Abstract
The genome expression of positive-stranded RNA viruses starts with translation rather than transcription. For some viruses, the genome is the only viral mRNA and expression is regulated primarily at the translational level and by limited proteolysis of polyproteins. Other virus groups also generate subgenomic mRNAs later in the reproductive cycle. For nidoviruses, subgenomic mRNA synthesis (transcription) is discontinuous and yields a 5′ and 3′ coterminal nested set of mRNAs. Nidovirus transcription is not essential for genome replication, which relies on the autoprocessing products of two replicase polyproteins that are translated from the genome. We now show that the N-terminal replicase subunit, nonstructural protein 1 (nsp1), of the nidovirus equine arteritis virus is in fact dispensable for replication but crucial for transcription, thereby coupling replicase expression and subgenomic mRNA synthesis in an unprecedented manner. Nsp1 is composed of two papain-like protease domains and a predicted N-terminal zinc finger, which was implicated in transcription by site-directed mutagenesis. The structural integrity of nsp1 is essential, suggesting that the protease domains form a platform for the zinc finger to operate in transcription.
Positive stranded (+) RNA viruses, as their name implies, possess genomes of mRNA polarity. In contrast to other RNA viruses, including retroviruses, +RNA-virus particles do not contain the viral polymerase (1). Because of these properties, and unlike all other forms of life, +RNA viruses start their genome expression with RNA translation. The viral genome enters the cellular translation machinery and directs the expression of a set of protein functions, always including a unique RNA-dependent RNA polymerase (RdRp) activity. This RdRp, with the probable involvement of viral and cellular factors, utilizes the genome as template for replication, which proceeds via a minus-strand RNA intermediate.
To regulate gene expression, +RNA viruses have evolved sophisticated (post)translational control mechanisms including cap-independent initiation of translation at internal ribosome entry sites (IRESs), read-through translation of nonsense codons, ribosomal frameshifting, and limited proteolytic processing of precursor polyproteins. In addition, many +RNA-virus genomes contain 3′ proximal ORFs that remain silent upon genome translation. These ORFs encode structural polypeptides and sometimes also auxiliary proteins, which are not essential for genome replication (2–4) and can be expressed only after the synthesis of subgenomic (sg) mRNAs. The latter process exclusively serves to regulate gene expression and thus, functionally, it is transcription. Consequently, sg-mRNA synthesis should be distinguished from replication, which amplifies an RNA molecule with a dual role as genome and mRNA.
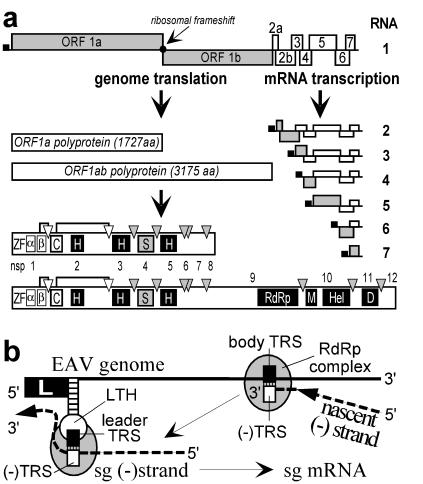
Although RNA signals with specific functions in transcription have been identified in a number of viruses, the viral-protein functions involved remain understood poorly. We have been studying the genome replication and expression of +RNA viruses by using equine arteritis virus (EAV), which belongs to the family Arteriviridae (5). Based on a similar polycistronic genome organization, common transcriptional and (post)translational strategies, and a conserved array of nonstructural domains, the arteriviruses have been united with the Coronaviridae in the order Nidovirales (Fig. 1A; refs. 5–7). One of the most striking features of the nidovirus life cycle is the synthesis of an extensive nested set of sg mRNAs. The protein functions required for the synthesis of genomic and sg RNAs are encoded by two ORFs (1a and 1b) in the 5′ proximal three-quarters of the genome (4). The ORF1b polyprotein, which includes the putative RdRp activity, can be translated only after ribosomal frameshifting (6). In EAV, this strategy yields ORF1a and ORF1ab polyproteins of 187 and 345 kDa, respectively, which are processed autocatalytically into 12 nonstructural proteins (nsps 1 to 12; Fig. 1A) by proteases residing in nsp1, nsp2, and nsp4 (reviewed in ref. 8). The nidovirus nsps are described commonly as “replicase,” although they may include domains that are not involved in genome replication per se (see below).
Figure 1.
(A) Schematic diagram of EAV genome organization and expression. The nested set of sg mRNAs (RNAs 2 to 7) is shown, with the leader sequence represented as a black box and the ORF(s) expressed from each mRNA depicted in gray. (Lower) The ORF1a and ORF1ab replicase polyproteins and their processing maps are depicted with protease domains and corresponding cleavage sites indicated. Abbreviations for conserved domains: ZF, nonstructural protein- (nsp) 1 zinc finger; α, nsp1 papain-like cysteine protease (PCP) α; β, nsp1 PCPβ; C, nsp2 cysteine protease; H, hydrophobic domain; S, nsp4 serine protease; RdRp, RNA-dependent RNA polymerase; M, predicted metal-binding domain; Hel, helicase; D, conserved nidovirus-specific domain. (B) Model for nidovirus discontinuous minus-strand synthesis, yielding sg minus-strand RNAs that function as a template for sg mRNAs in transcription (9, 10). Discontinuous minus-strand synthesis involves attenuation of the RdRp complex at the body transcription-regulating sequence (TRS), translocation of the nascent minus strand to the leader TRS in the genomic template [exposed by the leader TRS hairpin (LTH)], base pairing between the minus-sense body TRS and plus-sense leader TRS, and reinitiation of RNA synthesis to complete the sg minus strand with the complement of the leader sequence.
To express structural proteins and some nsps, nidoviruses employ a nested set of 4 to 8 sg mRNAs. With exception of the smallest transcript, these sg mRNAs are structurally polycistronic (Fig. 1A), although generally only their most 5′ proximal ORF is translated. The sg transcripts share a common 5′ “leader” sequence (211 nt in EAV) that is identical to the 5′ end of the genome (Fig. 1A). The sequences downstream of the mRNA leader (the mRNA “bodies”) represent different but 3′ coterminal parts of the genomic 3′ terminal region. Most likely, genome replication and transcription proceed through different minus-strand intermediates (9, 10). For genome replication, a full-length minus-strand template is used. In contrast, sg mRNAs were proposed to be synthesized from sg minus-strand templates generated by discontinuous minus-strand synthesis (Fig. 1B) in a process resembling similarity-assisted RNA recombination (10–12). During sg minus-strand synthesis, leader and body sequences that are noncontiguous in the full-length minus strand are “fused” at conserved sequence elements, termed TRSs. In the genomic plus strand, TRSs are present at the 3′ end of the leader and at the 5′ end of each of the sg-RNA bodies. As suggested originally for coronaviruses (13, 14), EAV discontinuous transcription was shown to depend on base pairing between the body TRS complement in the nascent minus strand and the leader TRS of the genomic positive-strand template (Fig. 1B; ref. 10). The TRS base-pairing interaction may be promoted by a predicted stem–loop structure in the 5′ end of the EAV genome, the LTH, which appears to “expose” the leader TRS at the top of its loop (ref. 10; Fig. 1B).
The first product of EAV replicase autoprocessing is nsp1 (15), a 260-aa protein that cotranslationally releases itself by a papain-like cysteine protease (PCP) β in its C-terminal half. We now show that EAV nsp1 is fully dispensable for genome replication but crucial for sg-RNA synthesis. Consequently, transcription is linked directly to and controlled by a protein, the expression of which is determined at the level of genome replication and translation. These findings are compatible with the evolution of transcription in a primitive self-replicating system in which the regulation of gene expression was confined initially to the (post)translational levels.
Methods
Mutagenesis of the EAV Full-Length cDNA Clone.
Site-directed PCR mutagenesis (16) was used to engineer mutations. After complete sequence analysis, DNA fragments containing mutations were cloned back into infectious cDNA clone pEAV030 (17) or into pEAV030-derived replicons. Mutant Δnsp1 (Fig. 2) was constructed by mutating the original ORF1a ATG start codon to TAG (TATG→CTAG mutation at position 224–227) and deleting nucleotide T-297 to C-1004 of the EAV genome. The sequence 5′-TCCATG-3′ was inserted at the position of the deletion, thereby providing an ATG codon immediately upstream of (and in frame with) the nsp2-coding sequence. The DITRAC replicon (for discontinuous transcription complementation) was based on construct pEnsp10 (18), a derivative of pEΔBal (18) in which nucleotide 10,023 to nucleotide 11,638 of the EAV genome (ORFs 2b-5) were replaced with the encephalomyocarditis virus (EMCV) IRES (19) and the coding sequence for EAV nsp10. We now replaced the nsp10-coding sequence with the nsp1-coding sequence (nucleotides 225–1,004 of the EAV genome) and introduced the 5′ deletion of construct Δnsp1 (see above and Fig. 4A). In control-construct pDICAT, the chloramphenicol acetyl transferase (CAT) reporter gene (17) was inserted downstream of the IRES.
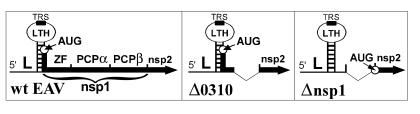
Figure 2.
Construction of knockout mutant Δnsp1. Depicted is a schematic overview of the important elements in the 5′ end of the EAV genome: the genomic leader sequence (L), LTH, ORF1a and its translation-initiation codon (circle), the nsp1 zinc finger (ZF) and PCP domains (see Fig. 5), and the nsp2 coding region. The replicase ORF is shown as a solid bar.
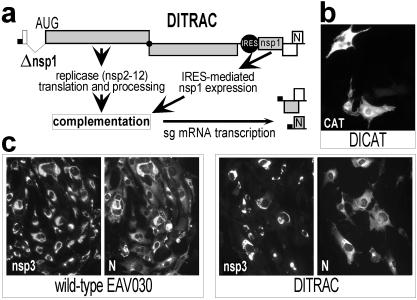
Figure 4.
Trans-complementation of the function of nsp1 in transcription. (A) Outline of the DITRAC replicon and complementation assay. The expression of nsp1 from the 5′ end of the genome was inactivated (see Fig. 2A), and an IRES-nsp1 cassette was inserted in the structural gene region. Complementation of nsp1 function should restore sg-RNA synthesis from the DITRAC 3′ end. (B) Reporter-gene expression from the IRES in control-replicon DICAT, which is fully negative for sg-mRNA synthesis (Fig. 3). (C) Trans-complementation of nsp1 function. Cells were transfected with wild-type EAV030 RNA or with DITRAC, fixed at 24 h after electroporation, and double stained for nsp3 and the nucleocapsid protein to monitor genome replication and transcription, respectively. The nucleocapsid protein is expressed from sg mRNA7 (see A).
Transfections and RNA Analysis.
In vitro-transcribed RNA was transfected into BHK-21 cells by electroporation (17). Intracellular RNA for reverse transcription (RT)–PCR analysis was isolated at 24 h after transfection by using the acidic phenol-extraction method (20). Northern blot analysis of intracellular RNA and RT-PCR analysis of plus- and minus-strand RNA1 and sg-RNA7 synthesis were described by van Marle et al. (10, 20). To examine the stability of the Δnsp1 deletion, we performed an additional RT-PCR and sequence analysis by using antisense oligonucleotide E054 (complementary to nucleotides 1,335–1,354 of the EAV genome) and the described sense leader primer E157 (20). For the analysis of the IRES-nsp1 cassette, an RT-PCR analysis was performed by using antisense oligonucleotide E296 (complementary to nucleotides 11,643–11,662 of the EAV genome) and sense primer EMCV083 (5′-CTAGGCCCCCCGAAC-3′), which maps to the 3′ end of the EMCV IRES.
Protein Analysis.
Immunofluorescence assays (IFAs; ref. 21) were performed with antisera specific for the EAV replicase (22–24), the ORF7-encoded nucleocapsid protein (25), and the CAT protein (purchased from 5 Prime→3 Prime). For immunoprecipitation analysis, transfected cells were labeled with [35S]methionine/[35S]cysteine from 8 to 12 h after transfection (20). Cell lysis and immunoprecipitation of labeled EAV nsps have been described before (22, 26).
Results
Construction of an EAV nsp1 Knockout Mutant.
To investigate the role of nsp1 in the EAV life cycle, we engineered an nsp1 knockout mutant of the EAV infectious cDNA clone (17). Because the 5′ end of the nsp1-coding sequence (nucleotides 225–262) was postulated previously to be part of the LTH stem (Fig. 2; ref. 10), we expected that deletion of the entire nsp1-coding sequence (nucleotides 225–1,004) would be deleterious for genome replication and/or transcription. The results obtained with a set of 5′ deletion constructs, which will be described in detail elsewhere, indeed confirmed the importance of RNA sequences that overlap with the 5′ end of ORF1a (unpublished data). Surprisingly, the in-frame deletion of nucleotides 297–1,004 (mutant Δ0310; Fig. 2) abolished sg-mRNA synthesis but did not affect genome replication (data not shown). Mutant Δ0310 expressed only the N-terminal 24 residues of nsp1, fused to nsp2 by means of a connecting Ser-Met dipeptide. To create a true nsp1 knockout mutant (Δnsp1; Fig. 2), in which the expression of all nsp1-related sequences was inactivated without compromising the predicted LTH, the ORF1a translation initiation codon (nucleotides 225–227) of mutant Δ0310 was replaced with UAG. Consequently, nucleotides 225–296 were no longer translated and replicase translation started with nsp2 from an engineered AUG codon upstream of the nsp2-coding sequence.
Nsp1 Is Dispensable for Genome Replication but Not Transcription.
Wild-type and Δnsp1 RNA were transfected into BHK-21 cells and the viral phenotype was analyzed by using three different assays. First, genome replication and sg-RNA synthesis were monitored indirectly by using our previously described double IFA (data not shown; ref. 17) for nonstructural and structural protein expression, in this case a double labeling with a rabbit (anti-nsp3) replicase antiserum and a mouse monoclonal antibody against the mRNA7-encoded nucleocapsid protein. Second, the expression and processing of the EAV replicase was monitored by immunoprecipitation analysis (Fig. 3A). Third, intracellular viral RNA was isolated and analyzed directly by using Northern blot analysis (Fig. 3B) and RT-PCR assays (Fig. 3C). The latter are the most sensitive method to detect the synthesis of genomic and sg plus and minus strands (20).
Figure 3.
Nsp1 is an essential factor for transcription but not replication of the EAV genome. (A) Immunoprecipitation analysis of nsp1 (29 kDa) and nsp2 (61 kDa) expression. (B) Northern blot analysis of intracellular RNA (24 h after transfection) with a probe complementary to the 3′ end of all EAV mRNAs (10). The positions of the EAV genome and six sg mRNAs are indicated. The band labeled E in the DITRAC lane represents an extra sg RNA transcribed from a cryptic TRS in the EMCV IRES (see text). (C) RT-PCR analysis of sg-RNA7 synthesis. The generation of minus- (Upper) or plus-stranded (Lower) sg RNA7 in transfected cells was tested by RNA7-specific RT-PCR. One of the primers was located in the RNA7 body and the other in the leader sequence, thereby generating an RNA7-specific PCR product of 540 bp (arrowhead). Mutant EAV030F, which generates about 500-fold reduced levels of sg RNAs (17, 20), was included to confirm the sensitivity of the assay.
According to the assays described above, replication of the somewhat smaller Δnsp1 genomic RNA was similar to that of the wild-type EAV genome (Fig. 3). The immunoprecipitation analysis confirmed that Δnsp1 did not produce nsp1 and yielded a 61-kDa nsp2 protein that comigrated with its wild-type counterpart. The replicase IFA revealed that Δnsp1, although it was fully replication-competent, did not express the RNA7-encoded nucleocapsid protein (data not shown). The Northern blot analysis (Fig. 3B) confirmed the efficient replication of the Δnsp1 genome, whereas synthesis of sg mRNAs could not be detected. Finally, a sensitive RT-PCR aimed at the amplification of the sg-RNA7 leader-body junction region (Fig. 3C) was used to confirm that Δnsp1 was completely deficient in the synthesis of sg RNA7, which normally is the most abundant sg-RNA species of EAV. Thus, we concluded that nsp1 expression was not essential for genome replication, but that either the RNA sequence between nucleotides 297 and 1,004 (which was lacking from Δnsp1) or the nsp1 protein itself played a pivotal role in transcription.
Nsp1 Can Trans-Activate EAV Transcription.
To investigate the role of EAV nsp1 in sg-mRNA synthesis, we designed an assay to trans-complement the Δnsp1 mutation on the basis of the previously developed autonomous replicon EΔBal (18). EΔBal is unable to produce infectious progeny because of a large deletion in the ORF2b-5 region. Nevertheless, it expresses a full-length replicase, is not impaired in genome replication, and transcribes sg RNAs from its 3′ end. The site of the ORF2b-5 deletion in EΔBal can be used to insert a gene under the translational control of the EMCV IRES (18), allowing the direct expression of the inserted gene from the 3′ end of the replicon RNA. We now created the Δnsp1 deletion in the 5′ end of the EΔBal replicon, thereby inactivating its transcription (data not shown). As an expression and specificity control, we first inserted the CAT reporter gene downstream of the IRES element (replicon DICAT). As expected, DICAT produced abundant CAT expression (Fig. 4B) in the absence of sg-mRNA synthesis (Fig. 3), thus confirming that the IRES-driven gene indeed was expressed from the 3′ end of the replicon. Subsequently, we engineered nsp1 trans-complementation construct DITRAC by replacing the CAT gene downstream of the DICAT IRES element with the nsp1-coding sequence (Fig. 4A). Thus, DITRAC carried the Δnsp1 deletion in its 5′ end and could express nsp1 from the 3′ proximal IRES, which indeed was confirmed by immunoprecipitation (Fig. 3A).
The ability of IRES-driven nsp1 to trans-activate sg-mRNA synthesis (Fig. 4A) was detected initially by an IFA aimed at the detection of the mRNA7-encoded nucleocapsid protein in DITRAC-transfected cells (Fig. 4C). Next, the generation of sg mRNAs 6 and 7 was confirmed by Northern blot analysis of intracellular RNAs from DITRAC-transfected cells (Fig. 3B). Finally, the synthesis of sg RNA6 (data not shown) and -7 (Fig. 3C) was confirmed by RT-PCR and sequence analysis. These results clearly demonstrated that the transcription defect of the nsp1 knockout mutant could be trans-complemented by expression of nsp1 from the 3′ end of the genome.
The IFA data suggested that complementation levels in individual DITRAC-transfected cells were variable, and that some cells remained nucleocapsid protein-negative even after 72 h. Also, the synthesis of an sg mRNA from the RNA2 body TRS, which was retained in the DITRAC RNA, was not apparent. Furthermore, an extra sg transcript was generated from a cryptic TRS-like sequence within the IRES-nsp1 cassette, a phenomenon that has been observed more frequently after insertion of sequences in the 3′ end of the EAV genome (unpublished data). Most likely, the explanations for these (as-yet) poorly understood properties of DITRAC are of a purely technical nature. They could, for instance, be related to the mutations in the DITRAC 5′ end and/or the insertion of the IRES-nsp1 cassette immediately downstream of the RNA2 TRS (Fig. 3B). Also, DITRAC may be somewhat instable, although sequence rearrangements could not be detected by RT-PCR analysis of intracellular RNA from transfected cells (see Methods). Alternatively, the incomplete complementation could be linked to the artificial mode of nsp1 expression.
Arterivirus nsp1 Contains a Putative N-Terminal ZF.
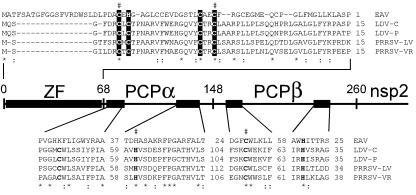
A renewed computer-assisted comparison of arterivirus nsp1 sequences revealed a weak conservation in their N-terminal domain upstream of the previously identified PCPα and PCPβ proteases (Fig. 5). The PCPβ enzyme is responsible for cleavage of the nsp1/nsp2 site (15). The upstream-located PCPα is defective proteolytically in EAV but cleaves the junction between the PCPα- and PCPβ-containing subunits in other arteriviruses (8, 27). The alignment of the N-terminal domains revealed only a few conserved positions, four of which are occupied by Cys or Cys/His residues in an arrangement that is typical for the zinc-coordinating residues of a ZF. Different variants of this structure are ubiquitous in transcription factors (28). The putative nsp1 ZF has the C3H formula in EAV, but is of the C4 type in other arteriviruses.
Figure 5.
Identification of a ZF motif in the N-terminal domain of the arterivirus replicase. The sequences of lactate dehydrogenase elevating virus [LDV-C (44) and LDV-P (45)], porcine reproductive and respiratory syndrome virus [PRRSV-LV (46) and PRRSV-VR2332 (47)], and EAV (6) were compared. Alignments of the complete ZF domain and selected conserved regions of PCPα and PCPβ containing the active-site Cys and His residues (bold) are shown. Invariant (*) and conserved (:) positions are highlighted. Conserved His and Cys residues in the nsp1 ZF that are proposed to be involved in zinc binding are shown in reverse shading, and mutated residues (Table 1) are indicated with “#.”
Mutagenesis Suggests a Pivotal Role for the nsp1 ZF in Transcription.
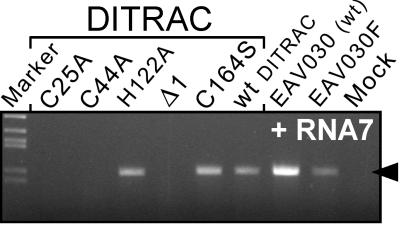
To analyze the role of specific nsp1 domains in transcription, we first engineered a set of three DITRAC derivatives containing 3′ truncated nsp1 genes downstream of the IRES (Table 1). All these mutants retained the ZF domain. The smallest deletion (Δ1) removed only the 47 C-terminal codons of nsp1, including the PCPβ catalytic His residue (His-230). The second deletion (Δ2) removed the entire PCPβ. The largest deletion (Δ3) left only the N-terminal 124 residues of nsp1. None of these C-terminally truncated nsp1 proteins was able to trans-activate transcription (Fig. 6; Table 1), suggesting that the full-length nsp1 protein is required for trans-activation of sg-RNA synthesis.
Table 1.
Results of mutagenesis of the nsp1 gene in the context of the DITRAC trans-complementation replicon
| Construct | Nsp1 mutation | Affected nsp1 domain | Trans-complementation of sg-mRNA synthesis |
|---|---|---|---|
| DITRAC | none | none | yes |
| DITRACΔ1 | amino acids 214–260 deleted | PCPβ | no |
| DITRACΔ2 | amino acids 155–260 deleted | PCPβ | no |
| DITRACΔ3 | amino acids 125–260 deleted | PCPα, PCPβ | no |
| DITRAC/C25A | Cys-25 to Ala | ZF | no |
| DITRAC/C44A | Cys-44 to Ala | ZF | no |
| DITRAC/H122A | His-122 to Ala | PCPα | yes |
| DITRAC/C164S | Cys-164 to Ser | PCPβ | yes |
Figure 6.
RT-PCR analysis of sg-mRNA7 synthesis by DITRAC derivatives containing either a truncated nsp1 gene or specific point mutations (Table 1) in the ZF, PCPα, or PCPβ domains. See Fig. 3C for RT-PCR details.
Subsequently, the functions of the three nsp1 domains were probed by mutagenesis of conserved residues (Fig. 6; Table 1). In two mutants, predicted zinc-coordinating residues of the ZF domain (Cys-25 and Cys-44) were replaced by Ala. In the third mutant, His-122, which resides in the most conserved region of PCPα (Fig. 5), was mutated to Ala. This residue is the counterpart of the active-site His in the PCPα of other arteriviruses. His-122 likely is associated with an unknown nonproteolytic activity of the inactivated PCPα protease of EAV, the catalytic Cys residue of which was replaced in the course of evolution (27). Finally, we replaced the PCPβ active-site Cys (Cys-164) with Ser, a mutation that was shown previously to block PCPβ proteolytic activity completely (15). The mutations were tested in the DITRAC background (Table 1) and analyzed for sg-mRNA7 synthesis. Strikingly, both ZF-Cys mutants were completely transcription negative (Fig. 6), supporting the theoretical identification of this nsp1 motif and strongly suggesting its involvement in discontinuous transcription. This result was highly specific, because both PCP active-site mutants still efficiently trans-activated sg-mRNA synthesis. Thus, we conclude that the two protease domains of nsp1 are not involved enzymatically in transcription; rather, nsp1 may use the combined PCP domains as a unique structural platform for its ZF domain.
Discussion
EAV nsp1 Is a “Discontinuous Transcription Factor.”
The progression of the RNA-virus life cycle is secured through the coordination of three basic, but complex processes: the replication, expression, and encapsidation of the viral genome (29–32). Viral proteases were implicated previously in coupling picornavirus-genome translation and replication (33), in the temporal regulation of alphavirus minus- and plus-strand RNA synthesis (34, 35), and in coupling expression of two birnavirus-genome segments (36). We have demonstrated here that in the nidovirus EAV, yet another protease, nsp1, directly connects the two main levels at which the expression of a nidovirus genome is regulated: the level of genome translation and replicase processing and that of sg-RNA synthesis. To our knowledge, nsp1 is the first example of such a +RNA virus “transcription factor,” although the exact role of the protein remains to be elucidated. Often the coupling of two processes results in a switch from one process to the other, but the up-regulation of EAV transcription is not accompanied by a shutoff of genome translation and/or replication. Still, the precise effect of nsp1 on the latter two processes remains to be examined and we cannot exclude that they are influenced by nsp1 in a more subtle manner.
The recent characterization of EAV nsp10-mutant 030F (17, 18, 20) had revealed already that nidovirus sg-mRNA synthesis could be impaired selectively (about 500-fold) without affecting genome replication. The mechanism underlying this phenotype remains to be established, but it is most plausible that nsp10 is a constitutive helicase of both replicative and transcriptional complexes (18). An EAV mutant that lacked nsp10 was found to be impaired completely in both replication and transcription (18), and thus nsp10 clearly falls into a different category than nsp1. We have shown here that as long as the 5′ proximal RNA signals were not affected, the complete nsp1-coding sequence could be deleted from the EAV genome without a detectable effect on genome replication. However, at the same time, the synthesis of sg RNAs was abolished (Fig. 3). The DITRAC trans-complementation assay (Fig. 4) provided the ultimate proof that nsp1 is required for transcription.
Nsp1 may be part of the EAV transcription complex or it may affect its composition or performance indirectly, for instance, by recruiting a cellular factor. The discontinuous minus-strand synthesis model (9) defines several potential targets for transcriptional regulation. Clearly, body and leader TRSs play a central role in this process, because their function may rely on specific protein–protein or protein–RNA interactions that are dispensable for genome replication. Such interactions could direct attenuation of minus-strand synthesis at the body TRS, release of the nascent transcript, or targeting and base paring of the incomplete minus strand, with or without the RdRp complex, to the leader TRS (10).
EAV nsp1 Is a Multidomain Protease.
EAV nsp1 is composed of (at least) three domains (ZF, PCPα, and PCPβ) that are all required for trans-activation of sg-RNA synthesis, although their precise roles remain to be established. Apparently, PCPβ not only controls the release of nsp1 by cleaving the nsp1–2 junction but also, along with PCPα, supplies nonproteolytic activities involved in transcriptional control. RNA-binding properties have been described for other viral proteases such as the poliovirus 3C(D)pro (33) and the hepatitis C virus NS3pro (37). Also, highly specialized protein-binding proteins appear to have evolved from proteolytic enzymes by accepting active-site and other mutations (38). Likewise, the PCPα and PCPβ domains of arteriviruses may have developed RNA- and/or protein-binding activities essential for sg-RNA synthesis.
Protease domains are not common in transcription factors (39), but the presence of a ZF domain in nsp1 is hardly surprising. ZF-containing proteins form one of the most ubiquitous protein classes in eukaryotes, binding to e.g., nascent RNA, template DNA, or subunits of the transcription machinery (28). Although the EAV nsp1 ZF remains to be characterized experimentally, zinc binding was demonstrated already for a distant relative: the coronavirus papain-like protease 1 (40). The conserved coronavirus ZF was predicted to adopt a variant of the Zn-ribbon fold, a conserved architecture also found in the transcription elongation factor TFIIS and related proteins (41). Accordingly, a role for coronavirus papain-like proteases in RNA synthesis was suggested. Our results strengthen this hypothesis and indicate that all nidoviruses may employ ZF-associated proteases to control transcription.
A Link Between the Evolution of Transcription and Proteases?
Nsp1 belongs to the so-called +RNA-virus accessory proteases (8). In contrast to main proteases, these are not involved directly in proteolytic processing of key replicative proteins like the RdRp and helicase. Accessory proteases are predominantly of the papain-like type and are found mostly in the N-terminal region of +RNA-virus polyproteins (42). They seem to be expendable for the basic process of genome replication but indispensable for virus reproduction, and seem able to influence different types of virus–host interactions. Here, we have demonstrated a crucial role for an accessory protease in transcriptional control. Why are accessory proteases so widespread among +RNA viruses?
The logic behind this choice may be multifaceted: proteases can (i) specifically control processes by recognizing a limited number of targets; (ii) be used as a platform for specific interactions with other molecules (see above); and (iii) autonomously ensure their own controlled expression. These combined properties make them suitable “building blocks” for +RNA viruses, which heavily rely on (post)translational regulation of gene expression. The polyproteins of many +RNA viruses, like nidoviruses (8), include multiple accessory proteases upstream of the core replicative domains (42). This organization suggests that their replicases have evolved in a complex manner by adding proteolytically autonomous building blocks to an existing polyprotein that already contained the key replicative domains.
In +RNA viruses, sg-mRNA synthesis depends on both translation and replication of the genome, but not vice versa. Thus, transcription probably has evolved after these two more basic biosynthetic processes. It is reasonable to speculate that, like a number of contemporary +RNA viruses, the ancestral nidovirus derived all its proteins from a single polyprotein. Although the nidovirus ancestor did not employ sg-mRNA synthesis, its replicative machinery essentially was primed for it, because genome replication and transcription are assumed to be enzymatically very similar in +RNA viruses. Given the low fidelity of RNA synthesis in RNA viruses, specific transcription signals may have evolved readily from sequences involved in genome replication. Their efficient use probably required the development of specialized factors, e.g., in the form of a protease acquired by intra- or intergenomic RNA recombination. Thus, the possibilities for regulated gene expression were diversified considerably and provided the “sg mRNA -competent” ancestral virus with a selective advantage.
Although entirely speculative and scarce on details, the above scenario takes into account a number of important peculiarities of the +RNA-virus life cycle. Because not all +RNA viruses that employ sg mRNAs encode proteases, alternative systems to control transcription also must have evolved. An intriguing remaining issue is the moment at which the postulated invention of transcription may have occurred. This question is related ultimately to the enigma of the time scale of RNA-virus origin and evolution. Many features of +RNA viruses suggest that they originated from primitive self-replicating entities in the primordial RNA/protein world (43). Thus, the invention of +RNA-virus transcription may well have been an early event with important implications for the evolution of both viral and cellular systems.
Acknowledgments
We thank Udeni Balasuriya and James MacLachlan for EAV-specific antisera, Guido van Marle, Richard Molenkamp, Sasha Pasternak, and Willy Spaan for helpful comments, and Sietske Rensen and Yvonne van der Meer for technical assistance. M.A.T. was supported by Grant 348-003 from the Council for Chemical Sciences of the Netherlands Organisation for Scientific Research (CW-NWO) to E.S. A.E.G. was funded with federal funds from the National Cancer Institute, National Institutes of Health, under Contract no. NO1-CO-56000. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
Abbreviations
- +RNA
positive-stranded RNA
- RdRp
RNA-dependent RNA polymerase
- IRES
internal ribosome entry site
- sg
subgenomic
- EAV
equine arteritis virus
- nsp
nonstructural protein
- PCP
papain-like cysteine protease
- TRS
transcription-regulating sequence
- LTH
leader TRS hairpin
- ZF
zinc finger
- DITRAC
discontinuous transcription complementation
- EMCV
encephalomyocarditis virus
- CAT
chloramphenicol acetyl transferase
- RT
reverse transcription
- IFA
immunofluorescence assay
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041390398.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041390398
References
- 1.Baltimore D. Bacteriol Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frolov I, Schlesinger S. J Virol. 1994;68:1721–1727. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peremyslov V V, Hagiwara Y, Dolja V V. J Virol. 1998;72:5870–5876. doi: 10.1128/jvi.72.7.5870-5876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molenkamp R, van Tol H, Rozier B C D, van der Meer Y, Spaan W J M, Snijder E J. J Gen Virol. 2000;81:2491–2496. doi: 10.1099/0022-1317-81-10-2491. [DOI] [PubMed] [Google Scholar]
- 5.Snijder E J, Meulenberg J J M. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 6.den Boon J A, Snijder E J, Chirnside E D, de Vries A A F, Horzinek M C, Spaan W J M. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziebuhr J, Snijder E J, Gorbalenya A E. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 9.Sawicki S G, Sawicki D L. Adv Exp Med Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 10.van Marle G, Dobbe J C, Gultyaev A P, Luytjes W, Spaan W J M, Snijder E J. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brian D A, Spaan W J M. Semin Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy P D, Simon A E. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 13.Spaan W J M, Delius H, Skinner M, Armstrong J, Rottier P J M, Smeekens S, van der Zeijst B A M, Siddell S G. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai M M C, Baric R S, Brayton P R, Stohlman S A. Proc Natl Acad Sci USA. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snijder E J, Wassenaar A L M, Spaan W J M. J Virol. 1992;66:7040–7048. doi: 10.1128/jvi.66.12.7040-7048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landt O, Grunert H-P, Hahn U. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 17.van Dinten L C, den Boon J A, Wassenaar A L M, Spaan W J M, Snijder E J. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dinten L C, van Tol H, Gorbalenya A E, Snijder E J. J Virol. 2000;74:5213–5223. doi: 10.1128/jvi.74.11.5213-5223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Marle G, van Dinten L C, Luytjes W, Spaan W J M, Snijder E J. J Virol. 1999;73:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Meer Y, van Tol H, Krijnse Locker J, Snijder E J. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snijder E J, Wassenaar A L M, Spaan W J M. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Dinten L C, Wassenaar A L M, Gorbalenya A E, Spaan W J M, Snijder E J. J Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen K W, van der Meer Y, Roos N, Snijder E J. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maclachlan N J, Balasuriya U B, Hedges J F, Schweidler T M, McCollum W H, Timoney P J, Hullinger P J, Patton J F. J Vet Diagn Invest. 1998;10:229–236. doi: 10.1177/104063879801000302. [DOI] [PubMed] [Google Scholar]
- 26.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.den Boon J A, Faaberg K S, Meulenberg J J M, Wassenaar A L M, Plagemann P G W, Gorbalenya A E, Snijder E J. J Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke N D, Berg J M. Science. 1998;282:2018–2022. doi: 10.1126/science.282.5396.2018. [DOI] [PubMed] [Google Scholar]
- 29.Novak J E, Kirkegaard K. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 30.Gamarnik A V, Andino R. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nugent C I, Johnson K L, Sarnow P, Kirkegaard K. J Virol. 1999;73:427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barton D J, Morasco B J, Flanegan J B. J Virol. 1999;73:10104–10112. doi: 10.1128/jvi.73.12.10104-10112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirako Y, Strauss J H. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemm J A, Rumenapf T, Strauss E G, Strauss J H, Rice C M. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birghan C, Mundt E, Gorbalenya A E. EMBO J. 2000;19:114–123. doi: 10.1093/emboj/19.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallinari P, Brennan D, Nardi C, Brunetti M, Tomei L, Steinkuhler C, De Francesco R. J Virol. 1998;72:6758–6769. doi: 10.1128/jvi.72.8.6758-6769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawlings N D, Barrett A J. Methods Enzymol. 1994;244:19–61. doi: 10.1016/0076-6879(94)44004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muise A M, Ro H S. Biochem J. 1999;343:341–345. [PMC free article] [PubMed] [Google Scholar]
- 40.Herold J, Siddell S G, Gorbalenya A E. J Biol Chem. 1999;274:14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B, Jones D N, Kaine B P, Weiss M A. Structure (London) 1998;6:555–569. doi: 10.1016/s0969-2126(98)00058-6. [DOI] [PubMed] [Google Scholar]
- 42.Gorbalenya A E, Snijder E J. Perspect Drug Discovery Des. 1996;6:64–86. doi: 10.1007/BF02174046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorbalenya A E. Semin Virol. 1992;3:359–371. [Google Scholar]
- 44.Godeny E K, Chen L, Kumar S N, Methven S L, Koonin E V, Brinton M A. Virology. 1993;194:585–596. doi: 10.1006/viro.1993.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer G A, Kuo L, Chen Z, Faaberg K S, Plagemann P G W. Virology. 1995;209:637–642. doi: 10.1006/viro.1995.1296. [DOI] [PubMed] [Google Scholar]
- 46.Meulenberg J J M, Hulst M M, de Meijer E J, Moonen P L J M, den Besten A, de Kluyver E P, Wensvoort G, Moormann R J M. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelsen C J, Murtaugh M P, Faaberg K S. J Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]