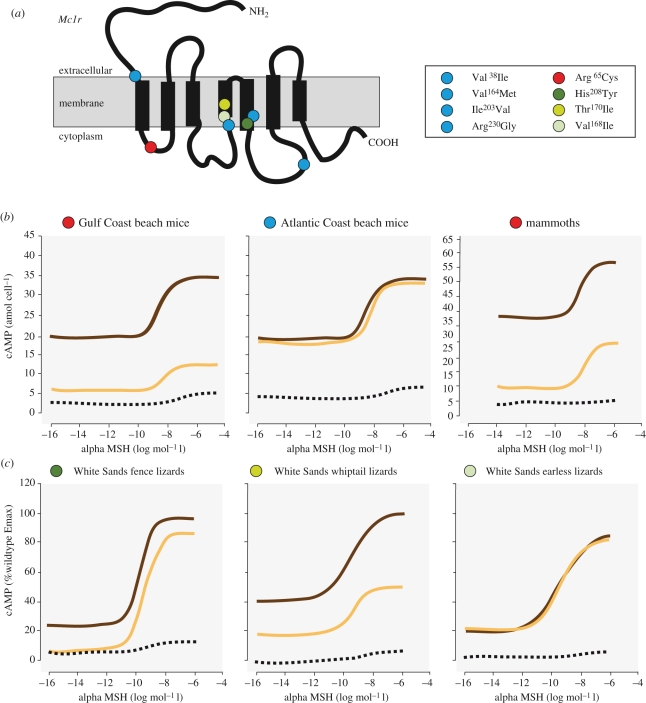
Figure 3.
Mutations in Mc1r change receptor signalling in some, but not all, species with light coloration. (a) Schematic of the Mc1r protein showing the position of amino acid variants in beach mice from Florida's Gulf (red) and Atlantic (blue) coasts, mammoths (red) and blanched lizards from White Sands, New Mexico (shades of green). Functional analysis of Mc1r alleles in (b) beach mice and mammoths and (c) lizards. Intracellular cyclic adenosine monophosphate (cAMP) accumulation was measured in response to increasing concentrations of the agonist alpha melanocyte-stimulating hormone (alpha MSH). For each taxon, the response curves for the dark Mc1r allele (brown), light allele (yellow) and control (black) are shown. Some mutations, but not all, cause a decrease in receptor signalling associated with lighter pigmentation (data taken from Hoekstra et al. 2006; Römpler et al. 2006; Steiner et al. 2009; Rosenblum et al. 2010).
By contrast, despite their similarities in colour pattern, the Atlantic Coast beach mice do not harbour the 65Cys mutation (Hoekstra et al. 2006), suggesting that the light pigment phenotype has a different mutational basis. Mc1r sequence comparison between the mainland and the Atlantic Coast beach mice revealed four new amino acid mutations. However, none of these mutations overlaps with previously described mutations known to affect coloration, none is fixed in an Atlantic Coast subspecies or strongly correlated with light coloration and none alone affected receptor function measured using in vitro assays (figure 3b; Steiner et al. 2009). Therefore, although it remains formally possible that Mc1r is responsible for light coats in Atlantic Coast mice (e.g. via a change in the regulatory region), results to date strongly suggest that different genes are responsible for the convergent phenotypes of the Atlantic and Gulf coasts beach mouse populations.
4. Same gene and mutation in different species produce similar phenotypes
Although different genetic paths can be taken within a species, the same mutation can sometimes contribute to phenotypic convergence between wildly different species (figure 1b). Several examples involve Mc1r (figure 3). In a recent ancient DNA study, the complete coding sequence of the Mc1r gene was sequenced from an approximately 43 000-year-old mammoth (Mammuthus primigenius) bone excavated in Siberia (Römpler et al. 2006). The sequenced individual was polymorphic at three amino acid sites, including the exact same mutation (C to T nucleotide and Arg to Cys amino acid) at the homologous position identified in the Gulf Coast beach mice (Arg65Cys). To functionally verify the effect of this mutation, Römpler et al. constructed expression vectors containing the two different Mc1r alleles and showed a difference in receptor signalling, similar in magnitude to that observed in beach mice (figure 3b). These results raise the possibility that Pleistocene mammoths were polymorphic for hair colour; in fact, both dark and light coloured mammoth hair has been recovered from permafrost mummies. However, the ecological relevance of mammoth colour variation remains unclear.
Although functional validation of Mc1r mutations is rare, other studies have shown that mutations at homologous sites of Mc1r are statistically associated with colour differences in divergent taxa. For example, bananaquits (Mundy et al. 2004), Japanese quail (Nadeau et al. 2006) and chickens (Ling et al. 2003) all share the same non-synonymous substitution (Glu92Lys), probably leading to Mc1r's constitutive activation and ultimately a melanic phenotype. Similarly, the Asp119Asn mutation is associated with melanism in Monarch flycatchers from the Solomon Island (Uy et al. 2009), domestic pigs (Kijas et al. 1998) and several strains of sheep (Vage et al. 1999). Finally, melanic arctic skuas (Mundy 2005) and melanic pocket mice (Nachman et al. 2003) both have an Arg233His mutation. In all of these cases, because the species sharing a common mutation are so divergent, mutational convergence probably represents the independent origin and subsequent selection of the same mutation.
5. Same gene but different mutations produce similar phenotypes
Similar phenotypes can also be produced by different mutations in the same gene. This can be accomplished in two distinct ways—the mutations either have the same effect on gene function (figure 1c) or have distinct effects on gene function but still produce ecologically equivalent phenotypes (figure 1d). Here we highlight two recent studies performed in lizards and mice—involving either coding-region mutations that directly affect protein function or regulatory mutations that modify gene expression—that illustrate how different mutations can produce ecologically equivalent phenotypes.
(a). Mutations in the coding region
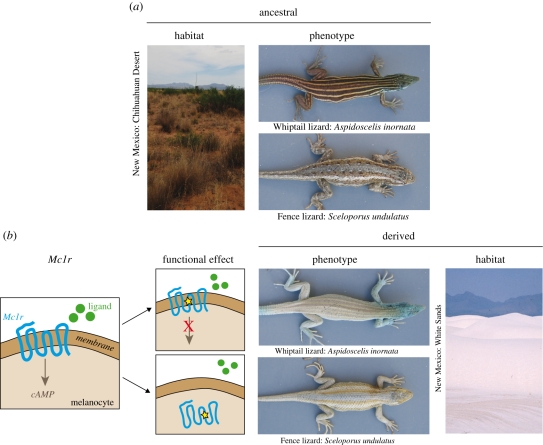
The White Sands of New Mexico represent a geologically young habitat (approx. 6000 years old) comprised of stark white gypsum sands. Three species of lizards inhabiting these sand dunes have evolved blanched dorsal coloration compared with their conspecifics in the surrounding Chihuahuan Desert, most likely as an adaptation for crypsis (Rosenblum 2006). Comparison of Mc1r sequences between light and dark forms of each species revealed a derived amino acid change in the blanched morph of all three species (Rosenblum et al. 2004). Cell-based functional assays have shown that there is no measurable effect of the derived Mc1r mutation in one species, the common lesser earless lizard (Holbrookia maculata), despite a strong statistical association between Mc1r mutation and colour (figure 3c; Rosenblum et al. 2010). This result demonstrates two points. First, functional verification of statistical associations is necessary before genes can be implicated in phenotypic change. Second, the blanched coloration in earless lizards is most likely caused by changes in a different gene(s), although, as is the case of the Atlantic Coast beach mice, further investigation is needed to rule out changes in Mc1r expression.
However, in the other two White Sands lizard species, the eastern fence lizard (Sceloporus undulatus) and the little striped whiptail (Aspidoscelis inornata), the derived amino acid replacement causes a change in Mc1-receptor activity (figure 4; Rosenblum et al. 2010). Moreover, the amino acid replacements in each species generate blanched phenotypes via entirely different functional mechanisms. In one species, the functional effect of the Mc1r mutation is similar to that observed in beach mice. Specifically, in whiptail lizards, the Thr170Ile mutation causes a reduction in Mc1r signalling similar in magnitude to the effects of the Arg65Cys mutation observed in Gulf Coast beach mice and mammoths (figure 3c). Thus in the comparison between whiptail lizards and beach mice, different mutations in the same gene produce similar blanched phenotypes via similar effects on gene function, in this case diminished receptor signalling (figure 1c). In fence lizards, the Mc1r mutation also results in a partial loss of function, affecting the transmembrane domain of the receptor. However, this mutation (His208Tyr) compromises the receptor's function via a different mechanism: the mutation in blanched fence lizards prevents Mc1r from efficiently integrating into the melanocyte membrane (as opposed to integrating efficiently but then impeding proper signal transduction; Rosenblum et al. 2010). Thus, in the comparison between fence and whiptail lizards, different mutations in the same gene produce similar blanched phenotypes, but through different functional mechanisms (figure 1d).
Figure 4.
Genetic convergence and functional divergence in lizards. (a) In the Chihuahuan Desert of New Mexico, both the little striped whiptail (A. inornata) and the eastern fence lizard (S. undulatus) have a dark dorsal colour that closely matches the local soil colour. (b) Compared with their dark counterparts, a blanched phenotype has independently evolved in whiptail and fence lizards that colonized the dunes of White Sands. In both species, the derived phenotype results from an amino acid mutation in Mc1r (schematized). In the whiptail lizard, the derived mutation impairs Mc1r signalling activity (top right panel). In the fence lizard, a different mutation diminishes the efficiency of Mc1r integration into the melanocyte membrane (bottom right panel). Phenotypic convergence (blanched colour) is reflected at the genetic level (mutations in the same gene, Mc1r), but not at the functional level (mutations have different effects on protein function).
(b). Mutations in the regulatory region
Far from the white sandy Florida beaches where its sister species, P. polionotus, evolved a light coat colour, deer mice (P. maniculatus) living on the pale dunes of the Nebraska Sand Hills also have dorsal coats that match their local habitat. Like beach mice, this adaptive pigmentation change has evolved recently (less than 8000 years ago), and visual predators are likely the agent of selection favouring a cryptic, light coat colour (figure 5; Dice 1947). The golden colour of Sand Hills mice is caused primarily by a change in the type and distribution of pigments on individual hairs (figure 5c). Like the hairs of most rodents, Peromyscus dorsal hairs contain a short, subterminal band of light phaeomelanic pigment on an otherwise dark eumelanic background. In Sand Hills mice, this light band is markedly wider than in their darker conspecifics.
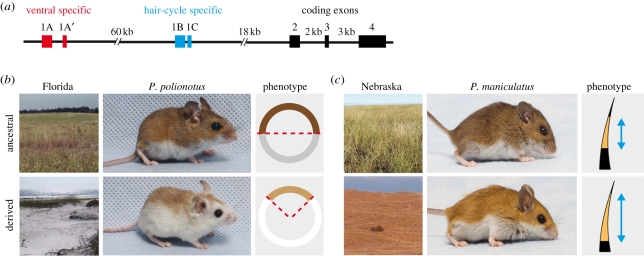
Figure 5.
Genetic convergence and functional divergence in deer mice. (a) The Agouti gene produces two transcriptional isoforms. Both contain exons 2 to 4, but they differ in the first, untranslated exons—the ventral-specific isoform contains exons 1A and 1A′, whereas the hair-cycle-specific isoform contains exons 1B and 1C. In populations of two sister species, (b) P. polionotus and (c) P. maniculatus, derived mutations in the Agouti gene result in an overall lighter coat colour, which is cryptic in novel habitat (the sand dunes of the Florida Coast or Nebraska's Sand Hills, respectively). However, Agouti mutations act through different changes in gene expression: in beach mice, changes in Agouti expression may primarily modify colour pattern, whereas in Sand Hills mice, Agouti mutations mainly modify pigment type and distribution on individual hairs (i.e. a wider phaeomelanin band). These results suggest that different isoforms might be targeted—the ventral Agouti isoform in beach mice and the hair-cycle-specific isoform in Sand Hills mice.
Laboratory crosses show that this ‘wideband’ phenotype (McIntosh 1956) is inherited as a single dominant allele (Dodson 1982) and that there is a perfect association between this phenotype and nucleotide variation in the Agouti gene, but not at other loci (Linnen et al. 2009). Moreover, Agouti mRNA levels are higher in P. maniculatus carrying the wideband allele than in their wild-type counterparts. This expression difference is maintained in wideband/wild-type heterozygotes, demonstrating that a cis-acting mutation(s) in, or linked to, the Agouti gene is causal (Linnen et al. 2009). These findings are consistent with Agouti's role in producing banded hairs. Specifically, when Agouti is expressed, melanocytes at the base of hair follicles switch from eumelanin to phaeomelanin production. As hairs grow, a pulse of Agouti expression results in the formation of phaeomelanin bands (Vrieling et al. 1994). In Sand Hills mice, phaeomelanin bands are widened because this pulse of Agouti expression during hair growth is both longer and higher than in wild-type mice (Linnen et al. 2009).
Nebraska P. maniculatus and Florida P. polionotus (beach mice) thus represent an example of phenotypic convergence, in which overall light colour is an adaptation for crypsis in light-coloured environments. However, in this case, the same gene, Agouti, causes light coloration through distinct mutations leading to distinct functional changes (recall that a change in Agouti expression also contributes to light coloration in beach mice). Moreover, contrary to the case of the White Sands lizards, the functional change in both mice species appears to alter patterns of gene expression, not protein function. These changes in Agouti expression produce ecologically equivalent, but distinct, phenotypes: although the light beach mouse pelage is due in large part to a pronounced change in pigment patterning (i.e. a change in dorsal–ventral coloration), the light Sand Hills coat is produced by an altered distribution of pigments on individual hairs (figure 5). One simple hypothesis that explains how expression changes in the same gene produce these different phenotypic outcomes is that the causal mutations affect the two different Agouti mRNA isoforms (figure 5a). While the causal Agouti mutations in both P. polionotus and P. maniculatus remain to be identified, association studies in both species are underway.
6. Discussion
Together, these studies on adaptive colour evolution performed in mice, mammoths and lizards illustrate how convergence at one level (i.e. the gene) does not necessarily imply convergence at other mechanistic levels (e.g. the mutation, gene function or gene expression). Evaluating convergence across levels is thus not merely academic: even when the same gene is involved, differences in the functional effects of different mutations can have important organism-level consequences. As we discuss here, extending the study of convergence to these additional levels has the potential to reveal novel insights about both the proximate and ultimate mechanisms underlying phenotypic convergence.
7. Convergence at the genetic level: lessons from studies of Mc1r
Just as pigmentation is an excellent model system in which to study genetic convergence, the case studies we present here highlight that Mc1r, with its simple genetic structure (i.e. a single coding exon) and well-understood protein function, is emerging as a model gene for understanding convergence at the mechanistic level. To date, Mc1r studies in a wide range of taxa have unequivocally demonstrated that the same gene can be responsible for convergent phenotypes. Beyond demonstrating widespread convergence at the gene level, these studies allow for an investigation of how often the same mutation versus a different mutation with an equivalent functional effect gives rise to similar phenotypes.
(a). Mutational convergence
For convergence to span all levels of biological organization, the same mutation must occur more than once, have the same functional and phenotypic consequences on different genetic backgrounds and be positively selected in different populations or species experiencing similar ecological pressures (figure 1b). Examples of mutational convergence (e.g. mice and mammoths) suggest that evolution is highly repeatable. However, we must be cautious in claiming convergence at the mutational level because observing the same causal mutation in multiple taxa does not necessarily imply that the mutation arose more than once. Two alternatives to mutational convergence include: (i) the causal mutation was present in an ancestral population (e.g. Colossimo et al. 2005; Barrett & Schluter 2008) or (ii) the causal mutation arose in one lineage and was transferred to another lineage via hybridization and introgression (e.g. Anderson et al. 2009). Although these two alternatives might still represent convergence at the phenotypic level (i.e. there have been independent bouts of selection), they are not caused by independent mutational events.
Fortunately, we can distinguish between ancestral variation, introgression and mutational convergence by examining evolutionary histories for both the causal alleles and the populations/species in which they occur. Specifically, both ancestral variation and introgression predict that haplotypes bearing the causal mutation will form a monophyletic group, whereas mutational convergence predicts that haplotypes will not be monophyletic. For Mc1r, several studies identified the exact same mutation in taxa distantly related enough that shared ancestral variation and introgression are highly unlikely, providing examples of ‘complete’ convergence (figure 1b). Mechanisms for the same mutation appearing repeatedly in different taxa could be: mutational hotspots (e.g. Chan et al. 2010), few mutational options for achieving particular phenotypes (e.g. Bull et al. 1997) and/or a relatively large net selection coefficient for a particular mutation on multiple genetic backgrounds.
(b). Mutational divergence (but functional convergence)
Different mutations—either in the same gene or in different genes—can lead to the same functional effect on protein function and/or expression and thus to the same phenotypic change. Mutational divergence but functional convergence is illustrated by the identification, in mammals and lizards, of different Mc1r amino acid substitutions that result in the same effect on Mc1r signalling activity (figure 3). Perhaps the simplest, and most common, type of functional convergence at the genetic level is a complete disruption of gene function caused by separate mutations (e.g. a premature stop codon or deletion that produces a non-functional protein product). For example, independent deletions in the OCA2 gene lead to repeated loss of protein function and thus pigmentation in cavefish (Protas et al. 2006), and deletions in Agouti repeatedly lead to melanism in deer mice (Kingsley et al. 2009; discussed subsequently). Surprisingly, despite the large number of potential knock-out mutations in Mc1r (i.e. it is a large mutational target), such mutations have not been reported in natural populations. Three potential explanations include: Mc1r null alleles (i) exist in nature but have not yet been described, (ii) are likely to be completely recessive and therefore more likely to be lost to drift, and (iii) have negative pleiotropic effects on fitness (e.g. in addition to its role in pigmentation, Mc1r affects pain tolerance; Mogil et al. 2005). Distinguishing among these alternatives will require identifying additional Mc1r alleles in natural populations and characterizing their phenotypic effects (on colour and other traits) in both heterozygotes and homozygotes. By focusing on the genetic bases of functional convergence (i.e. determining what types of mutations exist in nature and are selectively favoured and what types of mutations do not appear in the wild due to their patterns of dominance or pleiotropic effects), we can gain insight into constraints acting at the genetic level to shape the evolution of genes and populations.
(c). Mutational divergence (and functional divergence)
As the functional dissection of genetic and phenotypic convergence in lizard Mc1r and mouse Agouti vividly illustrates, the same genes can create ecologically equivalent traits via different functional mechanisms. Moreover, these examples show that separate mutations can have divergent functional effects on gene/protein function (Mc1r in lizards) and on gene expression (Agouti in mice). Importantly, mutations with different functional effects, even when they occur in the same gene, can lead to differences in trait expression. For example, in the White Sands lizards, Mc1r mutations with distinct functional effects—impaired protein signalling or membrane integration efficiency—can lead to blanched coloration. However, the functional consequences of the mutations lead to differences in allelic dominance: light colour is dominant in one species, but largely recessive in the other (Rosenblum et al. 2010). These differences in dominance can affect the chance that a new allele is lost due to drift, the rate of allele frequency change driven by selection and the spatial distribution of alleles in nature. In addition to dominance, other important genetic parameters are allele-specific, such as epistasis and pleiotropy, which also contribute to net selection coefficients. Therefore, understanding the functional effects of mutations can inform our understanding of the likelihood a particular allele will be favoured and thus be involved in adaptive phenotypic change.
8. Divergence at the genetic level: comparisons among pigmentation genes
(a). Comparison of Mc1r and Agouti
Like Mc1r, Agouti has also been implicated in pigmentation differences in several organisms (Kingsley et al. 2009 and references therein), and interesting parallels—and differences—emerge by comparing phenotypic convergence caused by these two loci. First, just as we observe functional convergence between two different mutations in Mc1r in lizards and mice, independent changes that similarly affect Agouti function have been identified in deer mice. For example, convergence at the functional level for Agouti was reported in several populations of P. maniculatus: melanic phenotypes are repeatedly produced by independent deletions in Agouti (Kingsley et al. 2009). These deletions eliminate Agouti expression and function. As noted earlier, full loss-of-function mutations have not been described in nature for the more intensively studied Mc1r. Second, like Mc1r, we observed functional divergence between two different mutations in Agouti that contribute to adaptive colour variation (in beach and Sand Hills mice). Unlike Mc1r, however, these mutations alter gene expression, not protein function, suggesting that adaptive changes in expression may be more common in Agouti than in Mc1r. Together, these examples illustrate how comparison of convergence and divergence in mutation and function across different genes can shed light on the functional flexibility of the genes of interest, thereby allowing us to hypothesize about the relative role that different genes play in adaptive evolution. In contrast, because selection acts on mutations (i.e. alleles, not genes) that can have different functional consequences, and hence can vary in dominance and pleiotropy, which in turn depend on the genetic background where they occur, evolution may be exceedingly difficult to predict. Testing these alternatives requires that we identify genes (and mutations) that contribute to natural phenotypic variation in an unbiased manner (Kopp 2009).
(b). Other pigmentation genes
The examples discussed in this review, along with several other studies (reviewed in Hubbard et al. 2010), point to the recurrent role of a few genes (i.e. Mc1r, and to a lesser extent, Agouti) in producing colour differences among vertebrates. Although the repeated implication of Mc1r in colour variation may reflect real biological processes (e.g. constraint, dominance and minimal pleiotropy), it may also stem, at least in part, from ascertainment bias. Thus, in addition to performing detailed functional analyses of candidate pigmentation genes, we also need to perform genome-wide studies in a diverse group of taxa to identify new pigmentation alleles (Kopp 2009). In fact, the two approaches are very complementary: genomic approaches provide a non-biased way to evaluate the relative importance of different pigmentation genes in generating natural colour variation and allow us to test the predictions generated by functional studies of candidate genes. To date, several genes underlying differences in pigmentation have been identified using an unbiased genome-wide approach (table 1). These genomic studies show that a diversity of genes, beyond just Mc1r and Agouti, contribute to colour variation in natural populations.
Table 1.
Genes involved in vertebrate pigment variation identified by an unbiased genomic approach.
| species | phenotype | gene | reference |
|---|---|---|---|
| oldfield mouse (Peromyscus polionotus) | pigment pattern | Agouti | Steiner et al. (2007) |
| Mc1r | |||
| Corin | Jacobs-Palmer et al. (submitted) | ||
| cavefish (Astyanax species) | lack of pigment | Oca2 | Protas et al. (2006) |
| Mc1r | Gross et al. (2009) | ||
| threespine stickleback fish (Gasterosteus aculeatus) | reduced pigment | Kitlg | Miller et al. (2007) |
| Lake Malawi cichlid fish (Labeotropheus trewavasae, Metriaclima zebra, M. xantomachus, Tropheops sp.) | increased pigment | Pax7 | Roberts et al. (2009) |
| North American gray wolf (Canis lupus)a | increased pigment | K locus | Anderson et al. (2009) |
| Soay sheep (Ovis aries) | pigment pattern and type | Tyrp1 | Gratten et al. (2006) |
| Agouti | Beraldi et al. (2006) | ||
| human (Homo sapiens) | pigment type | Slc24A5 | Stokowski et al. (2007) |
| Slc45A2 | Sabeti et al. (2007) | ||
| Slc24A4 | Han et al. (2008) | ||
| Tyrp1 | Sulem et al. (2007, 2008) | ||
| IRF4 | |||
| Mc1r | |||
| Agouti | |||
| pigment type | Kitlg | Sulem et al. (2007, 2008) | |
| Slc24A4 | Han et al. (2008) | ||
| Oca2 | |||
| TPCN2 | |||
| IRF4 | |||
| Mc1r | |||
| Agouti |
aThe K locus was identified by a QTL study as a contributor to pigmentation differences in dogs (Kerns et al. 2007). Introgression of this locus in wolf has been shown subsequently.
9. Conclusion
Biologists have long wondered how often the same genes are responsible for the repeated evolution of similar traits. However, the studies we highlight here demonstrate how pinpointing the mutations—not just the genes—underlying convergent phenotypes allows us to test for convergence at levels both below (mutational convergence) and above (functional convergence) the level of the gene. Expanding our view to these additional levels not only deepens our understanding of the proximate mechanisms that generate adaptive phenotypic variation, but also provides novel insights into why (e.g. mutational target size, mutational hotspots, dominance, epistasis and pleiotropy) some evolutionary outcomes are more common than others. Of course, convergence is not limited to the levels of biological organization we discuss here. For example, between the levels of mutation and gene, the same or different nucleotide mutations can produce convergent amino acid substitutions, and above the level of the gene, the same or different genes in the same developmental pathway can have similar impacts on gene interactions and ultimately phenotype. Likewise, phenotypic convergence can be examined at different levels—at a fine grain, we can ask how similar the distribution of pigments are on individual hairs, and at a coarse grain, we can ask how similar the phenotypes are in ecological function. Thus, while evolutionary change is still far from being predictable, the well-studied cases presented here show that evolution can sometimes be repeatable—often the same genes are targeted for adaptive change, but the precise mutations and their effects on protein function can differ.
Acknowledgements
The authors wish to thank B. Charlesworth and M. Bonsall for organizing a fantastic scientific meeting and this symposium volume. J. Losos and one anonymous reviewer kindly provided comments. C.R.L. was supported by a Ruth Kirschstein National Research Service Award from the National Institutes of Health; V.S.D. was supported by a Portuguese Foundation for Science & Technology Fellowship. This research was largely funded by the National Science Foundation (grants to E.B.R. and H.E.H.).
Footnotes
One contribution of 18 to a Discussion Meeting Issue ‘Genetics and the causes of evolution: 150 years of progress since Darwin’.
References
- Anderson T. M., et al. 2009Molecular and evolutionary history of melanism in North American gray wolves. Science 323, 1339–1343 (doi:10.1126/science.1165448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt J., Reznick D.2008Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 23, 26–32 (doi:10.1016/j.tree.2007.09.011) [DOI] [PubMed] [Google Scholar]
- Barrett R. D. H., Schluter D.2008Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44 (doi:10.1016/j.tree.2007.09.008) [DOI] [PubMed] [Google Scholar]
- Barsh G. S.1996The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 12, 299–305 (doi:10.1016/0168-9525(96)10031-7) [DOI] [PubMed] [Google Scholar]
- Beraldi D., McRae A. F., Gratten J., Slate J., Visscher P. M., Pemberton J. M.2006Development of a linkage map and mapping of phenotypic polymorphisms in a free-living population of Soay sheep (Ovis aries). Genetics 173, 1521–1537 (doi:10.1534/genetics.106.057141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge T. A., Gillespie R. G.2004Convergent evolution of behavior in an adaptive radiation of Hawaiian web-building spiders. Proc. Natl Acad. Sci. USA 101, 16 228–16 233 (doi:10.1073/pnas.0407395101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower A. V.1994Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl Acad. Sci. USA 91, 6491–6495 (doi:10.1073/pnas.91.14.6491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J. J., Badgett M. R., Wichman H. A., Huelsenbeck J. P., Hillis D. M., Gulati A., Ho C., Molineux I. J.1997Exceptional convergent evolution in a virus. Genetics 147, 1497–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S. J., Klebig M. L., Michaud E. J., Sweet H. O., Davisson M. T., Woychik R. P.1994Molecular analysis of reverse mutations from nonagouti (a) to black-and-tan (a(t)) and white-bellied agouti (Aw) reveals alternative forms of agouti transcripts. Genes Dev. 8, 481–490 (doi:10.1101/gad.8.4.481) [DOI] [PubMed] [Google Scholar]
- Chan Y. F., et al. 2010Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305 (doi:10.1126/science.1182213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colossimo P. F., et al. 2005Widespread parallel evolution in sticklebacks by repeated fixation of ecotdysplasin alleles. Science 307, 1928–1933 (doi:10.1126/science.1107239) [DOI] [PubMed] [Google Scholar]
- Conway Morris S.2003Life's solution: inevitable humans in a lonely universe. Cambridge, UK: Cambridge University Press [Google Scholar]
- Dice L.1947Effectiveness of selection by owls of deer mice (Peromyscus maniculatus) which contrast in color with their background. Contrib. Lab. Vert. Biol. Univ. Michigan 34, 1–20 [Google Scholar]
- Dice L., Blossom P. M.1937Studies of mammalian ecology in southwestern North America, with special attention to the colors of the desert mammals. Publ. Carn. Inst. 485, 1–25 [Google Scholar]
- Dodson K. M.1982Genetic linkage relationships among several coat color mutations in the deer mouse (Peromyscus maniculatus). Thesis, Department of Biology, University of South Carolina [Google Scholar]
- Ellegren H., Sheldon B. C.2008Genetic basis of fitness differences in natural populations. Nature 452, 169–175 (doi:10.1038/nature06737) [DOI] [PubMed] [Google Scholar]
- Enshell-Seijffers D., Lindon C., Morgan B. A.2008The serine protease Corin is a novel modifier of the Agouti pathway. Development 135, 217–225 (doi:10.1242/dev.011031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N., Prud'homme B.2009The causes of repeated genetic evolution. Dev. Biol. 332, 36–47 (doi:10.1016/j.ydbio.2009.04.040) [DOI] [PubMed] [Google Scholar]
- Gould S. J.2002The structure of evolutionary theory. Cambridge, MA: Harvard University Press [Google Scholar]
- Gratten J., Beraldi D., Lowder B. V., McRae A. F., Visscher P. M., Pemberton J. M., Slate J.2006Compelling evidence that a single nucleotide substitution in Tyrp1 is responsible for coat-colour polymorphism in a free-living population of Soay sheep. Proc. R. Soc. B 274, 619–626 (doi:10.1098/rspb.2006.3762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B., Borowsky R., Tabin C. J.2009A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 5, e1000326 (doi:10.1371/journal.pgen.1000326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. L., et al. 2008A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 4, e1000074 (doi:10.1371/journal.pgen.1000074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey P. H., Pagel M. D.1991The comparative method in evolutionary biology. Oxford, UK: Oxford University Press [Google Scholar]
- Hoekstra H. E.2006Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 97, 222–234 (doi:10.1038/sj.hdy.6800861) [DOI] [PubMed] [Google Scholar]
- Hoekstra H. E., Coyne J. A.2007The locus of evolution: evo devo and the genetics of adaptation. Evolution 61, 995–1016 (doi:10.1111/j.1558-5646.2007.00105.x) [DOI] [PubMed] [Google Scholar]
- Hoekstra H. E., Hirschmann R. J., Bundey R. A., Insel P. A., Crossland J. P.2006A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313, 101–104 (doi:10.1126/science.1126121) [DOI] [PubMed] [Google Scholar]
- Hofreiter M., Schöneberg T.2010The genetic and evolutionary basis of colour variation in vertebrates. Cell. Mol. Life Sci. (doi:10.1007/s00018-010-0333-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A. H.1920Description of a new species of beach mouse in Florida. J. Mamm. 1, 237–240 (doi:10.2307/1373248) [Google Scholar]
- Hubbard J. K., Uy J. A., Hauber M. E., Hoekstra H. E., Safran R. J.2010Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet. 26, 231–239 (doi:10.1016/j.tig.2010.02.002) [DOI] [PubMed] [Google Scholar]
- Jackson I. J.1994Molecular and developmental genetics of mouse coat color. Annu. Rev. Genet. 28, 189–217 (doi:10.1146/annurev.ge.28.120194.001201) [DOI] [PubMed] [Google Scholar]
- Jackson I. J., Budd P., Horn J. M., Johnson R., Raymond S., Steel K.1994Genetics and molecular biology of mouse pigmentation. Pigm. Cell Res. 7, 73–80 (doi:10.1111/j.1600-0749.1994.tb00024.x) [DOI] [PubMed] [Google Scholar]
- Jacobs-Palmer E., Domingues V. S., Manceau M., Hoekstra H. E.Submitted Ascending a fitness peak: the fixation of three adaptive pigmentation alleles. [Google Scholar]
- Kerns J. A., et al. 2007Linkage and segregation analysis of black and brindle coat color in domestic dogs. Genetics 176, 1679–1689 (doi:10.1534/genetics.107.074237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijas J. M. H., Wales R., Tornsten A., Chardon P., Moller M., Andersson L.1998Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 150, 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley E. P., Manceau M., Wiley C. D., Hoekstra H. E.2009Melanism in Peromyscus is caused by independent mutations in agouti. PLoS ONE 4, e6435 (doi:10.1371/journal.pone.0006435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A.2009Metamodels and phylogenetic replication: a systematic approach to the evolution of developmental pathways. Evolution 63, 2771–2789 (doi:10.1111/j.1558-5646.2009.00761.x) [DOI] [PubMed] [Google Scholar]
- Ling M. K., Lagerstrom M. C., Fredriksson R., Okimoto R., Mundy N. I., Takeuchi S., Schioth H. B.2003Association of feather colour with constitutively active melanocortin 1 receptors in chicken. Eur. J. Biochem. 270, 1441–1449 (doi:10.1046/j.1432-1033.2003.03506.x) [DOI] [PubMed] [Google Scholar]
- Linnen C. R., Kingsley E. P., Jensen J. D., Hoekstra H. E.2009On the origin and spread of an adaptive allele in deer mice. Science 325, 1095–1098 (doi:10.1126/science.1175826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos J. B., Jackman T. R., Larson A., Queiroz K., Rodriguez-Schettino L.1998Contingency and determinism in replicated adaptive radiations of island lizards. Science 279, 2115–2118 (doi:10.1126/science.279.5359.2115) [DOI] [PubMed] [Google Scholar]
- Mackay T. F., Stone E. A., Ayroles J. F.2009The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10, 565–577 (doi:10.1038/nrg2612) [DOI] [PubMed] [Google Scholar]
- McIntosh W. B.1956Linkage in Peromyscus and sequential tests for independent assortments. Contrib. Lab. Vert. Biol. Univ. Michigan 73, 1–27 [Google Scholar]
- Miller C. T., Beleza S., Pollen A. A., Schluter D., Kittles R. A., Shriver M. D., Kingsley D. M.2007Cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell 131, 1179–1189 (doi:10.1016/j.cell.2007.10.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil J. S., et al. 2005Melanocortin-1 receptor gene variants affect pain and μ-opioid analgesia in mice and humans. J. Med. Genet. 42, 583–587 (doi:10.1136/jmg.2004.027698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen L. M., Hoekstra H. E.2008Natural selection along an environmental gradient: a classic cline in mouse pigmentation. Evolution 62, 1555–1570 (doi:10.1111/j.1558-5646.2008.00425.x) [DOI] [PubMed] [Google Scholar]
- Mullen L. M., Vignieri S. N., Gore J. A., Hoekstra H. E.2009Adaptive basis of geographic variation: genetic, phenotypic and environmental differences among beach mouse populations. Proc. R. Soc. B 276, 3809–3818 (doi:10.1098/rspb.2009.1146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy N. I.2005A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc. R. Soc. B 272, 1633–1640 (doi:10.1098/rspb.2005.3107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy N. I., Badcock N. S., Hart T., Scribner K., Janssen K., Nadeau N. J.2004Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science 303, 1870–1873 (doi:10.1126/science.1093834) [DOI] [PubMed] [Google Scholar]
- Nachman M. W., Hoekstra H. E., D'Agostino S. L.2003The genetic basis of adaptive melanism in pocket mice. Proc. Natl Acad. Sci. USA 100, 5268–5273 (doi:10.1073/pnas.0431157100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau N. J., Minvielle F., Mundy N. I.2006Association of a Glu92Lys substitution in MC1R with extended brown in Japanese quail (Coturnix japonica). Anim. Genet. 37, 287–289 (doi:10.1111/j.1365-2052.2006.01442.x) [DOI] [PubMed] [Google Scholar]
- Protas M. E., Patel N. H.2008Evolution of coloration patterns. Annu. Rev. Cell Dev. Biol. 24, 425–446 (doi:10.1146/annurev.cellbio.24.110707.175302) [DOI] [PubMed] [Google Scholar]
- Protas M. E., Hersey C., Kochanek D., Zhou Y., Wilkens H., Jeffery W. R., Zon L. I., Borowsky R., Tabin C. J.2006Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Genet. 38, 107–111 (doi:10.1038/ng1700) [DOI] [PubMed] [Google Scholar]
- Roberts R. B., Ser J. R., Kocher T. D.2009Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 326, 998–1001 (doi:10.1126/science.1174705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römpler H., Rohland N., Lalueza-Fox C., Willerslev E., Kuznetsova T., Rabeder G., Bertranpetit J., Schoneberg T., Hofreiter M.2006Nuclear gene indicates coat-color polymorphism in mammoths. Science 313, 62 (doi:10.1126/science.1128994) [DOI] [PubMed] [Google Scholar]
- Rosenblum E. B.2006Convergent evolution and divergent selection: lizards at the White Sands ecotone. Am. Nat. 167, 1–15 (doi:10.1086/498397) [DOI] [PubMed] [Google Scholar]
- Rosenblum E. B., Hoekstra H. E., Nachman M. W.2004Adaptive reptile color variation and the evolution of the Mc1r gene. Evolution 58, 1794–1808 [DOI] [PubMed] [Google Scholar]
- Rosenblum E. B., Römpler H., Schöneberg T., Hoekstra H. E.2010Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc. Natl Acad. Sci. USA 107, 2113–2117 (doi:10.1073/pnas.0911042107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C., et al. 2007Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (doi:10.1038/nature06250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J., Gratten J., Beraldi D., Stapley J., Hale M., Pemberton J. M.2009Gene mapping in the wild with SNPs: guidelines and future directions. Genetica 136, 97–107 (doi:10.1007/s10709-008-9317-z) [DOI] [PubMed] [Google Scholar]
- Steiner C. C., Weber J. N., Hoekstra H. E.2007Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 5, 1880–1889 (doi:10.1371/journal.pbio.0050219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner C. C., Römpler H., Boettger L. M., Schöneberg T., Hoekstra H. E.2009The genetic basis of phenotypic convergence in beach mice: similar pigment patterns but different genes. Mol. Biol. Evol. 26, 35–45 (doi:10.1093/molbev/msn218) [DOI] [PubMed] [Google Scholar]
- Stern D. L., Orgogozo V.2008The loci of evolution: how predictable is genetic evolution? Evolution 62, 2155–2177 (doi:10.1111/j.1558-5646.2008.00450.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. L., Orgogozo V.2009Is genetic evolution predictable? Science 323, 746–751 (doi:10.1126/science.1158997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe J. R., Hoekstra H. E.2008Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity 100, 158–170 (doi:10.1038/sj.hdy.6800937) [DOI] [PubMed] [Google Scholar]
- Stokowski R., et al. 2007A genomewide association study of skin pigmentation in a South Asian population. Am. J. Hum. Genet. 81, 1119–1132 (doi:10.1086/522235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P., et al. 2007Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 39, 1443–1452 (doi:10.1038/ng.2007.13) [DOI] [PubMed] [Google Scholar]
- Sulem P., et al. 2008Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 40, 835–837 (doi:10.1038/ng.160) [DOI] [PubMed] [Google Scholar]
- Sumner F. B.1929The analysis of a concrete case of intergradation between two subspecies. Proc. Natl Acad. Sci. USA 15, 110–120 (doi:10.1073/pnas.15.2.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uy J. A., Moyle R. G., Filardi C. E., Cheviron Z. A.2009Difference in plumage color used in species recognition between incipient species is linked to a single amino acid substitution in the melanocortin-1 receptor. Am. Nat. 174, 244–254 (doi:10.1086/600084) [DOI] [PubMed] [Google Scholar]
- Vage D. I., Klungland H., Lu D., Cone R. D.1999Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm. Genome 10, 39–43 (doi:10.1007/s003359900939) [DOI] [PubMed] [Google Scholar]
- Vignieri S. N., Larson J., Hoekstra H. E.2010The selective advantage of cryptic coloration in mice. Evolution. (doi:10.1111/j.1558-5646.2010.00976.x) [DOI] [PubMed] [Google Scholar]
- Vrieling H., Duhl D. M., Millar S. E., Miller K. A., Barsh G. S.1994Differences in dorsal and ventral pigmentation result from regional expression of the mouse agouti gene. Proc. Natl Acad. Sci. USA 91, 5667–5671 (doi:10.1073/pnas.91.12.5667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake D. B.1991Homoplasy—the result of natural selection, or evidence of design limitations. Am. Nat. 138, 543–567 (doi:10.1086/285234) [Google Scholar]
- Wang I. J., Shaffer H. B.2008Rapid color evolution in an aposematic species: a phylogenetic analysis of color variation in the strikingly polymorphic strawberry poison-dart frog. Evolution 62, 2742–2759 (doi:10.1111/j.1558-5646.2008.00507.x) [DOI] [PubMed] [Google Scholar]