Abstract
Amyloid precursor protein (APP), implicated in Alzheimer's disease, is a transmembrane protein of undetermined function. APP is cleaved by gamma-secretase that releases the APP intracellular domain (AICD) in the cytoplasm. In vitro and in vivo studies have implicated the role of AICD in cell signaling and transcriptional regulation of Gsk3β, KAI1, BACE1, EGFR, and other proteins. In this study, by overexpressing AICD in mouse neuroblastoma cell lines, we have demonstrated the alteration in the expressions of two proteins, patched homolog 1 (PTCH1), a receptor for sonic hedgehog signaling, and transient receptor potential cation channel subfamily C member 5 (TRPC5), a component of receptor-activated nonselective calcium permeant cation channel. Our results indicate the possibility of regulation by AICD in developmental processes as well as in the maintenance of calcium homeostasis at the transcription level.
1. Introduction
Alzheimer's disease (AD) is an irreversible, progressive neurodegenerative disorder that occurs gradually and results in memory loss, unusual behavior, personality changes, and a decline in thinking abilities. A fundamental abnormality that plays a pivotal role in the dysfunction and death of neurons in AD is altered proteolytic cleavage of APP. The function of the APP holoprotein is not yet established and mice lacking the APP gene show relatively minor neurological impairments. This subtle phenotype is probably due to compensatory effects mediated by two other members of the APP gene family: amyloid-precursor-like protein-1 and -2 (APLP1 and APLP2). This view is supported by evidence showing that the combined ablation of APP and APLP2, both APLP genes or all three family members together leads to early postnatal lethality [1]. Both the amyloidogenic and nonamyloidogenic pathways, that is, the cleavages of APP by β- and α-secretases, respectively, liberate the soluble ectodomain of APP (ectodomain shedding) and retain the C-terminal fragments (CTF) (CT99 and CT83, resp.). Subsequent cleavages by γ-secretase in the transmembrane domain generate the amyloidogenic Aβ peptide or the nonamyloidogenic p3 peptide along with the intracellular C-terminal domain of APP (AICD). Biochemical and genetic interaction screens have led to the identification of both extracellular and multiple intracellular binding partners, which seem to anchor the APP/APLP C-termini to a complex protein network at the cell surface, which may transduce various cellular responses [2, 3]. Notably, a highly conserved cytoplasmic—682YENPTY687—motif is present in all APP/APLP family members, which confers clathrin-mediated endocytosis and was shown to bind several multidomain adaptor proteins, including X11/Mints, Fe65 family proteins and mDab [4]. A number of type-I transmembrane proteins including Notch, p75NTR, CD44, ErbB4, neuregulin-1, and alcadein undergo a similar secretase mediated processing leading to ectodomain shedding and generation of intracellular domains (ICD's) [5]. Some of these ICD's are known to take part in cellular differentiation and development by nuclear signaling and transcriptional transactivation [6]. Like NICD (Notch intracellular domain), several recent studies have suggested that AICD has transactivation activity and can regulate transcription of multiple genes including APP, GSK-3β, KAI1, neprilysin, BACE, and EGFR [7–11]. Recently, it has been shown that AICD-mediated transcriptional regulation of EGFR by directly binding to the EGFR promoter [11].
The role of APP in neuronal development and in calcium homeostasis is well established [1, 12, 13]. The expression of APP in brain is developmentally regulated and it is expressed ubiquitously in differentiated neurons. APP is axonally transported and secreted forms of APP (sAPPs) are released from neurons in an activity-driven manner. Secreted APPs modulate neuronal excitability, counteract effects of glutamate on growth cone behaviors, and increase synaptic complexity [14]. Moreover, aberrant processing of APP can also cause neurodegeneration by impairing a neuroprotective function sAPPs which normally regulate calcium homeostasis [12, 15].
But the role of AICD, if any, in both developmental processes and in maintenance of calcium ion homeostasis is yet to be elucidated. In the present study, we intended to look into the possibility of AICD having any role in the transcriptional regulation of the components of sonic hedgehog pathway and calcium channel forming proteins. Initially, microarray analysis was done to screen the genes whose expression would alter upon AICD overexpression (data not shown).
2. Materials and Methods
2.1. Cloning of AICD in pGFP C1 Vector
For the overexpression of AICD in mammalian cell line, it was cloned in pGFP vector. Specific primers for AICD (Forward: 5′ACGCGTCGACAAGAAGAAACAGTACACATCC3′ and the Reverse: 5′CGGGATCCTAGTTCTGCATCTGCTCAAAGAAC3′) with adaptors (underlined), for the restriction enzymes (RE) SalI and BamH1, were synthesized (Integrated DNA Technologies) to amplify the domain using brain c-DNA library (Stratagen) as template. PCR products were digested with SalI and BamH1 (New England Biolabs) and ligated to pGFP C1 vector (BD Biosciences). Construct was confirmed both by DNA sequencing and restriction enzyme digestion.
2.2. Cell Culture and Transfection
Neuro 2A cells were obtained from National Cell Science Centre, Pune, India and were cultured in DMEM (HiMedia) supplemented with 10% fetal bovine serum (Invitrogen) at 37°C in 5% CO2 atmosphere under humidified condition. Transfection of cells with empty vector (pGFP C1) or AICD-GFP was performed using Lipofectamine 2000 Transfection Reagent (Invitrogen).
2.3. Protein Extraction
For extraction of proteins, PBS-washed pellets from cell lines were lysed on ice in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 15 mM EDTA, 0.5% Triton X-100) for 30 min in presence of Complete protease inhibitor (Roche Diagnostics) and centrifuged at 13,000 rpm for 15 minutes. Protein concentration was determined by Bradford protein estimation assay.
2.4. RNA Isolation, c-DNA Preparation and Real-Time PCR
RNA was isolated from Neuro 2A cells by RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturers protocol. RNA equivalent to 500 ng–1 μg was taken to synthesize the first strand cDNA using random hexamer primers and reverse transcriptase (Invitrogen). Real time RT-PCR reaction was carried out using Syber green 2X Universal PCR Master Mix (Applied Biosystems, USA) in ABI Prism 7500 sequence detection system. Each reaction was performed in triplicate using 100 ng of total RNA using corresponding primer sequences (primer sequences and PCR conditions were mentioned in the Table 1). For each gene, nontemplate control was used at the same condition to ascertain the baseline and threshold value for the analysis. The absolute quantification given by the software was in terms of Ct values. The relative quantification of a target gene in a sample compared to parental cell is expressed in terms of 2−ΔΔCt values after normalization with respect to internal control (β-actin gene).
Table 1.
Primer sequences and PCR conditions of real-time PCR.
| Name of the genes | PCR condition | PCR Cycle | Primer sequences (5′-3′) | Size (bp) |
|---|---|---|---|---|
| PTCHI | 95°C→10 min | 35 | GAAAAATGAGCAGAACCATGG TGTCTTCCTTCTGAACCCCTG | 102 |
| [95°C 30 sec, 50°C 30 sec, 60°C 1 min] | ||||
| 72°C→10 min | ||||
| TRPC5 | 95°C→10 min | 35 | TTCCAGCTCTCTTCACTGTGC AAGTCACAAGCCTCTCCCCAA | 102 |
| [95°C 30 sec, 50°C 30 sec, 60°C 1 min] | ||||
| 72°C→10 min | ||||
| Beta actin | 95°C→10 min | 35 | TCCTGTGGCATCCACGAAACT GAAGCATTTGCGGTGGACGAT | 315 |
| [95°C 30 sec, 55°C 30 sec, 60°C 1 min] | ||||
| 72°C→10 min | ||||
2.5. Western Blot
Proteins were separated on SDS-polyacrylamide gels and transferred onto PVDF membranes (Millipore Corporation), which were blocked by incubation in 5% dried milk in TBST (50 mM Tris-HCl, 150 mM NaCl, pH 7.5 containing 0.05% Tween 20). Membranes were probed with primary antibodies against PTCH (ab53715, Abcam plc, 1 : 1000); TRPC5 (ab58374, Abcam plc, 1 μg/mL); beta actin (loading control for whole cell extracts; Abcam plc, 1 : 5000). HRP-conjugated antibodies (Chemicon; 1 : 5000) were then added to the blots. Immunoreactive bands were detected with enhanced chemiluminescence reagent (Super Signal West Pico Substrate; Pierce) and signals were visualized by exposing the membranes to ECL Hyperfilm (Amersham Biosciences). Quantification of western blots was carried out using Quantity One software of Bio-Rad. At least three separate experiments were analyzed and band intensities were normalized to loading control. P-values were determined using unpaired t-tests.
Mean and standard deviation were calculated by Microsoft Office Excel 2007. The error bar represents standard error ((standard deviation/√n) n = sample size).
3. Results and Discussion
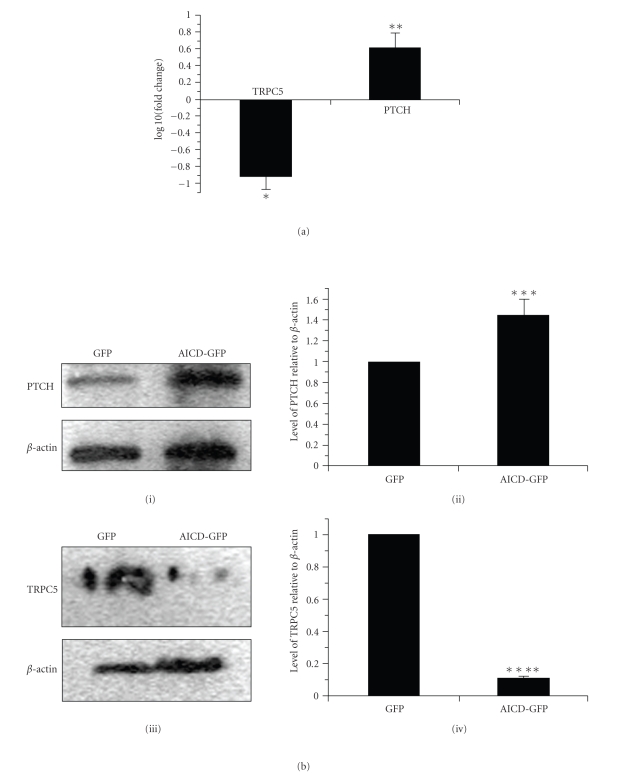
From microarray experiment we got the hint that AICD overexpression might alter the expression level of PTCH1 and TRPC5 (data not shown). To provide further evidence, the mRNA expression levels of both the proteins were checked by real-time PCR using total RNA from AICD-GFP overexpressed cells and compared those with the expression of only pGFP C1 transfected cells. About 2-fold increase and 3-fold decrease in the expressions of PTCH1 and TRPC5, respectively, was observed upon AICD overexpression (Figure 1(a)). The changes in the expressions of the genes at RNA level was further verified at protein levels also. By western blot analysis it was revealed that endogenous PTCH1 level was raised 1.5 fold (Figure 1(b), panels (i) and (ii)) and TRPC5 level was decreased 9 fold (Figure 1(b), panels (iii) and (iv)) in AICD-GFP overexpressing Neuro 2A cells compared to control.
Figure 1.
(a) RNA was isolated from Neuro 2A cells transfected with either GFP or AICD-GFP. The first strand cDNA was synthesized using random hexamer primers and reverse transcriptase. Using that c-DNA as template, expression of PTCH1 and TRPC5 were also checked by real-time PCR in both the cell lines. The relative quantification of both the genes in AICD-GFP transfected cell compared to only GFP transfected cell were expressed in terms of 2−ΔΔCt values after normalization with respect to internal control (beta-actin gene) and plotted in log scale (log10(Fold change)) * indicated P < .02 and ** indicated P < .05. (b) Proteins were prepared from both GFP and AICD-GFP transfected cells after 24 hours of transfection, run on SDS-PAGE and western blot was done with antibody against PTCH1 (i) as well as TRPC5 (iii) and beta actin as loading control in both the cases. Fold change was calculated by densitometry analysis taking beta-actin as loading control (ii) and (iv). *** indicated P < .05 and **** indicated P < .005.
PTCH1 acts as a receptor for sonic hedgehog (SHH) [16] and seem to have a tumor suppressor function, as inactivation of this protein is probably a necessary, if not sufficient step for tumorigenesis [17]. Defects in PTCH1 are known to be a cause of sporadic basal cell carcinoma (BCC) [18]. Hence overexpression of PTCH1 by AICD could be a protective measure of cells against tumorigenesis. This observation seems to be interesting because AICD is previously reported to modulate EGFR-mediated tumorigenesis by reducing the expression of EGFR [11].
On the other hand TRPC5 forms a receptor-activated nonselective calcium permeant cation channel. TRPC1 and TRPC5 are subunits of a heteromeric neuronal channel in mammalian brain [19]. TRPC5 is reported to colocalize with stathmin-like-2, a neuronal growth protein, within the vesicles and in the growth cone. A dominant-negative form of TRPC5 allowed significantly longer neurites and filopodia to form, suggesting that TRPC5 regulates neuronal growth [20]. It was also reported that influxes of calcium via voltage-gated channels play a role in neuronal outgrowth and suggested that TRPC5 is a candidate for the regulation of calcium waves [20]. A decrease in the expression of TRPC5 by AICD overexpression might affect neurite outgrowth.
4. Conclusion
In conclusion, our results suggest that overexpression of AICD can modulate both developmental and degenerative pathways in the cell. Whether the cell would take the survival or the degenerative route at the end depends on other regulatory parameters.
Acknowledgment
This work was supported by the SPGHGD (DAE) project, Government of India.
References
- 1.Heber S, Herms J, Gajic V, et al. Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. Journal of Neuroscience. 2000;20(21):7951–7963. doi: 10.1523/JNEUROSCI.20-21-07951.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Strooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. Journal of Cell Science. 2000;113(11):1857–1870. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- 3.Koo EH. The beta-amyloid precursor protein (APP) and Alzheimer's disease: does the tail wag the dog? Traffic. 2002;3:763–770. doi: 10.1034/j.1600-0854.2002.31101.x. [DOI] [PubMed] [Google Scholar]
- 4.Raychaudhuri M, Mukhopadhyay D. AICD and its adaptors—in search of new players. Journal of Alzheimer’s Disease. 2007;11(3):343–358. doi: 10.3233/jad-2007-11311. [DOI] [PubMed] [Google Scholar]
- 5.Heese K, Akatsu H. Alzheimer’s disease—an interactive perspective. Current Alzheimer Research. 2006;3(2):109–121. doi: 10.2174/156720506776383022. [DOI] [PubMed] [Google Scholar]
- 6.Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 2006;127(1):185–197. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 7.Cao X, Sudhof TC. transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293(5534):115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 8.Pardossi-Piquard R, Petit A, Kawarai T, et al. Presenilin-dependent transcriptional control of the Abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP. Neuron. 2005;46:541–554. doi: 10.1016/j.neuron.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 9.von Rotz RC, Kohli BM, Bosset J, et al. The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. Journal of Cell Science. 2004;117(19):4435–4448. doi: 10.1242/jcs.01323. [DOI] [PubMed] [Google Scholar]
- 10.Kim HS, Kim EM, Lee JP, et al. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. The FASEB Journal. 2003;17:1951–1953. doi: 10.1096/fj.03-0106fje. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y-W, Wang R, Liu Q, Zhang H, Liao F-F, Xu H. Presenilin/γ-secretase-dependent processing of β-amyloid precursor protein regulates EGF receptor expression. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(25):10613–10618. doi: 10.1073/pnas.0703903104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barger SW, Smith-Swintosky VL, Rydel RE, Mattson MP. β-Amyloid precursor protein mismetabolism and loss of calcium homeostasis in Alzheimer’s disease. Annals of the New York Academy of Sciences. 1993;695:158–164. doi: 10.1111/j.1749-6632.1993.tb23045.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Tang BL. The amyloid precursor protein and postnatal neurogenesis/neuroregeneration. Biochemical and Biophysical Research Communications. 2006;341(1):1–5. doi: 10.1016/j.bbrc.2005.12.150. [DOI] [PubMed] [Google Scholar]
- 14.Mattson MP, Furukawa K. Signaling events regulating the neurodevelopmental triad glutamate and secreted forms of β-amyloid precursor protein as examples. Perspectives on Developmental Neurobiology. 1998;5(4):337–352. [PubMed] [Google Scholar]
- 15.Mattson MP, Rydel RE, Lieberburg I, Smith-Swintosky VL. Altered calcium signaling and neuronal injury: stroke and Alzheimer’s disease as examples. Annals of the New York Academy of Sciences. 1993;679:1–21. doi: 10.1111/j.1749-6632.1993.tb18285.x. [DOI] [PubMed] [Google Scholar]
- 16.Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. American Journal of Human Genetics. 2000;67(5):1047–1054. doi: 10.1016/s0002-9297(07)62934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stone DM, Hynes M, Armanini M, et al. The tumour-suppressor gene patched encodes a candidate receptor for sonic hedgehog. Nature. 1996;384(6605):129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 18.Hahn H, Wicking C, Zaphiropoulos PG, et al. Mutations of the human homolog of drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 19.Strübing C, Krapivinsky G, Krapivinsky L, Clapham DE. TRPC1 and TRPC5 form a novel cation channel in mammalian brain. Neuron. 2001;29(3):645–655. doi: 10.1016/s0896-6273(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 20.Greka A, Navarro B, Oancea E, Duggan A, Clapham DE. TRPC5 is a regulator of hippocampal neurite length and growth cone morphology. Nature Neuroscience. 2003;6(8):837–845. doi: 10.1038/nn1092. [DOI] [PubMed] [Google Scholar]