Abstract
Recent advances in the quantitative genetics of traits in wild animal populations have created new interest in whether natural selection, and genetic response to it, can be detected within long-term ecological studies. However, such studies have re-emphasized the fact that ecological heterogeneity can confound our ability to infer selection on genetic variation and detect a population's response to selection by conventional quantitative genetics approaches. Here, I highlight three manifestations of this issue: counter gradient variation, environmentally induced covariance between traits and the correlated effects of a fluctuating environment. These effects are symptomatic of the oversimplifications and strong assumptions of the breeder's equation when it is applied to natural populations. In addition, methods to assay genetic change in quantitative traits have overestimated the precision with which change can be measured. In the future, a more conservative approach to inferring quantitative genetic response to selection, or genomic approaches allowing the estimation of selection intensity and responses to selection at known quantitative trait loci, will provide a more precise view of evolution in ecological time.
Keywords: selection, evolution, estimated breeding value, environmental change, climate change, response to selection
1. Introduction
Can we observe evolutionary change in quantitative traits by natural selection in natural populations in real time? By ‘evolutionary change’ I mean genetic change over generations and by ‘real time’ I mean across the few decades for which some long-term ecological research projects have run. In this article, I will briefly review the prospects for this ambition, particularly highlighting some ways in which quantitative genetic approaches are short-circuited by ecological heterogeneity.
Recent years have seen a burst of activity in the area of quantitative genetic analysis of wild animal populations, enabled by two separate kinds of development. First, for several long-term, individual-based, ecological studies, it is possible to derive a multi-generational pedigree of individuals, either from field cues or using molecular techniques (Pemberton 2008), allowing estimation of coefficients of kinship between large numbers of individuals. Second, the use of tools from animal breeding, particularly a special kind of mixed model, the ‘animal model’, and restricted maximum likelihood, allows efficient estimation of genetic parameters from the unbalanced data and complex pedigree structures typical of field projects (Kruuk 2004). Pre-existing ambitions to estimate selection on traits and predict and measure evolution have increased alongside the developments outlined above. In principle, if a trait is shown to be under selection, and it is heritable, then surely it should respond? It is the realities of this process that I address in this paper.
The need to understand what is and is not genetic change owing to evolution by natural selection is pressing. Environments are changing rapidly, not only due to climate change, but also for other anthropogenic reasons. Populations of organisms are also changing rapidly, for example in terms of phenology and life-history traits (e.g. Olsen et al. 2004; Thackeray et al. in press) and a number of authors have emphasized the abundance and importance of evolutionary change as a cause of phenotypic change (Hendry & Kinnison 1999; Hairston et al. 2005; Kinnison & Hairston 2007; Pelletier et al. 2009). But ecological effects are often large and they are also capable of persisting across generations (for review see Rossiter 1996). Many study organisms do not lend themselves to the most diagnostic test of genetic change, the common garden experiment, leaving very substantial room for doubt about whether a change in mean phenotype is due to genetic change in response to natural selection.
2. Mechanisms underlying trait change
If the mean value of a trait changes over time in a population that is monitored over multiple years, there are four non-mutually exclusive processes that could cause such change.
(a). Individual phenotypic plasticity
Individual phenotypic plasticity is any response by an individual to a change in the environment. Plasticity is an important, if not the most important, kind of response to environmental change. Plasticity is most easily illustrated by so-called labile traits that can be measured repeatedly over individual lifetimes (Nussey et al. 2007). Thus, offspring birth weight is a plastic trait in female red deer (Cervus elaphus) because after a warm spring, calves are born at higher weight (Albon et al. 1987). This example has been studied in particular depth and illustrates a number of key features. Plasticity is a feature of individuals (here, mothers) and shows variation between individuals, with some individuals reacting more or less to the environmental change than others, and thus having different slopes of reaction norm (Nussey et al. 2005a). Plasticity can also influence ‘non-labile’ traits that occur only once in a lifetime, but may affect the rest of that lifetime (Nussey et al. 2007). For example, in many long-lived species there are strong ‘cohort effects’ in which growing conditions in the first year of birth (acting through such traits as birth weight) dictate later performance in terms of reproduction and survival (Albon et al. 1987; Forchhammer et al. 2001). Non-labile and labile effects can also interact with each other—for example, red deer females born at high density (i.e. conditions of greater food limitation) are less plastic in offspring birth weight than those born at lower density (Nussey et al. 2005a). Since early development is often key to downstream performance, the maternal environment may itself be an important axis of plastic change. All these effects may cause a population to change systematically with environmental change, but this is not evolution, because there is no change in the genetics of the population. Finally, individual plastic responses may or may not be adaptive, i.e. increase fitness, and many may be constraining and reduce fitness.
Of course, individual phenotypic plasticity may be inherited, i.e. there may be inherited variation in the way that individuals or populations respond to environmental variation (a genotype-by-environment reaction). Genetic analysis of plasticity in lay date phenology has been analysed in two great tit (Parus major) populations with variable results, depending on the intricacies of the specific models used, including the details of the environmental axis used (Nussey et al. 2005b; Charmantier et al. 2008; Husby et al. in press). This is an active area of research for the future, but it is a hard one, and demonstrating evolutionary change in plasticity will be subject to all the issues laid out further below.
(b). Selection
Selection can lead to a within-generation change in the phenotypic mean of a population. Whether this also results in a longer term evolutionary change (see §1d below) is another matter. It is not necessarily the case that the phenotypic variation subjected to selection has an underlying genetic basis. Variation may be generated by individual phenotypic plasticity (above). Alternatively, many populations have structure in terms of age, stage or sex which is correlated with phenotypic traits such as size. If ecological conditions allow more individuals to survive, and young animals are smaller than adults, a population may show a sustained decline in body size; conversely, if ecological conditions promote survival of older individuals, a population may increase in body size. An example is provided by the Soay sheep on St Kilda. Here, selection is always for larger body size, but recent mild winters have weakened the strength of selection and allowed more small lambs to survive, contributing to a decline in the mean body size in the population over time (Pelletier et al. 2007). This source of change is easily detected when the sex and age of individuals is known, as on St Kilda, but is easily overlooked where there is incomplete knowledge of individual life histories and traits.
(c). Immigration of individuals with different phenotypes
In principle, a trait mean could also change if a population is subject to immigration by individuals from other populations which, for non-genetic reasons, have different trait values. For example, if early life nutrition affects body size, then a population with poor habitat, and hence nutrition, may receive immigrants from surrounding populations with better nutrition. Phenotypic change may occur in the focal population, but it is not due to evolution by natural selection. Although this effect seems unlikely in the well-studied populations under discussion here, it could be an important force in vagile species and where individual monitoring is not possible.
(d). Evolutionary change
Evolutionary change is a sustained change, over generations, in the genetic make-up of a population. Evolutionary change can occur through immigration, genetic drift or a response to natural selection, the latter being the focus here. Predicting natural selection and measuring a response to it are arguably the most challenging of the processes of change discussed here. Very naively, according to the breeder's equation, R = h2S (where R is the per generation response to selection, h2 is the heritability of a trait and S is the selection differential on that trait), an evolutionary response must follow if both h2 and S are not zero (Falconer & Mackay 1996; Lynch & Walsh 1998). Such predictions have been reasonably successful in the laboratory or on the farm, when single traits have been selected under constant environmental conditions. However, in the wild, selection is likely to operate simultaneously on a whole suite of traits, not just the one(s) we can measure. When the focal trait is genetically correlated with other traits under selection then a correlated response to selection can occur, requiring a multi-variate model of evolution (Lande & Arnold 1983). Even though these points have been known for nearly three decades and despite the long-term individual-based datasets that have accumulated in the interim, with a few honourable exceptions (see Charmantier et al. 2006) multi-variate selection studies have not been conducted. This is largely for practical reasons: many studies measure what they can but this does not actually amount to a great many traits; most studies are still very underpowered for the purpose of estimating genetic correlations; and finally it is genuinely difficult to anticipate the ideal suite of traits for such an analysis.
3. Response to selection
Recent studies have shown a number of interesting problems relating to the demonstration of evolutionary change in natural populations which lie exactly at the interface of evolution and ecology—mechanisms by which ecological effects confound predicted evolution. A number of authors long since pointed out the probability of such difficulties (e.g. Mitchell-Olds & Shaw 1987), but they have perhaps not been so obvious in the empirical literature until recently. To introduce these problems, consider the following: Gienapp et al. (2008) recently reviewed efforts to predict evolution of traits in natural populations using the breeder's equation and found that of 35 studies where evolution was predicted (given estimates of heritability and selection differentials), 12 studies showed phenotypic change in the predicted direction, 15 showed no trait change and eight showed change in the opposite direction to the predicted.
Given the ecological sources of change outlined above and the failure to accurately predict evolutionary change from the breeder's equation, there is clearly a compelling need to estimate evolutionary change—or at least direction—with better accuracy. This was fully realized in several of the studies reviewed by Gienapp et al. (2008). How can this be achieved for natural populations? The gold standard would be a common garden experiment with samples of a population before and after selection. Such studies have been done, for example in the case of guppies (Poecilia reticulate) subjected to novel predation regimes (Reznick et al. 1997) and in several other studies (reviewed by Hendry & Kinnison 1999), but this is impractical for many wild animal populations, and so authors have used another device from animal breeding, individual estimated breeding values (EBVs). These provide an estimate of an individual's genetic merit for the trait in question, based on its own trait value, that of its relatives and the population level parameters such as heritability (Falconer & Mackay 1996; Lynch & Walsh 1998). Putative responses to selection have been measured by examining temporal (or sometimes spatial) trends in EBVs (see examples below). As described later, estimating EBVs for wild populations and inferring evolution from them has its own problems, but for now they will help to illustrate some phenomena.
4. The role of environmental variation
Why are predictions of response to selection from the breeder's equation so unsatisfactory? Here, I adopt the profound simplification of considering just one or a few traits at a time in order to highlight three manifestations of ecological heterogeneity which overwhelm or complicate predictions from naive application of the breeder's equation.
(a). Counter gradient variation
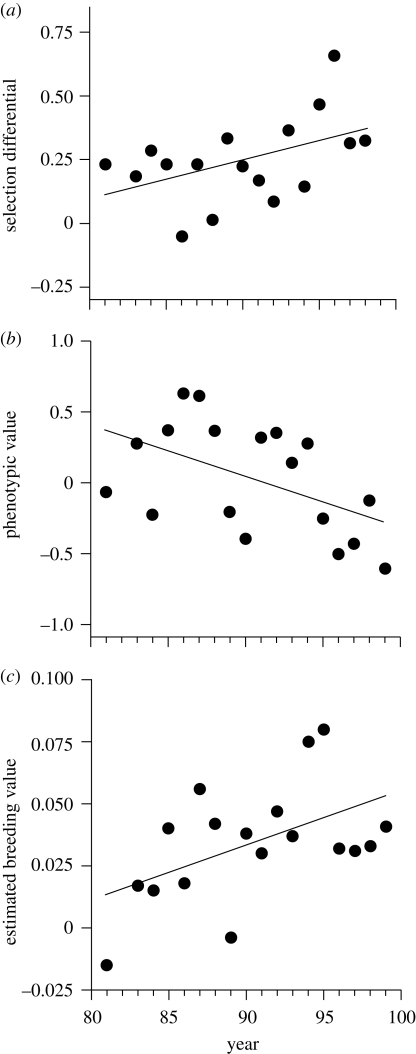
Ecological forces acting via plasticity, migration and selection can be strong and can induce relatively rapid changes in mean phenotype compared with those we might predict for evolutionary change. In consequence, ecological change can overwhelm predicted and actual evolutionary change. The case where ecological forces cause a phenotypic gradient in time or space which contrasts with an expected evolutionary gradient was called counter gradient variation by Conover & Schultz (1995). A couple of temporal examples from long-term studies of individuals will illustrate: Merilä et al. (2001) demonstrated strong selection on fledgling condition (a residual from a regression of body mass against tarsus length) in the collared flycatcher population (Ficedula hypoleuca) on the Swedish island of Götland (figure 1a). Given a heritability of 0.3, condition was expected to increase over time. In fact, condition declined over time (figure 1b) and ecological data were brought to bear, showing that food availability had almost certainly declined over the study period. However, EBVs for condition were not declining, and if anything were increasing (figure 1c—see also discussion of EBVs below), suggesting that the genetic merit for condition in the population was indeed responding to selection, but was being overwhelmed at the phenotypic level by individual plastic responses. Similarly, in Soay sheep on St Kilda, August body weight is heritable in most age classes (Wilson et al. 2007) and there is selection for higher body weight (Pelletier et al. 2007), and yet the sheep are getting smaller (Wilson et al. 2007; Ozgul et al. 2009). The August weight breeding values of the population were reported as showing a modest increase (Wilson et al. 2007), but the change has since been shown to be non-significant (Hadfield et al. 2010; see also EBV discussion below). Clearly, the potential evolutionary trend is being overwhelmed by ecological effects. As revealed by retrospective decomposition of trait variation over time using an age-structured Price equation, the sheep are not growing as fast as previously (Ozgul et al. 2009) and, simultaneously, selection for higher body weight has weakened (Pelletier et al. 2007).
Figure 1.
Counter gradient variation in fledgling condition in collared flycatchers on Götland (Merilä et al. 2001). (a) Selection favours higher condition and became stronger over the time series. (b) The phenotypic values for condition declined over the study period. (c) The EBVs over the same period appear to increase, though see text for discussion of the use of EBVs to determine genetic trends.
Counter gradient variation is just the most extreme outcome of the way in which an ecological trend may alter a trait's trajectory away from that expected from selection. Many changes in temporal or spatial ecological conditions seem likely to induce plastic responses or short-term changes in selection regime. In practice, therefore, the magnitude of many expected evolutionary responses must be modified by these processes.
The counter gradient idea extends far back into the evolutionary biology literature in a slightly different incarnation (Fisher 1958). This is the idea that evolution will itself lead to environmental deterioration, in the sense that improved competitors will inhabit an environment of increased competition, a hypothesis which predicts phenotypic (but not evolutionary) stasis (Cooke et al. 1990; Frank & Slatkin 1992) and may yet prove to be an important explanation for long-term stasis in some traits.
(b). Environmentally induced covariance between traits
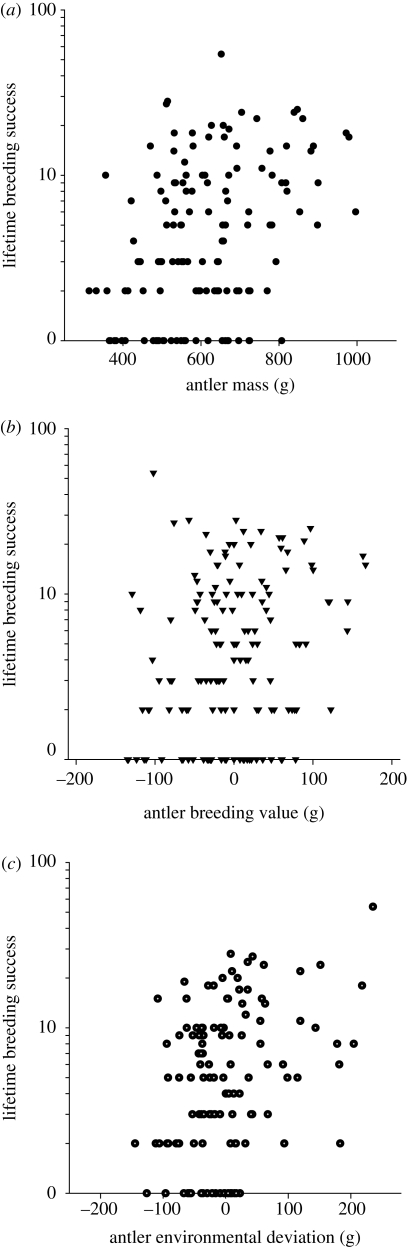
Predictions of evolutionary change from the breeder's equation can also go awry when relevant traits (traits under selection that are correlated with the focal trait) are omitted from the analysis (Mitchell-Olds & Shaw 1987). These can be traits in the classic sense (e.g. body size), but they can also be aspects of an individual's environment (Rausher 1992; Hadfield 2008). Price et al. (1988) put forward such an explanation for the situation where environmental heterogeneity causes a correlation between the trait of interest and fitness, through a third environmentally dependent trait such as condition. As an example, in the red deer on the Scottish island of Rum, cast antler weight is heritable and positively correlated with lifetime breeding success (LBS; figure 2a), but contrary to simple expectation, it has declined over time, another example of counter gradient variation, probably caused by increased density and thus reduced food availability, over the time period studied (Kruuk et al. 2002). Beyond this, the authors inspected correlations between individual LBS and both antler weight EBVs and antler weight ‘environmental deviations’—residual deviations in antler weight after accounting for EBVs. While LBS and breeding values are not associated (figure 2b), the relationship between LBS and environmental deviations is much more similar to the overall phenotypic correlation (figure 2c), suggesting that selection is not targeting the genetic component of antler size. This analysis is supported by a lack of genetic correlation between antler weight and LBS when investigated in a bivariate animal model (Kruuk et al. 2002). The authors postulate that both antler size and LBS are associated with a third, unmeasured trait which is associated with environmental heterogeneity, such as condition, which sets up a phenotypic, but not genetic, correlation between antler weight and LBS.
Figure 2.
Environmentally induced covariance between antler weight and lifetime breeding success in red deer on Rum (Kruuk et al. 2002). (a) Selection favours heavy antlers, since there is a strong correlation between antler weight and lifetime breeding success (LBS) of stags. (b) The correlation is less apparent when comparing LBS and EBVs of antler weight but (c) more apparent when comparing LBS and the individual environmental components of antler weight. The observations in (b) were supported by a lack of genetic correlation between antler weight and LBS in a bivariate animal model (Kruuk et al. 2002). See text for discussion of the use of EBVs in such analyses.
Variation in condition, loosely defined as nutritional state and hence profoundly influenced by ecological conditions, is likely to be extremely common in nature, so once again we see that this phenomenon, environmentally generated covariance between traits, may commonly dampen evolutionary response in nature.
(c). Correlated effects of a fluctuating environment
Environmental fluctuations (as distinct from a consistent trend in a single direction) represent a third way in which ecology interferes with simple predictions of response to selection. On St Kilda, variation in Soay sheep population density and weather conspires to create years which vary widely in environmental quality. In some winters virtually all lambs of the year survive; in other winters virtually all die, with continuous variation between these two extremes. Recent investigations of the quantitative genetic architecture of traits using random regression animal models, which allow quantitative genetic parameter estimates to vary with covariates such as ecological conditions, suggest that additive genetic variance is not constant in different environmental conditions. Thus, the additive genetic variance of a number of traits including birth weight, August weight, horn growth and parasite faecal egg count increases with good environmental conditions (Wilson et al. 2006; Robinson et al. 2009). These observations are consistent with the pattern found in a meta-analysis and lend support to the idea that benign conditions allow the expression of additional genetic variance (Charmantier & Garant 2005). Combined with environmentally determined changes in the total phenotypic variance, these observations mean that a heritability calculated over a period of years is not necessarily representative of heritability in any particular year.
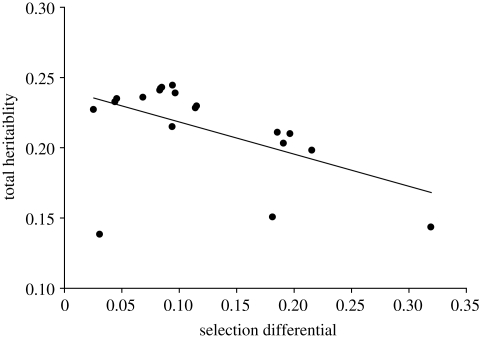
Furthermore, the strength of selection also varies over time, and this variation may have a systematic relationship with the expression of genetic variation. Continuing with the St Kilda example, selection on birth weight is greatest under harsh conditions and least under benign conditions (Wilson et al. 2006). This is exactly inverse to the pattern shown by the total heritability of birth weight (Wilson et al. 2006). So, when environmental conditions are good, birth weight is most heritable, but selection is weakest, and when environmental conditions are poor, selection is stronger but heritability is lowest (figure 3). This somewhat reduces the overall expectation of response to selection relative to what is expected from average values of heritability and selection for the whole time period (Wilson et al. 2006). Note that although selection and heritability are correlated here, this is not suggested to be a causal link.
Figure 3.
Inverse relationship between the strength of selection and heritability promoted by between-year variation in environmental conditions in Soay sheep birth weight on St Kilda (Wilson et al. 2006). Each year is represented by a data point. Total heritability refers to the combined heritability and maternal genetic effect. When conditions are good (left-hand side of plot) heritability is at its highest but selection is relatively weak. When conditions are poor (right-hand side of plot) selection is relatively strong but heritability is low.
In principle, spatial variation in environmental conditions can also generate correlations between the expression of additive genetic variance and selection. If spatial detail is not recognized, then just like the example of temporal variation in environment quality above, the response to selection may be inaccurately predicted. Spatial effects may be further complicated by dispersal: Garant et al. (2005) found that in the Wytham, Oxford, great tit study population, additive genetic variance and selection are spatially correlated, and that differences between areas are reinforced by localized recruitment patterns.
If, as these examples suggest, subtle associations exist between the environmental conditions experienced by individuals, the expression of additive genetic variance and covariance between traits and the strength of selection, then predicting and measuring evolutionary change is considerably more complex for natural populations than is widely appreciated.
The three phenomena outlined above, counter gradient variation, environmentally induced covariance and the correlated effects of fluctuating environments, have in common the feature that environmental heterogeneity sets up complex relationships which confound simple predictions of the response to selection. Mechanistically, they are linked by the facts that phenotypes are multi-variate, we cannot measure all the traits and environmental influences we would like to measure, and the expression of genetic and environmentally induced variation cannot be assumed to be constant under different conditions, either at an individual or a population level. Under these circumstances the breeder's equation, even in a multi-variate form, is too strong in assuming we have all the relevant information to make predictions (Lande 1979; Lande & Arnold 1983; Mitchell-Olds & Shaw 1987; Rausher 1992; van Tienderen & de Jong 1994). An alternative approach for making predictions for genetic change, which does not make so many strong assumptions, is to estimate the genetic covariance between a trait and fitness (Robertson 1966; Price 1970), as recommended in several recent papers (Hadfield 2008; Kruuk et al. 2008; Morrissey & Wilson submitted). This is a very hard prescription, because how best to measure fitness in the wild is a matter of debate and because many studies of wild individuals lack power, but that is really the point: no genetic covariance, no expected evolution by natural selection.
5. Better measurement of response to selection
In long-term studies of wild populations where common garden experiments are impractical, what evidence is sufficient to conclude that a trait has shown an evolutionary response to selection? For reasons described above, it is not sufficient to show selection and phenotypic change in a consistent direction, nor is it sufficient to show selection, heritability and phenotypic change in a consistent direction. It is necessary to show selection and heritability and a genetic response in the trait which cannot be explained by alternative forces, e.g. drift. There are two ways to approach this in future.
(a). Estimated breeding values
As outlined above, one approach to measuring response to selection is to measure the change in breeding values that has occurred over time or space. Since breeding values cannot be directly measured, they are commonly estimated from the data; for example, by using the animal model. There are a number of weaknesses of this approach which need to be recognized. First, breeding values are estimated using both the focal individual's phenotype and those of its relatives. In wild pedigrees there is generally a lot of variation in data quantity (in terms of known pedigree connections and phenotypic data) from one individual to the next. Consequently, for individuals with few known relatives, the EBV is primarily determined by the individual's own phenotypic value, which may principally reflect its environment and not its genetics (Postma 2006). Furthermore, there may be time-dependent systematic biases; for example, pedigree founders and currently alive individuals on average have fewer known relatives than individuals in middle generations, meaning that EBVs at the beginning or end of a time series are on average more biased than those elsewhere in the intervening generations (Postma 2006). Second, failure to acknowledge the sampling error and non-independence of EBVs has been criticized because of the resulting underestimation of error in tests of significance (Hadfield et al. 2010). When analysed with appropriate errors, the trend towards increased August weight EBV for Soay sheep body size (see above; Wilson et al. 2007) is not significantly different from zero.
How do these problems affect the findings outlined above? They do not change the central result that in flycatchers and several other examples, phenotypic change is in the opposite direction to that predicted by the breeder's equation, but they do mean that the suggested cryptic genetic change in the opposite direction requires reanalysis in the flycatchers and other studies, as outlined above for the Soay sheep body weight example. In the case of environmental covariance in red deer antler weight, the lack of association between EBV and LBS is already confirmed by the alternative and more conservative analysis that found no genetic correlation between antler weight and LBS (Kruuk et al. 2002). EBVs were not involved on the third phenomenon on the correlated effects of fluctuating environments.
We now have a more appropriate statistical framework for the use of EBVs when investigating response to selection, including distinguishing it from drift (Hadfield et al. 2010). A systematic attempt to reanalyse existing datasets to estimate responses to selection is under way (B. C. Sheldon & M. B. Morrissey 2009, personal communication).
(b). Evolutionary genomics
The principle alternative to the quantitative genetic approach to measuring response to selection on quantitative traits (i.e. EBVs) will come through genomic approaches. A temporal or spatial trend in an allele or haplotype which is associated with a quantitative trait that is known to be under selection and is in the predicted direction and which cannot be explained by drift would be strong evidence of a response to selection. Importantly, in contrast to a change in estimated breeding values, we would have a measured change in the frequency of identified stretches of DNA. Genomic approaches to understanding quantitative traits in wild populations are in their infancy (Slate 2005; Ellegren & Sheldon 2008; Kruuk et al. 2008; Slate et al. 2010) and there is a lot of work yet to be done in this area. Nevertheless, there are some highly promising pieces of progress: genes underlying quantitative traits in natural populations are beginning to be found, for example in sticklebacks (Gasterosteus aculeatus; e.g. Colosimo et al. 2004; Shapiro et al. 2004) and Darwin's finches (Geospiza spp.; Abzhanov et al. 2004, 2006). But the evidence we should aspire to is of selection on and response at a quantitative trait locus (QTL) within a population. One attempt to provide this combined evidence comes from Soay sheep: the coat colour locus Tyrosinase-related protein 1, TYRP1, is closely linked to a body size QTL, and selection favours the T allele conferring pale coat colour and (perversely) small body size when homozygous. The T allele and pale colour are increasing in the study population (Gratten et al. 2007, 2008), though this relationship is not yet significantly different from the predictions of drift.
More generally, genomic analyses of the various long-term wild animal field studies have the potential to contribute greatly to the whole field under discussion in this article. Quantitative genetics treats the genome as a black box and must infer estimates of genetic merit such as EBVs for the entire genome; genomics has the potential to identify the components of that merit more precisely and observe change (or stasis) in them over time and space.
6. Conclusions
The recent wave of quantitative genetics studies in natural populations, taking advantage of the developments of pedigrees and numerical methods such as the animal model, have made incisive progress in describing and understanding the genetic architecture of traits and the action of selection. However, it has also thrown up major complexities, including the one emphasized here: the environment is a noisy place and causes simple predictions that work in the laboratory or on the farm to go wrong in nature. This presents a major exciting challenge to the field which must be addressed if we are to demonstrate that phenotypic change has anything to do with evolutionary response (Gienapp et al. 2008; Kruuk et al. 2008; Sheldon 2010). Evolutionary response to environmental change, including anthropogenic change, may well be happening all around us, but in long-term studies of wild individuals it is very difficult to demonstrate, requiring simultaneous demonstration of selection, heritable variation and genetic response, the latter using appropriate analytical methods (Hadfield et al. 2010), demonstrating QTL marker frequency change that cannot be explained by drift or, in those rare cases where it is possible, a common garden experiment.
Acknowledgements
I thank all members of the Wild Evolution Group at Edinburgh and the Wild Animal Modelling Bi-Annual Meeting for the unending interest of their work and Michael Morrissey, Alastair Wilson, Jarrod Hadfield, Tim Coulson and two referees for comments.
Footnotes
One contribution of 18 to a Discussion Meeting Issue ‘Genetics and the causes of evolution: 150 years of progress since Darwin’.
References
- Abzhanov A., Protas M., Grant B. R., Grant P. R., Tabin C. J.2004Bmp4 and morphological variation of beaks in Darwin's finches. Science 305, 1462–1465 (doi:10.1126/science.1098095) [DOI] [PubMed] [Google Scholar]
- Abzhanov A., Kuo W. P., Hartmann C., Grant B. R., Grant P. R., Tabin C. J.2006The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature 442, 563–567 (doi:10.1038/nature04843) [DOI] [PubMed] [Google Scholar]
- Albon S. D., Cluttonbrock T. H., Guinness F. E.1987Early development and population-dynamics in red deer. 2. Density-independent effects and cohort variation. J. Anim. Ecol. 56, 69–81 [Google Scholar]
- Charmantier A., Garant D.2005Environmental quality and evolutionary potential: lessons from wild populations. Proc. R. Soc. B 272, 1415–1425 (doi:10.1098/rspb.2005.3117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A., Perrins C., McCleery R. H., Sheldon B. C.2006Evolutionary response to selection on clutch size in a long-term study of the mute swan. Am. Nat. 167, 453–465 [DOI] [PubMed] [Google Scholar]
- Charmantier A., McCleery R. H., Cole L. R., Perrins C., Kruuk L. E. B., Sheldon B. C.2008Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science 320, 800–803 (doi:10.1126/science.1157174) [DOI] [PubMed] [Google Scholar]
- Colosimo P. F., Peichel C. L., Nereng K., Blackman B. K., Shapiro M. D., Schluter D., Kingsley D. M.2004The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover D. O., Schultz E. T.1995Phenotypic similarity and the evolutionary significance of countergradient variation. Trends Ecol. Evol. 10, 248–252 (doi:10.1016/S0169-5347(00)89081-3) [DOI] [PubMed] [Google Scholar]
- Cooke F., Taylor P. D., Francis C. M., Rockwell R. F.1990Directional selection and clutch size in birds. Am. Nat. 136, 261–267 [Google Scholar]
- Ellegren H., Sheldon B. C.2008Genetic basis of fitness differences in natural populations. Nature 452, 169–175 (doi:10.1038/nature06737) [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C.1996Introduction to quantitative genetics. London, UK: Longman [Google Scholar]
- Fisher R. A.1958The genetical theory of natural selection. New York, NY: Dover Books [Google Scholar]
- Forchhammer M. C., Clutton-Brock T. H., Lindstrom J., Albon S. D.2001Climate and population density induce long-term cohort variation in a northern ungulate. J. Anim. Ecol. 70, 721–729 (doi:10.1046/j.0021-8790.2001.00532.x) [Google Scholar]
- Frank S. A., Slatkin M.1992Fishers fundamental theorem of natural selection. Trends Ecol. Evol. 7, 92–95 (doi:10.1016/0169-5347(92)90248-A) [DOI] [PubMed] [Google Scholar]
- Garant D., Kruuk L. E. B., Wilkin T. A., McCleery R. H., Sheldon B. C.2005Evolution driven by differential dispersal within a wild bird population. Nature 433, 60–65 (doi:10.1038/nature03051) [DOI] [PubMed] [Google Scholar]
- Gienapp P., Teplitsky C., Alho J. S., Mills J. A., Merilä J.2008Climate change and evolution: disentangling environmental and genetic responses. Mol. Ecol. 17, 167–178 (doi:10.1111/j.1365-294X.2007.03413.x) [DOI] [PubMed] [Google Scholar]
- Gratten J., Beraldi D., Lowder B. V., McRae A. F., Visscher P. M., Pemberton J. M., Slate J.2007Compelling evidence that a single nucleotide substitution in TYRP1 is responsible for coat-colour polymorphism in a free-living population of Soay sheep. Proc. R. Soc. B 274, 619–626 (doi:10.1098/rspb.2006.3762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J., Wilson A. J., McRae A. F., Beraldi D., Visscher P. M., Pemberton J. M., Slate J.2008A localized negative genetic correlation constrains microevolution of coat color in wild sheep. Science 319, 318–320 (doi:10.1126/science.1151182) [DOI] [PubMed] [Google Scholar]
- Hadfield J. D.2008Estimating evolutionary parameters when viability selection is operating. Proc. R. Soc. B 275, 723–734 (doi:10.1098/rspb.2007.1013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J. D., Wilson A. J., Garant D., Sheldon B. C., Kruuk L. E. B.2010The misuse of BLUP in ecology and evolution. Am. Nat. 175, 116–125 [DOI] [PubMed] [Google Scholar]
- Hairston N. G., Ellner S. P., Geber M. A., Yoshida T., Fox J. A.2005Rapid evolution and the convergence of ecological and evolutionary time. Ecol. Lett. 8, 1114–1127 (doi:10.1111/j.1461-0248.2005.00812.x) [Google Scholar]
- Hendry A. P., Kinnison M. T.1999Perspective: the pace of modern life: measuring rates of contemporary microevolution. Evolution 53, 1637–1653 (doi:10.2307/2640428) [DOI] [PubMed] [Google Scholar]
- Husby A., Nussey D. H., Visser M. E., Wilson A. J., Sheldon B. C., Kruuk L. E. B.In press Contrasting patterns of phenotypic plasticity in reproductive traits in two great tit (Parus major) populations. Evolution. [DOI] [PubMed] [Google Scholar]
- Kinnison M. T., Hairston N. G.2007Eco-evolutionary conservation biology: contemporary evolution and the dynamics of persistence. Funct. Ecol. 21, 444–454 (doi:10.1111/j.1365-2435.2007.01278.x) [Google Scholar]
- Kruuk L. E. B.2004Estimating genetic parameters in natural populations using the ‘animal model'. Phil. Trans. R. Soc. Lond. B 359, 873–890 (doi:10.1098/rstb.2003.1437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk L. E. B., Slate J., Pemberton J. M., Brotherstone S., Guinness F., Clutton-Brock T.2002Antler size in red deer: heritability and selection but no evolution. Evolution 56, 1683–1695 [DOI] [PubMed] [Google Scholar]
- Kruuk L. E. B., Slate J., Wilson A. J.2008New answers for old questions: the evolutionary quantitative genetics of wild animal populations. Annu. Rev. Ecol. Evol. Systemat. 39, 525–548 (doi:10.1146/annurev.ecolsys.39.110707.173542) [Google Scholar]
- Lande R.1979Quantitative genetic analysis of multivariate evolution, applied to brain-body size allometry. Evolution 33, 402–416 (doi:10.2307/2407630) [DOI] [PubMed] [Google Scholar]
- Lande R., Arnold S. J.1983The measurement of selection on correlated characters. Evolution 37, 1210–1226 (doi:10.2307/2408842) [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh B.1998Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer [Google Scholar]
- Merilä J., Kruuk L. E. B., Sheldon B. C.2001Cryptic evolution in a wild bird population. Nature 412, 76–79 (doi:10.1038/35083580) [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T., Shaw R. G.1987Regression analysis of natural selection—statistical inference and biological interpretation. Evolution 41, 1149–1161 (doi:10.2307/2409084) [DOI] [PubMed] [Google Scholar]
- Morrissey M. B., Wilson A. J.Submitted The danger of applying the breeder's equation outside of the context of artificial selection. J. Evol. Biol. [DOI] [PubMed] [Google Scholar]
- Nussey D. H., Clutton-Brock T. H., Albon S. D., Pemberton J., Kruuk L. E. B.2005aConstraints on plastic responses to climate variation in red deer. Biol. Lett. 1, 457–460 (doi:10.1098/rsbl.2005.0352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussey D. H., Postma E., Gienapp P., Visser M. E.2005bSelection on heritable phenotypic plasticity in a wild bird population. Science 310, 304–306 (doi:10.1126/science.1117004) [DOI] [PubMed] [Google Scholar]
- Nussey D. H., Wilson A. J., Brommer J. E.2007The evolutionary ecology of individual phenotypic plasticity in wild populations. J. Evol. Biol. 20, 831–844 (doi:10.1111/j.1420-9101.2007.01300.x) [DOI] [PubMed] [Google Scholar]
- Olsen E. M., Heino M., Lilly G. R., Morgan M. J., Brattey J., Ernande B., Dieckmann U.2004Maturation trends indicative of rapid evolution preceded the collapse of northern cod. Nature 428, 932–935 (doi:10.1038/nature02430) [DOI] [PubMed] [Google Scholar]
- Ozgul A., Tuljapurkar S., Benton T. G., Pemberton J. M., Clutton-Brock T. H., Coulson T.2009The dynamics of phenotypic change and the shrinking sheep of St. Kilda. Science 325, 464–467 (doi:10.1126/science.1173668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier F., Clutton-Brock T., Pemberton J., Tuljapurkar S., Coulson T.2007The evolutionary demography of ecological change: linking trait variation and population growth. Science 315, 1571–1574 (doi:10.1126/science.1139024) [DOI] [PubMed] [Google Scholar]
- Pelletier F., Garant D., Hendry A. P.2009Eco-evolutionary dynamics introduction. Phil. Trans. R. Soc. B 364, 1483–1489 (doi:10.1098/rstb.2009.0027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton J. M.2008Wild pedigrees: the way forward. Proc. R. Soc. B 275, 613–621 (doi:10.1098/rspb.2007.1531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma E.2006Implications of the difference between true and predicted breeding values for the study of natural selection and micro-evolution. J. Evol. Biol. 19, 309–320 (doi:10.1111/j.1420-9101.2005.01007.x) [DOI] [PubMed] [Google Scholar]
- Price G. R.1970Selection and covariance. Nature 227, 520–521 (doi:10.1038/227520a0) [DOI] [PubMed] [Google Scholar]
- Price T., Kirkpatrick M., Arnold S. J.1988Directional selection and the evolution of breeding date in birds. Science 240, 798–799 (doi:10.1126/science.3363360) [DOI] [PubMed] [Google Scholar]
- Rausher M. D.1992The measurement of selection on quantitative traits—biases due to environmental covariances between traits and fitness. Evolution 46, 616–626 (doi:10.2307/2409632) [DOI] [PubMed] [Google Scholar]
- Reznick D. N., Shaw F. H., Rodd F. H., Shaw R. G.1997Evaluation of the rate of evolution in natural populations of guppies (Poecilia reticulata). Science 275, 1934–1937 (doi:10.1126/science.275.5308.1934) [DOI] [PubMed] [Google Scholar]
- Robertson A.1966A mathematical model of culling process in dairy cattle. Anim. Prod. 8, 95–108 [Google Scholar]
- Robinson M. R., Wilson A. J., Pilkington J. G., Clutton-Brock T. H., Pemberton J. M., Kruuk L. E. B.2009The impact of environmental heterogeneity on genetic architecture in a wild population of Soay sheep. Genetics 181, 1639–1648 (doi:10.1534/genetics.108.086801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossiter M. C.1996Incidence and consequences of inherited environmental effects. Annu. Rev. Ecol. Systemat. 27, 451–476 (doi:10.1146/annurev.ecolsys.27.1.451) [Google Scholar]
- Shapiro M. D., Marks M. E., Peichel C. L., Blackman B. K., Nereng K. S., Jonsson B., Schluter D., Kingsley D. M.2004Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428, 717–723 (doi:10.1038/nature02415) [DOI] [PubMed] [Google Scholar]
- Sheldon B. C.2010Genetic perspectives on the micro-evolutionary consequences of climate change in birds. In Birds and climate change (eds Moller A. P., Berthold P., Fiedler W.). Oxford, UK: Oxford University Press [Google Scholar]
- Slate J.2005Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 14, 363–379 (doi:10.1111/j.1365-294X.2004.02378.x) [DOI] [PubMed] [Google Scholar]
- Slate J., Gratten J., Beraldi D., Stapley J., Hale M., Pemberton J. M.2010Gene mapping in the wild with SNPs: guidelines and future directions. Genetica 138, 467 (doi:10.1007/s10709-010-9445-0) [DOI] [PubMed] [Google Scholar]
- Thackeray S. J., et al. In press Trophic level asynchrony in rates of phenological change for marine, freshwater and terrestrial environments. Global Change Biol. [Google Scholar]
- van Tienderen P. H., de Jong G.1994A general model of the relation between phenotypic selection and genetic response. J. Evol. Biol. 7, 1–12 (doi:10.1046/j.1420-9101.1994.7010001.x) [Google Scholar]
- Wilson A. J., Pemberton J. M., Pilkington J. G., Coltman D. W., Mifsud D. V., Clutton-Brock T. H., Kruuk L. E. B.2006Environmental coupling of selection and heritability limits evolution. PLoS Biol. 4, 1270–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. J., Pemberton J. M., Pilkington J. G., Clutton-Brock T. H., Coltman D. W., Kruuk L. E. B.2007Quantitative genetics of growth and cryptic evolution of body size in an island population. Evol. Ecol. 21, 337–356 (doi:10.1007/s10682-006-9106-z) [Google Scholar]