Abstract
To investigate regulation of SERCA1a and SERCA2a calcium pump isoforms by phospholamban (PLB), the proteins were fused to fluorescent protein tags and their interactions were quantified by fluorescence resonance energy transfer (FRET) in live cells. For both SERCA1a or SERCA2a, FRET to PLB increased with increasing protein expression level to a maximum value corresponding to a probe separation distance of 64 angstroms. The data indicate the respective regulatory complexes assume the same overall quaternary conformation. However, FRET measurements also revealed that PLB has a 50% higher apparent affinity for SERCA1a relative to SERCA2a. The results suggest that despite structural similarities of the respective regulatory complexes, there is preferential binding of PLB to SERCA1a over SERCA2a. This apparent selectivity may have implications for biochemical studies in which SERCA1a is used as a substitute for SERCA2a. It may also be an important strategic consideration for therapeutic overexpression of SERCA isoforms in cardiac muscle.
Keywords: phospholamban, SERCA1a, SERCA2a, calcium ATPase, affinity, FRET, calcium handling, membrane proteins
Introduction
In cardiac muscle cells, the ion-motive ATPase SERCA2a sequesters calcium (Ca) by transporting it across the membrane of the sarcoplasmic reticulum (SR). The rate of uptake of Ca from the cytoplasm largely determines the rate of muscle relaxation during the diastolic phase of the cardiac cycle. In addition, the amount of Ca sequestered in the SR affects the size of subsequent Ca release events. Thus, SERCA2a activity is an important determinant of both cardiac relaxation (lusitropy) and contraction (inotropy). SERCA’s central role in cardiac function is underscored by its association with heart disease. Impaired cardiac Ca handling is correlated with heart failure 1, so SERCA is considered a valuable target for therapeutic intervention 2. SERCA2a is dynamically regulated by a small membrane protein phospholamban (PLB)3, which inhibits pump activity by decreasing its apparent affinity for Ca. SERCA2a and PLB are also expressed in slow skeletal muscle fibers but they are mostly excluded from fast skeletal (type II) fibers. These fast fibers predominantly express kinetically faster calcium pump SERCA1a, without phospholamban 4. Nevertheless, SERCA1a is capable of functional regulation by PLB 5, and because of its ready availability from fast skeletal muscle preparations it is often used as a substitute for SERCA2a in tests of PLB regulatory function 6. Exogenous SERCA1a has also been used to supplement SERCA2a in order to improve dysfunctional cardiac calcium uptake in heart failure 7; 8. Under these conditions, both SERCA1a and (endogenous) SERCA2a are available for binding and functional regulation by PLB.
In the present study we quantify the relative affinities of SERCA1a and SERCA2a for PLB in order to determine how different SERCA isoforms might compete for available PLB. We have previously used fluorescence resonance energy transfer (FRET) microscopy to measure relative dissociation constants of PLB-PLB binding (Kd1) and PLB-SERCA binding (Kd2) in live AAV-293 cells 9,10. This strategy is applied here to quantify and compare dissociation constants for PLB-SERCA1a (Kd2-1a) and PLB-SERCA2a (Kd2-2a). FRET also provides a measure of donor-acceptor probe separation distance in the PLB-SERCA regulatory complex 9,10. The results give insight into the relative conformations and binding affinity of the SERCA1a and SERCA2a regulatory complexes.
Results
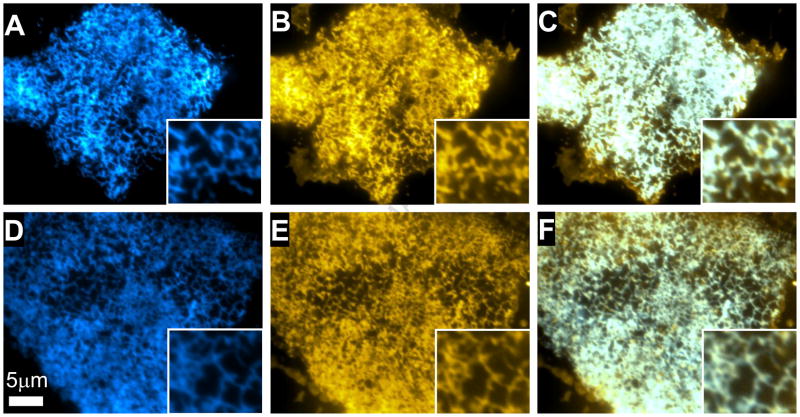
TIRF imaging of live AAV-293 cells co-expressing mCerulean (Cer)-SERCA1a/YFP-PLB or Cer-SERCA2a/YFP-PLB showed cyan fluorescence distributed in a reticulated pattern consistent with the expected ER localization of SERCA (Fig. 1A, D). No difference in the subcellular localization of SERCA1a and SERCA2a was observed. As previously shown, YFP-PLB was localized predominantly in the ER, with an additional fraction visible in the plasma membrane 11 (Fig. 1B, E). Colocalization of YFP-PLB and Cer-SERCA1a or Cer-SERCA2a is shown in Fig. 1C, F.
Figure 1.
TIRF microscopy showed reticulate subcellular distribution of fluorescently labeled proteins. A) Cer-SERCA1a. B) YFP-PLB. C) Overlay of SERCA1a and PLB. D)Cer-SERCA2a. E) YFP-PLB F) Overlay of SERCA2a and PLB. Areas of colocalized Cer/YFP fluorescence appear white in the overlay images.
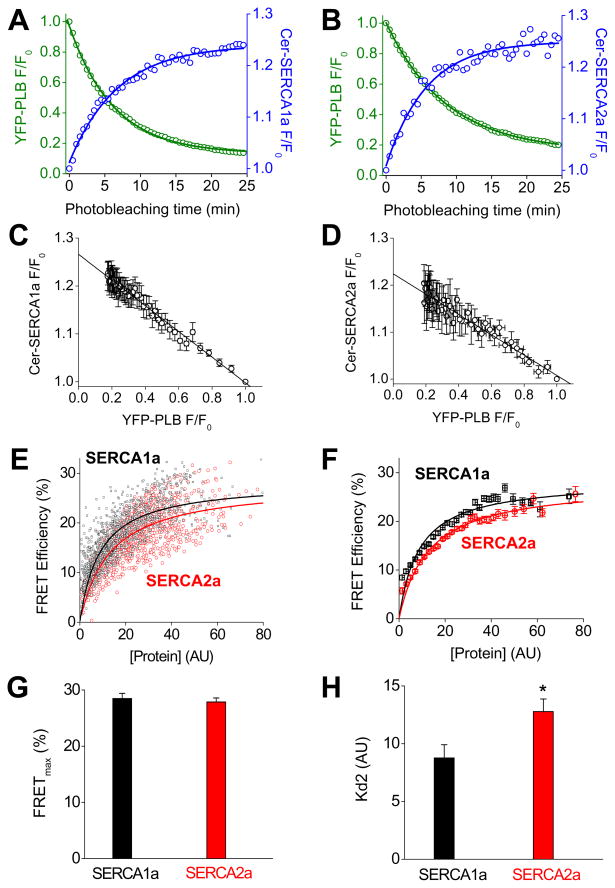
Progressive photobleaching of YFP-PLB fluorescence resulted in increased fluorescence of Cer-SERCA1a (Fig. 2A) or Cer-SERCA2a (Fig. 2B), indicating FRET from Cer-SERCA isoforms to YFP-PLB. The inverse relationship between YFP-PLB and Cer-SERCA1a or Cer-SERCA2a was highly linear (Fig. 2C,D), consistent with a heterodimeric regulatory complex 9. This is in contrast to the highly nonlinear donor/acceptor relationships previously observed for PLB 9 and phospholemman 12 homooligomers. which are consistent with oligomers with 3 or more subunits. The linear regression y-intercept (Fig. 2C,D) also provides a measure of average FRET for SERCA1a (27%) and SERCA2a (22%). It is not clear from this simple measurement whether the observed difference is due to different conformations of PLB-SERCA regulatory complex or different amounts of that complex (i.e. differential binding affinities). To dissect these independent factors we surveyed large populations of cells expressing fluorescently-tagged proteins and compared each cell’s FRET efficiency with its YFP-PLB fluorescence intensity, an index of protein expression 9; 10. For SERCA1a (Fig. 2E, black points) and SERCA2a (Fig. 2E, red points) FRET efficiency increased with increasing protein expression, a relationship that can be approximated by a hyperbolic fit of the form y = (FRETmax) × /(Kd2+×). The parameter FRETmax is the maximal FRET, taken to represent the intrinsic FRET of the bound complex, and Kd2 represents the protein concentration at which half-maximal FRET is achieved (dissociation constant), in arbitrary units. These data are pooled and averaged in Fig. 2F to aid comparison of SERCA1a-PLB and SERCA2a-PLB FRET concentration dependence. Multiple independent experiments were performed to assess the dependence of FRET on protein concentration, each using a population of >200 cells. Mean FRETmax and Kd2 values are summarized in Fig. 2G,H and Table 1. The mean SERCA1a FRETmax value was 28.5 ± 0.9 %, which is similar to previous results 9; 10; SERCA2a FRETmax was 27.9 ± 0.7 %. The measured FRETmax values were not significantly different (Fig. 2G). Notably, the apparent Kd2 (Fig. 2H) for SERCA2a-PLB was 50% higher than for SERCA1a-PLB (p < 0.01), suggesting PLB has a higher affinity for SERCA1a than to its native regulatory target SERCA2a.
Figure 2.
A) Progressive acceptor photobleaching of YFP-PLB (green) resulted in an increase in Cer-SERCA1a fluorescence (blue) indicating FRET. B) Photobleaching of YFP-PLB (green) resulted in an increase in Cer-SERCA2a fluorescence (blue). C) The relationship between the normalized fluorescence of Cer-SERCA1a and YFP-PLB during progressive photobleaching was linear, consistent with a heterodimeric regulatory complex. D) The relationship between the normalized fluorescence of Cer-SERCA1a and YFP-PLB during progressive photobleaching was also linear. E) FRET increased with protein expression level. Hyperbolic fitting showed a right-shifted concentration dependence for Cer-SERCA2a (red) compared to Cer-SERCA1a (black). F) A comparison of pooled data for Cer-SERCA1a (black) and Cer-SERCA2a (red). G) Summary of FRETmax values obtained by fitting reported as mean ± SE. H) Summary of Kd2 values obtained by fitting reported as mean ± SE. * indicates p-value <0.01. The data suggest PLB binds more avidly to SERCA1a than SERCA2a.
Table 1.
Summary of FRET results
| SERCA1a | SERCA2a | |
|---|---|---|
| Mean FRET (%) | 26.6 ± 0.2 | 22.4 ± 0.4 |
| FRETmax (%) | 28.5 ± 0.9 | 27.9 ± 0.7 |
| Probe distance (Å) | 64.3 ± 0.5 | 64.6 ± 0.4 |
| Apparent Kd2 (AU) | 8.8 ± 1.1 | 12.9 ± 1.1 |
| Sample size (# cells) | 1346 | 1110 |
Discussion
The FRETmax values obtained for SERCA1a and SERCA2a correspond to a Cer-YFP probe separation distance of 64.3 ± 0.5 Å and 64.6 ± 0.4 Å, respectively. These values are similar to our previous FRET measurements using a different SERCA1a construct fused to CFP instead of Cer (59 Å) 9; 10. That both isoforms yield the same FRETmax suggests that they assume the same gross conformation, in which the fluorescent proteins (fused to PLB and SERCA N-termini) are on opposite sides of the regulatory complex. The errors associated with distance measurements are calculated from the errors in FRETmax (Table 1). Such errors are an indicator of the precision (reproducibility) of the measurements, and are not to be taken as a indicator of the accuracy of the measurements. In particular, the absolute distance between the fluorescent probes is subject to additional ambiguities (eg. relative dipole orientations). Regulatory complex structural equivalence 3 13 is in harmony with the apparent functional equivalence observed in vitro and in vivo. Specifically, PLB binding results in the same degree of inhibition of calcium affinity for both isoforms 7. However, despite similar quaternary structure and equivalent inhibitory function of the respective regulatory complexes, SERCA1a-PLB and SERCA2a-PLB apparent Kd2 values differed by 50%. This surprising result suggests that PLB binds more avidly to the fast-twitch skeletal isoform SERCA1a than to its natural regulatory partner SERCA2a. We considered whether calcium pump structural heterogeneity could influence the measured affinity of PLB for SERCA isoforms. SERCA2a has a higher calcium affinity than SERCA1a 14, and compared to SERCA1a it may have a larger steady-state E1/E2 ratio at basal calcium concentrations in live AAV-293 cells. This might cause the observed lower affinity of PLB for SERCA2a, since PLB is proposed to bind only to the E2 (calcium-free) substate 13 15. However, the difference in affinities persisted in the presence of intracellular Ca chelator BAPTA-AM (not shown), a treatment that should stabilize the population of SERCA pumps in the E2 substate. This suggests the difference in affinities is intrinsic to the SERCA isoforms and is not an indirect effect of pump enzymatic substate structural heterogeneity.
The observed differential binding of PLB to SERCA isoforms is noteworthy, and may be a strategic consideration for studies using skeletal SERCA1a as an experimental model for SERCA2a. Moreover, the data suggest that when both targets are available, the relative binding of PLB to each isoform depends on their respective binding affinities. PLB and SERCA2a have both been localized to slow muscle fibers, while SERCA1a is mostly expressed in fast muscle fibers 4; PLB is rarely expressed together with both SERCA1a and SERCA2a. However, a small number of “hybrid” fibers expressing both SERCA1a and SERCA2a can be detected in skeletal muscle (1.5%)16, and this percentage is markedly increased in humans by muscle unloading (bed rest) 17. Hybrid fibers are also increased in chronically stimulated fibers 16 or by treatment with β2 agonist 18. Nevertheless, the present results may be most directly relevant for gene therapy approaches in which SERCA1a overexpression is used to enhance calcium handling in cardiac muscle 7 8. Based on its greater affinity for PLB, one may expect a greater tonic inhibition of exogenous SERCA1a relative to endogenous SERCA2a, but this putative effect may be masked in SERCA overexpression by the increased pump/PLB ratio. Indeed, SERCA1a transgenic murine trabeculae show increased baseline developed force. Moreover one may speculate that increased PLB binding by overexpressed SERCA1a would result an increase in adrenergic reserve. This may be a desirable outcome in therapies for heart failure, a condition characterized by blunted a adrenergic response. However, the magnitude and significance of such a possible effect cannot be determined from the present experiments.
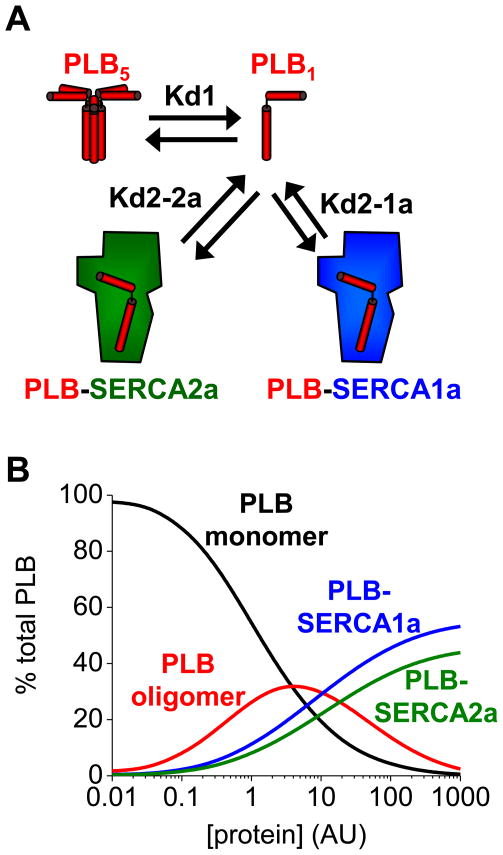
A model of PLB regulation of pump isoforms is depicted schematically in Fig. 3A, in which the oligomer (PLB5), SERCA1a, and SERCA2a compete for the pool of monomers (PLB1). The equilibrium allocation of PLB among these complexes is determined by the relative dissociation constants of oligomerization (Kd1) and the respective regulatory complexes (Kd2). To integrate our experimental observations, the linked equilibria were simulated in a simple computational model using the Kd1 and Kd2 values measured by FRET. To facilitate comparison of affinities, the model simulated equimolar concentrations of PLB, SERCA1a, and SERCA2a. Fig. 3B shows that the binding of PLB to itself, SERCA1a, and SERCA2a depends on the level of expression and the relative affinities of the respective protein complexes. At low concentrations, PLB is mostly in the monomer form (black line), but as the concentration approaches Kd1 (~2.2 AU) 10, the oligomer increases (red line). Oligomerization is favored over binding to SERCA1a or SERCA2a until the concentration approaches Kd2. At the highest concentrations (≫Kd2), first-order binding of PLB to SERCA isoforms outcompetes second-order oligomerization. Because of the difference in respective binding affinities, the PLB-SERCA1a complex exceeds the PLB-SERCA2a complex at all concentrations. Table 2 gives the results of simulations mimicking conditions of normal ventricle, heart failure (decreased PLB and SERCA), and heart failure treated with exogenous SERCA1a/2a. The model comports with several known features of in vivo regulation. First, cardiac SERCA2a is not fully regulated (Table 2), despite a large excess of PLB in ventricles. This is consistent with the high pentamer/monomer ratio in the myocardium and the observation that a 2.6-fold overexpression of PLB is required to fully saturate SERCA regulation 19. This may be attributed to the relatively higher affinity of oligomerization vs. SERCA-binding (Kd1<Kd2). Moreover, it is known that PLB is partially depolymerized by SERCA20, and the model predicts pentamer/monomer ratio is reduced by overexpression of SERCA. We conclude that the distribution of PLB among possible complexes is determined the availability of the binding partners and the relative affinity of competing interactions. Specifically, PLB binds to SERCA1a in excess of what is expected from the simple molar balance of regulatory targets.
Figure 3.
A) A model of PLB regulation of SERCA when both SERCA2a and SERCA1a are accessible. The relative distribution of PLB among the pentamer (PLB5), monomer (PLB1), and SERCA1a/2a regulatory complexes depends on the dissociation constants for the oligomer (Kd1) and the regulatory complexes (Kd2). B) A computational model of PLB interactions shows the concentration dependence of the distribution of PLB among oligomer (red), monomer (black), and regulatory complexes with SERCA1a (blue) and SERCA2a (green). An equimolar system is simulated; the protein concentration indicated corresponds to the measured YFP-PLB concentration (AU), as in Fig. 2E.
Table 2.
Results of computational modeling
| Equimolar Model | ventricle | Heart Failure | HF + | SERCA1a | |
|---|---|---|---|---|---|
| PLB distribution | |||||
| Monomer | 14% | 9% | 13% | 10% | |
| Oligomer | 28% | 82% | 80% | 50% | |
| SERCA1a | 33% | 0% | 0% | 33% | |
| SERCA2a | 25% | 9% | 8% | 7% | |
| SERCA distribution | |||||
| regulated SERCA1a | 33% | - | - | 55% | |
| unregulated SERCA1a | 67% | - | - | 45% | |
| regulated SERCA2a | 25% | 60% | 51% | 45% | |
| unregulated SERCA2a | 75% | 40% | 49% | 55% | |
Materials and Methods
Molecular biology and cell culture
Dog SERCA2a or rabbit SERCA1a were fused to the C-terminus of mCerulean (Cer); dog PLB was fused to the C-terminus of enhanced yellow fluorescent protein (YFP) 9. Plasmids encoding Cer-SERCA2a or Cer-SERCA1a were cotransfected with a 20-fold molar excess of YFP-PLB in AAV-293 cells as previous described 4.
Fluorescence microscopy
Widefield and TIRF microscopy were performed as previously described 9; 11. Through-objective TIRF excitation was achieved with a 449 nm diode laser (for Cer-SERCA) or 514 nm Ar laser (for YFP-PLB). The laser incident angle was adjusted to create an evanescent field that illuminated the plasma membrane in contact with the surface substrate and a thin section of endoplasmic reticulum.
FRET measurements
FRET was measured by epifluorescence microscopy with progressive acceptor photobleaching 4, quantifying each cell’s transfer efficiency (E) from the relationship E = 1 − (Iprebleach/Ipostbleach) , where Iprebleach and Ipostbleach are the intensity of the donor fluorescence before and after selective photobleaching of the acceptor. YFP-PLB fluorescence intensity was used as an index of protein concentration. FRET dependence on protein expression level was fit to a hyperbolic curve of the form y = (FRETmax) × /(Kd2 + ×), where × = protein concentration in arbitrary units (AU). The parameter FRETmax was taken to represent the intrinsic FRET of the regulatory complex, and Kd2 was the regulatory complex dissociation constant. FRET was also measured with a “3-cube” method (E-FRET) 21. FRET was calculated according to
where IAA is the intensity of fluorescence emission detected in the donor channel (472/30 nm) with excitation of 427/10 nm; IAA is acceptor channel (542/27 nm) emission with excitation of 504/12 nm; IDA is the “FRET” channel, with 542/27 nm emission and excitation of 427/10 nm; a and d are cross-talk coefficients determined from acceptor-only or donor-only samples, respectively. We obtained a value for a of 0.82 for mCerulean , and a value for d of 0.082 for YFP. G is the ratio of the sensitized emission to the corresponding amount of donor recovery, which was 3.2 for this setup. Probe separation distance (r) was calculated from the relationship described by Förster 22, R = (R0)[(1/E) −1)1/6, where E is the measured FRETmax value after subtraction of 4% nonspecific FRET, as previously described 9 (also, Hou and Robia, manuscript in preparation). R0 is the Förster radius, which is 53.3 Å for the mCerulean-YFP pair 23. A Student’s t-test was used to determine the significance of measured average FRETmax and Kd2. To determine the effect of removal of calcium, cells were washed with calcium-free phosphate buffered saline (PBS) and incubated for 20 minutes in 5 μM BAPTA-AM in PBS at 37cC. Data acquisition was completed within 30 minutes of exposure of cells to calcium-free conditions.
Computational modeling of PLB complexes
The balanced equilibria of PLB binding to itself, SERCA1a, and SERCA2a were simulated using the shareware kinetics modeling software Tenua. The linked equilibria were modeled as Pentamer ↔ 5(monomer); monomer + SERCA1a ↔ [PLB-SERCA1a complex]; monomer + SERCA2a ↔ [PLB-SERCA2a complex]. Forward and reverse rate constants were estimated from oligomer and regulatory complex subunit exchange rates 11, oligomerization dissociation constant (Kd1) 10, and the apparent PLB dissociation constants of SERCA1a and SERCA2a (Kd2-1a and Kd2-2a, respectively) determined in the present study. These dissociation constants are expressed in arbitrary units, as the absolute membrane concentrations of labeled proteins in the present cell expression system are unknown. For the purposes of this analysis, the native cardiac SERCA2a concentration was set equal to the apparent dissociation constant obtained for the PLB-SERCA2a interaction. This assumption is compatible with cardiac PLB overexpression studies that showed SERCA2a was not fully saturated with PLB in vivo 19. Values for the relative membrane concentrations of PLB and SERCA isoforms were taken from published measurements of the relative expression of PLB, SERCA1a, and SERCA2a in porcine ventricle, soleus, and extensor digitorum longus 24. A hypothetical model in which the total moles of PLB, SERCA1a, and SERCA2a are equal was also simulated to facilitate comparison of relative affinities.
Acknowledgments
SERCA2a was a gift of Iain K Farrance. Plasmid pmCerulean-C1 was obtained from Addgene (David Piston). The authors thank Eileen M. Kelly for technical assistance and Mike Autry, Peter Ayittey, and Ilya Bezprozvanny for helpful advice.
Sources of funding- This work was supported by the National Institute of Biomedical Imaging and BioEngineering (EB006061) and the National Heart, Lung and Blood Institute (HL092321).
Abbreviations
- PLB
phospholamban
- SERCA
sarco(endo)plasmic reticulum calcium ATPase
- FRET
fluorescence resonance energy transfer
- CaMKII
calcium/calmodulin-dependent protein kinase II
- Cer
mCerulean
- YFP
enhanced yellow fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gwathmey JK, Copelas L, MacKinnon R, Schoen FJ, Feldman MD, Grossman W, Morgan JP. Abnormal intracellular calcium handling in myocardium from patients with end-stage heart failure. Circ Res. 1987;61 :70–6. doi: 10.1161/01.res.61.1.70. [DOI] [PubMed] [Google Scholar]
- 2.Sipido KR, Vangheluwe P. Targeting sarcoplasmic reticulum Ca2+ uptake to improve heart failure: hit or miss. Circ Res. 106:230–3. doi: 10.1161/CIRCRESAHA.109.210740. [DOI] [PubMed] [Google Scholar]
- 3.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4 :566–77. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 4.Jorgensen AO, Jones LR. Localization of phospholamban in slow but not fast canine skeletal muscle fibers. An immunocytochemical and biochemical study. J Biol Chem. 1986;261 :3775–81. [PubMed] [Google Scholar]
- 5.Ferrington DA, Yao Q, Squier TC, Bigelow DJ. Comparable levels of Ca-ATPase inhibition by phospholamban in slow-twitch skeletal and cardiac sarcoplasmic reticulum. Biochemistry. 2002;41 :13289–96. doi: 10.1021/bi026407t. [DOI] [PubMed] [Google Scholar]
- 6.Reddy LG, Autry JM, Jones LR, Thomas DD. Co-reconstitution of phospholamban mutants with the Ca-ATPase reveals dependence of inhibitory function on phospholamban structure. J Biol Chem. 1999;274 :7649–55. doi: 10.1074/jbc.274.12.7649. [DOI] [PubMed] [Google Scholar]
- 7.Lalli MJ, Yong J, Prasad V, Hashimoto K, Plank D, Babu GJ, Kirkpatrick D, Walsh RA, Sussman M, Yatani A, Marban E, Periasamy M. Sarcoplasmic reticulum Ca(2+) atpase (SERCA) 1a structurally substitutes for SERCA2a in the cardiac sarcoplasmic reticulum and increases cardiac Ca(2+) handling capacity. Circ Res. 2001;89 :160–7. doi: 10.1161/hh1401.093584. [DOI] [PubMed] [Google Scholar]
- 8.O'Donnell JM, Pound K, Xu X, Lewandowski ED. SERCA1 expression enhances the metabolic efficiency of improved contractility in post-ischemic heart. J Mol Cell Cardiol. 2009;47 :614–21. doi: 10.1016/j.yjmcc.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly EM, Hou Z, Bossuyt J, Bers DM, Robia SL. Phospholamban oligomerization, quaternary structure, and sarco(endo)plasmic reticulum calcium ATPase binding measured by fluorescence resonance energy transfer in living cells. J Biol Chem. 2008;283:12202–11. doi: 10.1074/jbc.M707590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou Z, Kelly EM, Robia SL. Phosphomimetic mutations increase phospholamban oligomerization and alter the structure of its regulatory complex. J Biol Chem. 2008;283 :28996–9003. doi: 10.1074/jbc.M804782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robia SL, Campbell KS, Kelly EM, Hou Z, Winters DL, Thomas DD. Forster transfer recovery reveals that phospholamban exchanges slowly from pentamers but rapidly from the SERCA regulatory complex. Circ Res. 2007;101 :1123–9. doi: 10.1161/CIRCRESAHA.107.159947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bossuyt J, Despa S, Han F, Hou Z, Robia SL, Lingrel JB, Bers DM. Isoform-specificity of the Na/K-ATPase association and regulation by phospholemman. J Biol Chem. 2009 doi: 10.1074/jbc.M109.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toyoshima C, Asahi M, Sugita Y, Khanna R, Tsuda T, MacLennan DH. Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proc Natl Acad Sci U S A. 2003;100 :467–72. doi: 10.1073/pnas.0237326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dode L, Andersen JP, Leslie N, Dhitavat J, Vilsen B, Hovnanian A. Dissection of the functional differences between sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) 1 and 2 isoforms and characterization of Darier disease (SERCA2) mutants by steady-state and transient kinetic analyses. J Biol Chem. 2003;278 :47877–89. doi: 10.1074/jbc.M306784200. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Stokes DL, Rice WJ, Jones LR. Spatial and dynamic interactions between phospholamban and the canine cardiac Ca2+ pump revealed with use of heterobifunctional cross-linking agents. J Biol Chem. 2003;278 :48348–56. doi: 10.1074/jbc.M309545200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang KM, Hu P, Wang SW, Wright LD, Wechsler AS, Spratt JA, Briggs FN. Fast- and slow-twitch isoforms (SERCA1 and SERCA2a) of sarcoplasmic reticulum Ca-ATPase are expressed simultaneously in chronically stimulated muscle fibers. Pflugers Arch. 1997;433 :766–72. doi: 10.1007/s004240050343. [DOI] [PubMed] [Google Scholar]
- 17.Salanova M, Schiffl G, Blottner D. Atypical fast SERCA1a protein expression in slow myofibers and differential S-nitrosylation prevented by exercise during long term bed rest. Histochem Cell Biol. 2009;132 :383–94. doi: 10.1007/s00418-009-0624-y. [DOI] [PubMed] [Google Scholar]
- 18.Zhang KM, Hu P, Wang SW, Wright LD, Wechsler AS, Spratt JA, Briggs FN. The beta2-agonist salbutamol affects the expression of phospholamban and both isoforms of SERCA in canine skeletal muscle and blocks changes in these induced by neuromuscular stimulation. Pflugers Arch. 1998;435 :511–7. doi: 10.1007/s004240050546. [DOI] [PubMed] [Google Scholar]
- 19.Brittsan AG, Carr AN, Schmidt AG, Kranias EG. Maximal inhibition of SERCA2 Ca(2+) affinity by phospholamban in transgenic hearts overexpressing a non-phosphorylatable form of phospholamban. J Biol Chem. 2000;275 :12129–35. doi: 10.1074/jbc.275.16.12129. [DOI] [PubMed] [Google Scholar]
- 20.Reddy LG, Jones LR, Thomas DD. Depolymerization of phospholamban in the presence of calcium pump: a fluorescence energy transfer study. Biochemistry. 1999;38 :3954–62. doi: 10.1021/bi981795d. [DOI] [PubMed] [Google Scholar]
- 21.Zal T, Gascoigne NR. Photobleaching-corrected FRET efficiency imaging of live cells. Biophys J. 2004;86 :3923–39. doi: 10.1529/biophysj.103.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Förster T. Intermolecular energy migration and fluorescence. Ann Phys (Leipzig) 1948;2 :55–75. [Google Scholar]
- 23.Rizzo MA, Springer G, Segawa K, Zipfel WR, Piston DW. Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc Microanal. 2006;12 :238–54. doi: 10.1017/S1431927606060235. [DOI] [PubMed] [Google Scholar]
- 24.Vangheluwe P, Schuermans M, Zador E, Waelkens E, Raeymaekers L, Wuytack F. Sarcolipin and phospholamban mRNA and protein expression in cardiac and skeletal muscle of different species. Biochem J. 2005;389 :151–9. doi: 10.1042/BJ20050068. [DOI] [PMC free article] [PubMed] [Google Scholar]