Abstract
Organisms are predisposed to different types in DNA damage. Multiple mechanisms have evolved to deal with the individual DNA lesions. Translesion synthesis is a special pathway that enables the replication fork to bypass blocking lesions. Proliferative Cell Nuclear Antigen (PCNA), which is an essential component of the fork, undergoes posttranslational modifications, particularly ubiquitylation and sumoylation that are critical for lesion bypass and for filling of DNA gaps which result from this bypass. A special ubiquitylation system, represented by the Rad6 group of ubiquitin conjugating and ligating enzymes, mediates PCNA mono- and polyubiquitylation in response to fork stalling. The E2 SUMO conjugating enzyme Ubc9 and the E3 SUMO ligase Siz1 are responsible for PCNA sumoylation during undisturbed S phase and in response to fork stalling as well. PCNA monoubiquitylation mediated by Rad6/Rad18 recruits special polymerases to bypass the lesion and fill in the DNA gaps. PCNA polyubiquitylation achieved by ubc13-mms2/Rad 5 in yeast mediates an error-free pathway of lesion bypass likely through template switch. PCNA sumoylation appears required for this error-free pathway, and it plays an antirecombinational role during normal replication by recruiting the helicase Srs2 to prevent sister chromatid exchange and hyper-recombination.
1. Introduction
DNA damage is an unavoidable aspect of existence that results from both endogenous metabolism and exogenous insults. These include reactive oxygen species and DNA replication errors, in addition to ionizing radiation, UV light and mutagenic chemicals. There are multiple specialized DNA repair pathways that correct various DNA lesions, such as abasic nucleotides [1], mismatched bases [2], single strand defects or lesions [3], and double strand breaks (DSBs) [4]. In certain conditions, the DNA damage may persist unrepaired until a replication fork collides with it. This is seen often with DNA interstrand cross-linking (ICL) lesions which are some of the most toxic types of DNA damage. ICL lesions are usually repaired in S-Phase, after replication forks encounter them [5]. Other types of DNA damage observed at the single strand level, resulting from UV exposure or certain chemicals, can also block the replication fork.
The arrested fork usually deals with such collision utilizing a potentially mutagenic process named Translesion Synthesis (TLS) [6, 7]. This type of DNA synthesis ensures relatively uninterrupted replication even in the face of DNA injury. TLS was initially described in prokaryotes, and termed Post-Replication Repair (PRR) [8]. In E Coli TLS leaves behind single stranded gaps that are repaired at a subsequent cell cycle stage. Similar gaps have been described in Eukaryotes as well [9, 10]. When a replication fork collides with an ICL or a single strand lesion, a one-ended DSB may form. This could trigger homology-based repair, also termed Homologous Recombination (HR), which is another pathway the fork utilizes to repair DNA lesions particularly ICLs [11]. The classic DNA polymerases that mediate undisturbed replication cannot bypass most ssDNA lesions, and several alternative (atypical) polymerases that mediate lesion bypass in TLS have been described. The key regulator of TLS is the replication sliding clamp Proliferative Cell Nuclear Antigen (PCNA). The PCNA trimer holds multiple proteins that participate in both normal replication and TLS. The role of PCNA in TLS is governed by posttranslational modifications that occur to it in response to an arrested fork. In this brief paper we describe the process of PCNA ubiquitylation and sumoylation in response to replication fork stalling and the impact of these modifications on TLS.
2. PCNA Ubiquitylation in Response to Fork Stalling Lesions
PCNA has been described as the coordinator of the replicating fork [12]. It mediates the recruitment of multiple factors required for DNA replication and repair. In a seminal paper, the Jentsch group described the involvement of PCNA ubiquitylation and sumoylation in TLS [13]. This Ubiquitylation is mediated by the Rad6 group TLS factors [13]. It had been known for a long time that Rad6 family was involved in TLS in Eukaryotes, and that most of its members display ubiquitin conjugating and ligating activities [14–16]. Protein ubiquitylation has emerged as a widespread process that impacts a myriad of cellular processes in eukaryotes [17]. This process starts with binding of the conserved 76 aa peptide ubiquitin to E1 ubiquitin activating enzyme. This transfers ubiquitin to an E2 conjugating enzyme, which interacts with an E3 ligase to transfer ubiquitin to a specific substrate. The E3 ligase is the determinant of the specificity of the substrate to which ubiquitin is attached. Rad6/Rad18 are the E2/E3 complex that mediates PCNA ubiquitylation at a conserved Lysine164 [13]. It has been shown that this modification increases affinity of TLS DNA polymerases to PCNA which contributes to lesion bypass [18]. This was initially demonstrated for the human TLS Polymerase eta (polη) but later was shown to apply to other eukaryotic polymerases of the Y family, including polymerases ι and κ which are not present in yeast [19, 20]. This enhanced affinity for PCNA by the Y family of TLS DNA polymerases is due to presence of ubiquitin-binding domains of the UBM or UBZ types [21] in these polymerases which interact with the monoubiquitin on PCNA (Figure 1). The major TLS polymerases in yeast include Rad30, Rev1, Rev3, and Rev7 (Rev7 is the regulatory unit, and Rev3 is the catalytic unit of Polymerase zeta.) Further discussion of the detailed role of each TLS polymerase can be found elsewhere [6, 7].
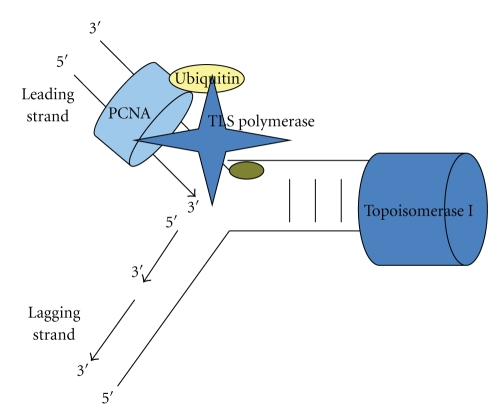
Figure 1.
Fork stalling results in monoubiquitylation of the replication clamp PCNA which increases its affinity to a TLS polymerase. The latter replaces the high-fidelity replicative polymerase and it can accommodate the lesion even with the risk of generating mutations in certain circumstances. This Polymerase binds PCNA through its PIP box and a ubiquitin binding domain.
PCNA undergoes further ubiquitylation on the same Lysine 164 to generate a ubiquitin chain [13]. This polyubiquitin chain formation is dependent on the presence of the monoubiquitin species mediated by Rad6/Rad18 [13]. The ubiquitin molecules within this chain bind each other through the lysine 63 (K63) of each ubiquitin. This type of ubiquitin chain is different from that of the canonical ubiquitin chains bound through K48 which are responsible for substrate degradation by the proteasome system [17]. Although a K63 ubiquitin chain could lead to protein degradation, it has mainly been implicated in non-degradation pathways such as vesicle excretion, signaling, the immune response, and different forms of DNA repair [17]. PCNA polyubiquitylation is mediated by another E2/E3 complex in the Rad6 pathway of PRR, namely ubc13-mms2/Rad 5 in yeast [13]. A mammalian ubc13-mms2 homologue has been known for years [15], and two mammalian homologues of Rad5 with E3 ligase and helicase activities (HLTF and SHPRH) were recently described [22–24]. PCNA polyubiquitylation in the setting of fork-blocking lesions results in a form of error-free repair believed to be mediated by template switching (Figure 2) [25]. The exact mechanism of such template switching is unknown, but it appears (at least in yeast) that there are two pathways: one is Rad52 dependent and the other is Rad52 independent [26]. A recent study indicated that Rad5-mediated template switch is appreciated as X-shaped DNA structure on 2-gel electrophoresis, and it appears to involve holiday junctions (recombination intermediates) and HR proteins [27]. Another potential scenario for this error-free pathway is through fork regression after stalling which has been demonstrated in vitro [28]. Rad5 appears to promote this fork regression. However, since competent replication checkpoint prevents such fork regression, it is believed to be unlikely mechanism to mediate this pathway [29]. Mutant Rad5 or ubc13 yeast strains lack such error-free repair [13, 25]. PCNA monoubiquitylation at K164 has been described across all eukaryotic cells, from budding [13, 30] and fission yeast [31] to humans [18], and includes chicken DT40 cells [32, 33] and Xenopus laevis egg extracts [34]. This monoubiquitylation is seen during normal replication in DT40 and mammalian cells [31, 32]. It is induced by DNA damage in human cells, and is observed only after DNA damage in budding yeast [13, 30]. PCNA polyubiquitylation is clearly seen in yeast after DNA damage, but has been difficult to demonstrate in mammalian systems, and often requires over-expression of the E3 ligases to be observed [22–24]. Evidence of the existence of an error-free TLS pathway mediated by PCNA polyubiquitylation is clearly demonstrated in yeast, and such pathway is believed to exist in higher eukaryotes including mammals. There appear to be other E3 ligases besides Rad18 that could result in PCNA monoubiquitylation, and although Rad18 is the major E3 ligase for PCNA monoubiquitylation post DNA damage, a Rad18 independent monoubiquitylation has been described [33]. A recent report revealed a contribution of the PCNA binding E3 ligase CRL4 to PCNA monoubiquitylation, in vitro and in vivo, in a steady state without DNA damage [35]. In preliminary experiments, we have observed that decreased expression of the DNA repair, protein and E3 ligase Pso4 decreases PCNA monoubiquitylation (Shaheen and Hromas: unpublished observation). Finally, PCNA monoubiquitylation has also been noted in G1 phase of cell cycle in response to ICL agents in a pathway that relies on NER and the TLS polymerase Zeta [36].
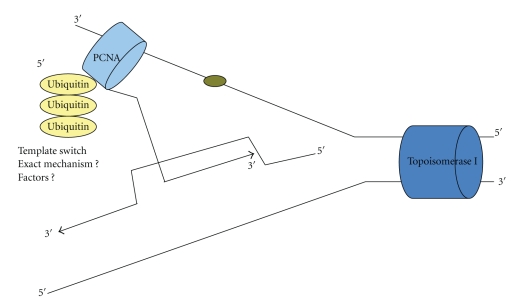
Figure 2.
PCNA polyubiquitination mediates an error-free repair believed to occur due to template switch. The factors that perform this process are unknown. Holiday Junctions (HJs) appear be intermediates in this pathway [27].
3. The Sumoylation of PCNA in Response to Fork Stalling
The Small Ubiquitin-like MOdifier (SUMO) is involved in multiple cellular functions including nuclear transport, signal transduction, transcription, and genome stability [37]. It has been shown that the ubiquitin and SUMO modifications compete for the same attachment site on a common substrate in certain circumstances, such as the case of the NF-κB inhibitor IκBα, where sumoylation was found to counteract the proteolytic effect of ubiquitylation [38]. In a screen for sumoylated proteins, PCNA was identified as a target of this modification [13]. PCNA from S. cerevisiae is sumoylated primarily on K164, the same site as ubiquitination, during S phase even in the absence of DNA damage. This is mediated by the SUMO-specific E2 conjugating enzyme Ubc9 and the SUMO E3 ligase Siz1. K127 on PCNA is also sumoylated independently of Siz1 although not nearly to the same extent as K164 [13]. The functional significance of PCNA sumoylation is described only in budding yeast, and its role in higher organisms is debatable. Although PCNA K164 sumoylation is seen in chicken DT40 cells [32] and Xenopus laevis egg extracts [34], it has yet to be reported in human cells. PCNA SUMO chains have been described but their function is uncertain [30].
It was first suggested that PCNA sumoylation favors the repair of fork blocking lesions by a mutagenic TLS pathway over an error-free recombination based pathway. This is shown through investigating the RAD6 pathway mutants which are highly sensitive to various DNA-damaging agents, but this sensitivity is partially rescued by blocking PCNA sumoylation, in a manner dependent on homologous recombination [30, 39]. A yeast antirecombination helicase (Srs2) was identified as a suppressor of mutant Rad6 phenotype, in a manner dependent on HR [40]. Subsequently, it was shown that Srs2 binds sumoylated PCNA through its sumo-interacting motif, and this may explain the function of sumoylated PCNA during physiologic replication in preventing deleterious hyper-recombination [41, 42]. The lack of PCNA sumoylation leads to increased spontaneous sister chromatid recombination during mitotic growth [41, 42]. A recent important yeast study [43] investigated the required factors to form sister chromatid junctions (expected to occur during template switching or HR) in the Rad18-Rad5 mediated damage-bypass, and it identified the SUMO conjugating enzyme Ubc9 and sumoylated PCNA as essential for this process, again dependent on Rad51 and so potentially on HR [43]. This research suggested the presence of two error-free pathways mediating lesion bypass: one is mediated by template switching (which may or may not involve recombination) and requires PCNA sumoylation and ubiquitination, and also it is genetically stable The other pathway operates when the first pathway is defective, and depends on Rad51 recombination and it is genetically unstable, and could lead to chromosomal translocation [43–45]. Another investigation demonstrated that sumoylation of PCNA could interfere with its association with another protein, Eco1 which is important for the establishment of chromatid cohesion during replication [46]. Blocking PCNA sumoylation partially rescues the temperature sensitivity of some Eco1 mutants [47]. It is also believed that the binding of the E2 SUMO ligase Ubc9 to PCNA-on a critical region may block the binding of other PCNA interacting proteins [12]. All the above may indicate that sumoylation of PCNA also plays a key regulatory role in recombination and thus genomic stability.
4. What Types of DNA Lesions Do Trigger PCNA Modification?
PCNA ubiquitylation and sumoylation are triggered by a wide variety of DNA lesions that block the replication fork, such as the ones caused by alkylating chemicals (e.g., methyl-methanesulfonate (MMS) and 4-nitroquinoline oxide), bulky adducts such as benzo[a]pyrene dihydrodiol epoxide (BPDE), hydrogen peroxide (which produces oxidative damage), UV light (producing photoproducts) and ICL agents [13, 30, 47]. Nucleotide depletion such as the one achieved by hydroxyurea (HU), which causes a fork stalling and subsequent collapse, can also induce PCNA modification. However, HU is a weaker trigger of PCNA monoubiquitylation when compared to ICL or alkylating agents [48]. PCNA modification is tightly connected to replication since chemicals that cause direct DNA DSBs, such as bleomycin, do not cause PCNA ubiquitination or sumoylation [47, 49]. The topoisomerase I inhibitor camptothecan does not trigger PCNA ubiquitylation even though it blocks fork progression, by collision with the camptothecin/Topo I complex [47, 49]. This led to the suggestion that the uncoupling of fork helicase activity and polymerase movement is the actual trigger of PCNA ubiquitylation [50]. This uncoupling creates ssDNA that binds the Replication Protein A (RPA), which recruits Rad18, to the stalled fork. RPA has been shown to be required for TLS to proceed across multiple types of DNA lesions [44, 46]. Camptothecin triggers some PCNA ubiquitylation in S. pombe [31], but this modification is minimally above the normal S-phase ubiquitin signal that is seen in this organism. Ionizing Radiation (IR) triggers PCNA ubiquitylation in budding and fission yeast [30, 31] but not in mammalian cells [18, 49]. As mentioned above, in S. cerevisiae, PCNA is sumoylated in S phase without DNA damage while PCNA ubiquitylation is noted during S phase in S. Pombe, and higher eukaryotes including humans [13, 30, 31, 47, 49].
5. The Relation between PCNA Ubiquitylation and the Kinetics of TLS
Traditionally, TLS is envisioned as an alternative replication process by which the stalled fork can bypass a lesion. PCNA ubiquitylation is imagined as facilitating this bypass by recruiting low-fidelity polymerases. However, recent lines of evidence suggest that PCNA ubiquitylation may play its major role in filling in the gaps generated in PRR by utilizing these TLS polymerases. One study, using 2-D gel and electron microscopy to probe repair intermediates, revealed that UV-irradiated S. cerevisiae cells uncouple leading and lagging strand replication at irreparable UV lesions, thus generating long ssDNA regions on one side of the fork [51]. Small ssDNA gaps accumulate along the replicated duplexes, likely resulting from repriming events on both leading and lagging strands. It was concluded that TLS and homologous recombination factors counteract gap accumulation without affecting fork progression [51]. Recent work revealed that limiting the mutagenic or error-free pathways of TLS to the G2/M phases of the cell-cycle promote efficiently lesion tolerance indicating that both branches of the DNA damage tolerance operate effectively after chromosomal replication, outside S phase [52]. Another elegant study using an inducible system of DNA damage bypass in S. cerevisiae demonstrated that TLS occur predominantly during S-phase but it is separable in time and space from genome replication [53]. The same study found that both during and after S phase, ultraviolet-radiation-induced lesions are bypassed predominantly via error-prone translesion synthesis whereas the error-free pathway functions as a backup system. The process of bypassing the lesion itself may rely more on other factors rather than on modified PCNA. For instance, using the genetically tractable chicken cell line DT40, it was shown that TLS at stalled replication forks requires both Rev1 translesion polymerase-interaction domain and ubiquitin-binding domain in its C terminus. Surprisingly, however, PCNA ubiquitylation was not required to maintain normal fork progression on damaged DNA. Conversely, PCNA ubiquitylation was essential for filling PRR gaps [54]. Rev1 may recruit other essential TLS components through its multifunctional domains required for lesion bypass. On the other hand, it was demonstrated that the level of Rev1 protein is extremely low during G1 and rises slowly throughout early and mid-S phase but begins to increase rapidly only in late S phase, reaching a maximum level in G2. Its level is then maintained at a high intracellular concentration throughout mitosis until after telophase [55]. DNA damage causes Rev1 to accumulate earlier in S phase without significantly affecting the level reached in G2/M phase. This is also suggestive of a role in PRR in G2 rather than S phase, as would be predicted [55]. This cell cycle regulation of Rev1 is seen mainly in yeast and has not been demonstrated in higher eukaryotes. Notice that, Rev1 catalytic activity may be dispensable for TLS [56], and this protein may play its major role in TLS as a scaffold that attracts other TLS polymerases [57]. Overall, the picture is still nebulous regarding the exact kinetics of TLS/PRR in eukaryotes. Future studies may shed light on the detailed mechanisms of lesion bypass.
With the sequence of events that build up at the stalled fork, it was shown that Rad18 binds ssDNA [58], but this binding is much weaker compared to the binding of RPA to ssDNA. Thus, it appears that RPA recruits Rad18 to ssDNA [47]. Rad18 in turn binds directly to Rad6 [58, 59] to initiates PCNA monoubiquitylation, and it also directly binds Rad5 [60], which together with MMS2/Ubc13 [16] mediate PCNA polyubiquitylation. Fluorescence-based biophysical methods revealed that mammalian Rad18 becomes immobilized in nuclear foci only in S phase cells, and that its physical association with Rad6 or Polymerase eta is appreciated only in these foci upon DNA damage [61].
6. Other Functions of the Rad6/Rad18 in DNA Repair
The 9-1-1 checkpoint clamp is a complex with structural similarity of PCNA. It is implicated in signaling from ssDNA at the stalled fork to the checkpoint proteins, particularly Chk1, to activate the replication checkpoint [62, 63]. One recent report identified 9-1-1 as a target of Rad6/Rad18 monoubiquitylation in budding yeast upon triggering DNA damage [64]. This ubiquitylation is involved in control of global gene regulation in a way reminiscent of the bacterial SOS response to DNA damage which enhances DNA repair gene transcription, translesion synthesis, and recombination [64]. Rad18 was also shown to be recruited to sites of DNA DSB probably through interaction of its Ubiquitin Binding domain with ubiquitin chains deposited at the DSB site [65]. It is shown that Rad18 contributes to homologous recombination repair of DSB probably through direct interaction with the recombinase RAD51C [65]. Furthermore, evidence implicates Rad18 in HR since chicken T40 deficient in Rad18 show aberrant gene conversion (the main form of HR) [66]. In addition, the HR pathway that gets activated in the absence of rad18 is a defective one and may lead to genetic instability [43, 66]. Rad18 appears to suppress an NHEJ pathway when DSB is induced at the fork level to promote repair by HR [67]. It seems from all the above that Rad6/Rad18 play key roles in coordinating several DNA damage response pathways through ubiquitylation of two DNA clamps, PCNA and 9-1-1, as well as other unidentified targets.
7. The Role of USP1 in PCNA Deubiquitylation
USP1 was identified in a screen for the ubiquitin protease which mediates the removal of monoubiquitin from ubiquitylated Fanconi anemia group D2 (Fancd2 is ubiquitylated in response to fork stalling, and it contributes to TLS, particularly in response to ICL lesions.) [68], Subsequently, USP1 was identified as a deubiquitylating enzyme for monoubiquitylated PCNA as well [48]. USP1 gets cleaved, and subsequently degraded by the proteasome system in response to UV light exposure, but not to alkylating or cross-linking agents [48, 68]. It is believed that there is a steady-state level of PCNA ubiquitylation by Rad6-Rad18 which is continuously antagonized by USP1, and when USP1 level goes down post-UV exposure, this leads to detectable PCNA ubiquitylation. Contrary to prediction, USP1 deletion leads to DNA damage sensitivity [69], and mice deficient in USP1 display DNA damage phenotype reminiscent of Fanconi anemia [70]. This defective DNA repair is associated with constitutively chromatin-bound monoubiquitylated FANCD2. In contrast, persistent PCNA monoubiquitylation has negligible impact on DNA repair or mutagenesis [69]. The molecular mechanism of this phenotype is uncertain. It is worth mentioning that PCNA ubiquitylation occurs earlier after UV light than that after chemical exposure, and it persists for a long time (at least 48 hours) after a single exposure to different DNA damaging agents ([48, 49], and our observation).
8. Does the Replication Checkpoint Activate PCNA Ubiquitylation?
The replication checkpoint gets activated in response to situations that cause replication fork stalling in S phase. ATR (Ataxia Telangiectasia and Rad3-related) protein plays a central role in activating this checkpoint. The exposed ssDNA at the fork recruits RPA, which, in turn, recruits ATRIP (ATR-Interacting Protein), and that brings in ATR, which phosphorylates and activates Chk1 [62, 63]. The PCNA-like replication clamp (the 9-1-1 complex), whose loader interacts with RPA, also contributes to ATR and Chk1 activation. ATR and Chk1 phosphorylate multiple proteins to among other functions stabilize the stalled fork, suppress the late-firing origins of replications, halt cell cycle progression, and induce repair pathways, [62, 63]. In yeast and lower eukaryotes, it has been shown that this checkpoint activation does not alter the status of PCNA ubiquitylation [31, 47, 50]. In other words, checkpoint activation in yeast could be prevented by suppressing ATR or Chk1 without impacting PCNA ubiquitylation in response to fork stalling. In mammalian systems, the picture is less clearer. One report demonstrated 60% reduction in PCNA monoubiquitylation triggered by chemical DNA damage when ATR or Chk1 levels were reduced by siRNA [20]. Another study failed to show a change in the PCNA-monoubiquitin level upon reducing ATR levels [49], but did find a reduction when RPA was reduced. Thus, RPA appears to be instrumental for PCNA ubiquitylation to be induced [47, 49]. PTIP/Swift, an adaptor protein for the checkpoint kinases ATM and ATR, appears to contribute to PCNA ubiquitylation in human cells and X. laevis egg extracts since depletion of PTIP/Swift results in a reduction in this modification [71]. The cell cycle inhibitor p21 binds PCNA, and its down-regulation is required for PCNA monoubiquitylation upon DNA damage [72]. On the other hand, a contradictory study showed that depleting p21 or p53 results in a decrease in PCNA monoubiquitylation post-UV exposure [73]. The mechanisms of these conflicting effects are unknown, but it is worth mentioning that one of these two studies exogenously over-expressed p21 and this could have contributed to the difference in outcome [72]. One report indicated that replication checkpoint proteins are dispensable for TLS to proceed in yeast [74]; however, if there is a deficiency of NER, then repair of ssDNA lesions is heavily tilted toward TLS, and checkpoint proteins enhance the repair by TLS in this situation [74, 75].
9. Summary and Future Directions
Research in TLS/PRR has progressed through multiple stages over the past 5 decades, from its description in bacteria and then eukaryotes, to discovering its mechanisms in bacteria and yeast. A key step in this field was the identification of Rad6 as essential for TLS in eukaryotes. This led to the report [13] that described the fundamental role of the Rad6 group in ubiquitylating PCNA. The identification of PCNA posttranslational modifications opened the door for a cascade of other studies into the role of modified PCNA in TLS and the mechanism of that. However, there are questions that remain to be answered about the details of the error-free pathway of TLS, and the timing of events that take place at the stalled fork. There is no doubt that ongoing research in this area will come up with explanations for all these questions that may or may not agree with current predictions. It is noteworthy to mention that research in this field has also enriched our understanding of the mechanisms of mutagenesis, and the implications of that in carcinogenesis and cancer therapy.
References
- 1.Robertson AB, Klungland A, Rognes T, Leiros I. Base excision repair: the long and short of it. Cellular and Molecular Life Sciences. 2009;66(6):981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G-M. Mechanisms and functions of DNA mismatch repair. Cell Research. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 3.Nouspikel T. DNA repair in mammalian cells: nucleotide excision repair: variations on versatility. Cellular and Molecular Life Sciences. 2009;66(6):994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annual Review of Genetics. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 5.Räschle M, Knipsheer P, Enoiu M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134(6):969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nature Reviews Molecular Cell Biology. 2005;6(12):943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann AR, Niimi A, Ogi T, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6(7):891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. Journal of Molecular Biology. 1968;31(2):291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 9.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Molecular and General Genetics. 1981;184(3):471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence C. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? BioEssays. 1994;16(4):253–258. doi: 10.1002/bies.950160408. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Heyer W-D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Research. 2008;18(1):99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moldovan G-L, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 14.Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96(5):645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 16.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. The EMBO Journal. 2000;19(13):3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochimica et Biophysica Acta. 2004;1695(1-3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 18.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Molecular Cell. 2004;14(4):491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 19.Plosky BS, Vidal AE, de Henestrosa ARF, et al. Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. The EMBO Journal. 2006;25(12):2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi X, Barkley LR, Slater DM, et al. Rad18 regulates DNA polymerase κ and is required for recovery from S-phase checkpoint-mediated arrest. Molecular and Cellular Biology. 2006;26(9):3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bienko M, Green CM, Crosetto N, et al. Biochemistry: ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310(5755):1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 22.Unk I, Hajdú I, Fátyol K, et al. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(10):3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unk I, Hajdú I, Fátyol K, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(48):18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motegi A, Liaw H-J, Lee K-Y, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(34):12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(44):15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gangavarapu V, Prakash S, Prakash L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae . Molecular and Cellular Biology. 2007;27(21):7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minca EC, Kowalski D. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Molecular Cell. 2010;38(5):649–661. doi: 10.1016/j.molcel.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blastyák A, Pintér L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Molecular Cell. 2007;28(1):167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297(5581):599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 30.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425(6954):188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 31.Frampton J, Irmisch A, Green CM, et al. Postreplication repair and PCNA modification in Schizosaccharomyces pombe . Molecular Biology of the Cell. 2006;17(7):2976–2985. doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arakawa H, Moldovan G-L, Saribasak H, Saribasak NN, Jentsch S, Buerstedde J-M. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biology. 2006;4(11):1947–1956. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson LJ, Ross A-L, Szüts D, et al. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Reports. 2006;7(9):927–932. doi: 10.1038/sj.embor.7400777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leach CA, Michael WM. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. Journal of Cell Biology. 2005;171(6):947–954. doi: 10.1083/jcb.200508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terai K, Abbas T, Jazaeri AA, Dutta A. CRL4Cdt2 E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Molecular Cell. 2010;37(1):143–149. doi: 10.1016/j.molcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase ζ . The EMBO Journal. 2006;25(6):1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson ES. Protein modification by SUMO. Annual Review of Biochemistry. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 38.Desterro JMP, Rodriguez MS, Hay RT. SUMO-1 modification of IκBα inhibits NF-κB activation. Molecular Cell. 1998;2(2):233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 39.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae . Molecular and Cellular Biology. 2004;24(10):4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiestl RH, Prakash S, Prakash L. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics. 1990;124(4):817–831. doi: 10.1093/genetics/124.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436(7049):428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 42.Papouli E, Chen S, Davies AA, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Molecular Cell. 2005;19(1):123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456(7224):915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 44.Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. Journal of Cell Biology. 2006;175(5):703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Motegi A, Kuntz K, Majeed A, Smith S, Myung K. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae . Molecular and Cellular Biology. 2006;26(4):1424–1433. doi: 10.1128/MCB.26.4.1424-1433.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moldovan G-L, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Molecular Cell. 2006;23(5):723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Molecular Cell. 2008;29(5):625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang TT, Nijman SMB, Mirchandani KD, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nature Cell Biology. 2006;8(4):339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 49.Niimi A, Brown S, Sabbioneda S, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(42):16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. Journal of Biological Chemistry. 2006;281(43):32081–32088. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- 51.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Molecular Cell. 2006;21(1):15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 52.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141(2):255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 53.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465(7300):951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Molecular Cell. 2008;30(4):519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 55.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G2/M phase rather than S phase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(24):8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ross A-L, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Research. 2005;33(4):1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo C, Fischhaber PL, Luk-Paszyc MJ, et al. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. The EMBO Journal. 2003;22(24):6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes and Development. 1994;8(7):811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 59.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. Journal of Biological Chemistry. 1997;272(37):23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 60.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. The EMBO Journal. 2000;19(13):3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watson NB, Nelson E, Digman M, Thornburg JA, Alphenaar BW, McGregor WG. RAD18 and associated proteins are immobilized in nuclear foci in human cells entering S-phase with ultraviolet light-induced damage. Mutation Research. 2008;648(1-2):23–31. doi: 10.1016/j.mrfmmm.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nature Reviews Molecular Cell Biology. 2010;11(3):208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 64.Fu Y, Zhu Y, Zhang K, Yeung M, Durocher D, Xiao W. Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell. 2008;133(4):601–611. doi: 10.1016/j.cell.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 65.Huang J, Huen MSY, Kim H, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nature Cell Biology. 2009;11(5):592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szüts D, Simpson LJ, Kabani S, Yamazoe M, Sale JE. Role for RAD18 in homologous recombination in DT40 cells. Molecular and Cellular Biology. 2006;26(21):8032–8041. doi: 10.1128/MCB.01291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saberi A, Hochegger H, Szuts D, et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Molecular and Cellular Biology. 2007;27(7):2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nijman SMB, Huang TT, Dirac AMG, et al. The deubiquitinating enzyme USP1 regulates the fanconi anemia pathway. Molecular Cell. 2005;17(3):331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 69.Oestergaard VH, Langevin F, Kuiken HJ, et al. Deubiquitination of FANCD2 is required for DNA crosslink repair. Molecular Cell. 2007;28(5):798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim JM, Parmar K, Huang M, et al. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Developmental Cell. 2009;16(2):314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Göhler T, Munoz IM, Rouse J, Blow JJ. PTIP/Swift is required for efficient PCNA ubiquitination in response to DNA damage. DNA Repair. 2008;7(5):775–787. doi: 10.1016/j.dnarep.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 72.Soria G, Podhajcer O, Prives C, Gottifredi V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 2006;25(20):2829–2838. doi: 10.1038/sj.onc.1209315. [DOI] [PubMed] [Google Scholar]
- 73.Avkin S, Sevilya Z, Toube L, et al. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Molecular Cell. 2006;22(3):407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 74.Pagès V, Santa Maria SR, Prakash L, Prakash S. Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes and Development. 2009;23(12):1438–1449. doi: 10.1101/gad.1793409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paulovich AG, Armour CD, Hartwell LH. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics. 1998;150(1):75–93. doi: 10.1093/genetics/150.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]