Abstract
microRNAs (miRNAs) have emerged as critical regulators of gene expression. These small, non-coding RNAs are believed to regulate more than a third of all protein coding genes, and they have been implicated in the control of virtually all biological processes, including the biology of stem cells. The essential roles of miRNAs in the control of pluripotent stem cells were clearly established by the finding that embryonic stem (ES) cells lacking proteins required for miRNA biogenesis exhibit defects in proliferation and differentiation. Subsequently, the function of numerous miRNAs has been shown to control the fate of ES cells and to directly influence critical gene regulatory networks controlled by pluripotency factors Sox2, Oct4, and Nanog. Moreover, a growing list of tissue-specific miRNAs, which are silenced or not processed fully in ES cells, have been found to promote differentiation upon their expression and proper processing. The importance of miRNAs for ES cells is further indicated by the exciting discovery that specific miRNA mimics or miRNA inhibitors promote the reprogramming of somatic cells into induced pluripotent stem (iPS) cells. Although some progress has been made during the past two years in our understanding of the contribution of specific miRNAs during reprogramming, further progress is needed since it is highly likely that miRNAs play even wider roles in the generation of iPS cells than currently appreciated. This review examines recent developments related to the roles of miRNAs in the biology of pluripotent stem cells. In addition, we posit that more than a dozen additional miRNAs are excellent candidates for influencing the generation of iPS cells as well as for providing new insights into the process of reprogramming.
Keywords: ES cells, iPS cells, miRNAs, reprogramming, Sox2, Oct4
Introduction
The seminal finding that lin-4, and later let-7, are developmental regulators of Caenorhabditis elegans sparked the exciting discovery that a class of small RNAs play critical roles in the regulation of gene expression (Lee, et al. 1993; Reinhart, et al. 2000). Subsequently, homologs of let-7 were identified in higher organisms, including mammals (Pasquinelli, et al. 2000) and, soon thereafter, it was recognized that these small RNAs belong to a large family of non-coding RNAs known as microRNAs (miRNAs) (Lagos-Quintana, et al. 2001; Lau, et al. 2001; Lee, Ambros. 2001). miRNAs have emerged as key regulators of gene silencing that act by targeting specific mRNAs for degradation or by suppressing their translation. Remarkably, miRNAs are believed to regulate more than a third of all protein coding genes, and they have been shown to directly influence virtually all biological processes, including stem cell self-renewal and lineage specification during development, as well as diseases, such as cancer (Kanellopoulou, et al. 2005; Murchison, et al. 2005; Wang, et al. 2007; Kato, Slack. 2008; Liu, et al. 2008; Stefani, Slack. 2008; Wang, et al. 2008b; Cordes, Srivastava. 2009; Friedman, Jones. 2009).
In this review, we discuss the varied roles played by miRNA in the maintenance of embryonic stem (ES) cell self-renewal and pluripotency. We also discuss several recent studies that have begun to probe the roles of miRNAs during the reprogramming of somatic cells into induced pluripotent stem (iPS) cells. Finally, we discuss why more than a dozen additional miRNAs are likely to influence the process of reprogramming. Not discussed in this review are studies dealing with miRNAs in adult stem cells. Readers interested in this topic are referred to several excellent reviews (Zhao, Srivastava. 2007; Lakshmipathy, Hart. 2008; Gangaraju, Lin. 2009; Li, Jin. 2010).
Biogenesis of miRNAs
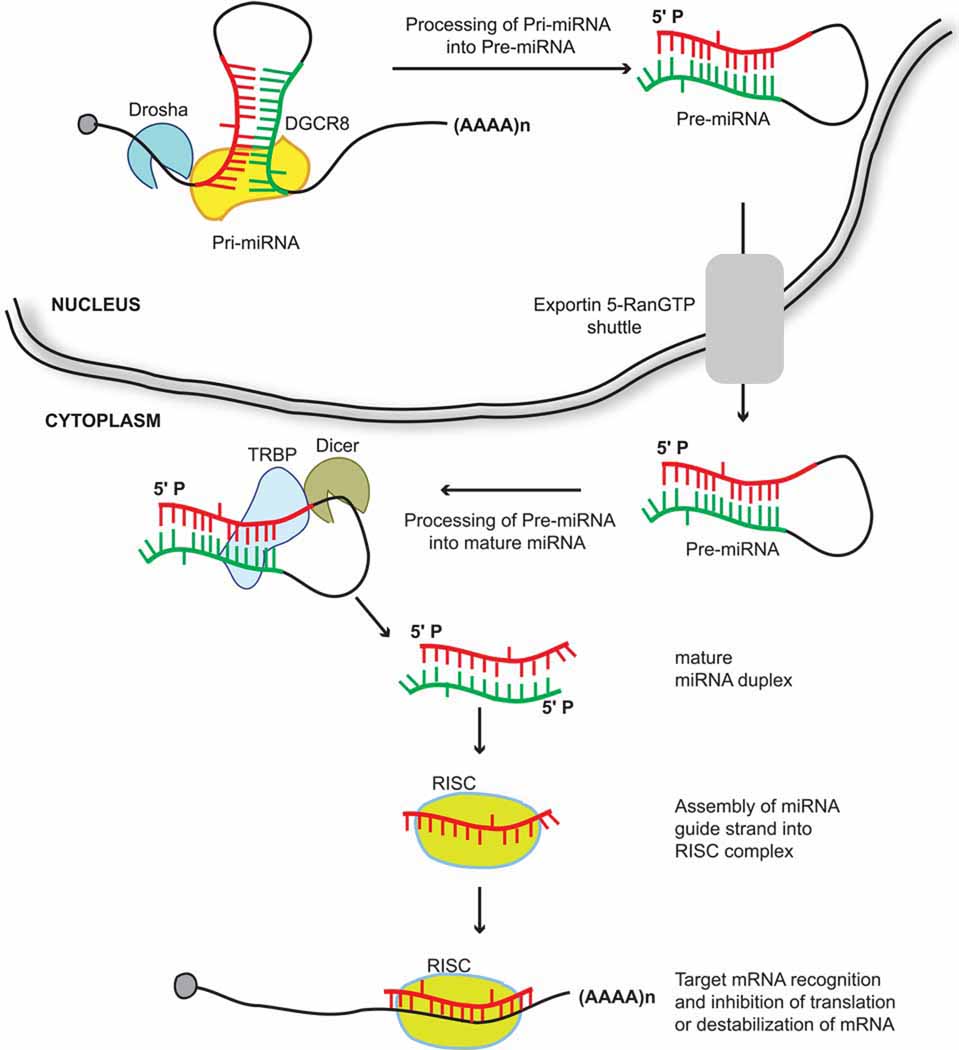
miRNAs provide an additional level of gene regulation by functioning at the post-transcriptional level. Location of miRNA coding sequences can be either intragenic or intergenic, and miRNAs are most often transcribed by RNA polymerase II (Bartel. 2004; Rodriguez, et al. 2004). Generation of mature miRNAs involves both nuclear and cytoplasmic steps (Figure 1). Once transcribed, primary-miRNA (pri-miRNA) transcripts in the nucleus are recognized and cleaved into ~70-nucleotide precursor-miRNAs (pre-miRNAs) by a microprocessor complex, which contains RNase III enzyme Drosha and RNA binding protein DiGeorge syndrome Critical Region gene 8 (DGCR8) (Lee, et al. 2002; Lee, et al. 2003; Zeng, Cullen. 2003; Kim. 2005). Pre-miRNAs are transported into the cytoplasm by Exportin 5, followed by Dicer-mediated processing into ~22-nucleotide mature miRNA duplexes (Lee, et al. 2003; Yi, et al. 2003; Lund, et al. 2004). Like Drosha, Dicer functions in concert with RNA binding protein Transactivating Region Binding Protein (TRBP) (Chendrimada, et al. 2005; Haase, et al. 2005). One of the miRNA duplex strands serves as a guide strand and is incorporated into the Argonaute-containing RISC complex, while the other strand is released and degraded (Maniataki, Mourelatos. 2005). The eight-nucleotide seed sequence in the 5’ terminus of the miRNA guide strand is critical for target recognition, and the guide strand directs the RISC complex to target mRNA sequences (Brennecke, et al. 2005; Lewis, et al. 2005). Target recognition by miRNA is often mediated by imperfect base pairing with a region that lies in the 3’ untranslated region (UTR) of mRNA. Imperfect base pairing with their targets enables miRNAs to target multiple genes simultaneously. Recent studies have demonstrated that miRNAs can also target regions outside of the 3’ UTR, such as regions in 5’ UTR and the amino acid coding sequence of mRNA (Lytle, et al. 2007; Tay, et al. 2008a).
Figure 1. Schematic representation of the miRNA biogenesis.
miRNA biogenesis involves both nuclear and cytoplasmic steps. Following transcription, pri-miRNAs are processed into pre-miRNAs by a microprocessor complex containing Drosha and DGCR8. Pre-miRNAs are exported from the nucleus into the cytoplasm by Exportin 5 – RanGTP. In the cytoplasm, pre-miRNAs are processed into mature miRNA duplex by Dicer, which functions in concert with TRBP. One of the strands from the mature miRNA duplex, the guide strand, is loaded into the Argonaute-RISC complex. The guide strand then directs the RISC complex to mRNA target sequences to mediate gene silencing.
Roles of miRNAs in the establishment of the ES cell phenotype
ES cells have a unique ability to replicate indefinitely (self-renewal), yet they are capable of forming all cell types of the body (pluripotency). These properties of ES cells, and their reprogrammed counterparts, iPS cells, offer immense potential for the field of regenerative medicine. However, before ES cells and iPS cells are used clinically, a thorough understanding of the mechanisms that control pluripotent stem cell identity would be extremely valuable. Recent work has demonstrated that miRNAs play key roles in controlling the fate of ES cells. Contribution of miRNA pathways to ES cell identity has been studied using Dicer null and DGCR8 null ES cells, which lack all mature miRNAs (Kanellopoulou, et al. 2005; Murchison, et al. 2005; Wang, et al. 2007). Dicer is necessary for maturation of both miRNAs and small interfering RNAs, whereas DGCR8 is not required for siRNA processing (Denli, et al. 2004; Gregory, et al. 2004; Han, et al. 2004; Landthaler, et al. 2004; Wang, et al. 2007). Dicer null ES cells exhibit slow proliferation rates and defective differentiation (Kanellopoulou, et al. 2005; Murchison, et al. 2005). When these cells are induced to differentiate in embryoid bodies, Oct4 expression is only partially decreased, and endodermal and mesodermal markers, which are typically expressed by differentiated ES cells, are not detectable (Kanellopoulou, et al. 2005). DGCR8 null ES cells also exhibit defective differentiation (Wang, et al. 2007). When these cells were subjected to conditions that normally induce ES cell differentiation, they exhibited abnormal activation of multiple markers of differentiation, coupled with the inability to silence expression of pluripotency markers. DGCR8 null ES cells accumulate in the G1 phase of the cell cycle, indicating that DGCR8 is necessary for normal ES cell proliferation and cell cycle progression (Wang, et al. 2007). Together, these studies demonstrated the requirement of mature miRNAs for the maintenance of ES cell self-renewal and pluripotency. Additionally, several independent studies have identified distinct miRNAs expressed in ES cells and their differentiated counterparts, reinforcing the role of ES cell-specific miRNAs in regulating ES cell identity (Houbaviy, et al. 2003; Bar, et al. 2008; Laurent, et al. 2008).
Convergence of miRNAs and a key ES cell gene regulatory network
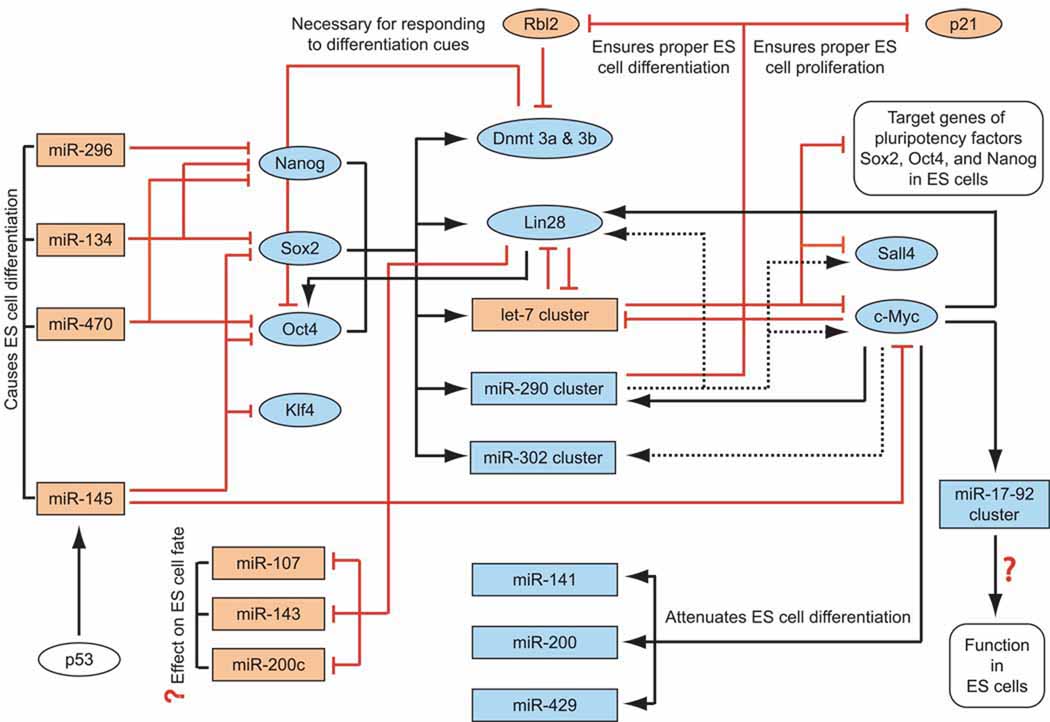
Recently, significant progress has been made towards understanding the contribution of specific miRNAs in establishing the ES cell phenotype (summarized in Figure 2). It is becoming evident that miRNAs are an integral part of the gene networks regulated by pluripotency factors Sox2, Oct4, and Nanog. Genome-wide binding assays of Sox2, Oct4, and Nanog demonstrated that these transcription factors co-occupy promoters of the majority of miRNAs that are preferentially expressed in mouse ES cells, including the miR-290 cluster and miR-302 cluster miRNAs (Marson, et al. 2008). Moreover, these transcription factors not only bind to the promoters of the miR-290 cluster and the miR-302 cluster, but they have also been shown to regulate the expression of these miRNAs (Barroso-delJesus, et al. 2008; Card, et al. 2008; Marson, et al. 2008).
Figure 2. Regulatory networks of miRNAs and proteins involved in control of ES cell self-renewal and differentiation.
Black arrow: direct binding and/or activation of miRNA/protein expression
Black dashed arrow: indirect activation of miRNA/protein expression
Red lines with a vertical stub: direct inhibition of miRNA/protein expression or evidence for direct inhibition as suggested by binding of the protein to the promoter region
Blue colored ovals (proteins/protein coding genes) and rectangles (miRNAs/miRNA coding genes): expressed only in ES cells / expressed abundantly in ES cells compared to differentiated cells
Gold colored ovals (proteins/protein coding genes) and rectangles (miRNAs/miRNA coding genes): expressed only in differentiated cells / expressed abundantly in differentiated cells compared to ES cells
Sox2, Oct4, and Nanog, in addition to regulating the expression of numerous proteins, regulate the expression of several miRNAs in ES cells, including the miR-290 cluster and miR-302 cluster. Conversely, several tissue-specific miRNAs such as miR-296, miR-134, miR-470, and miR-145 inhibit the expression of Sox2, Oct4, and Nanog in ES cells. ESCC miRNAs, which include select miRNAs from the miR-290 cluster and the miR-302 cluster, and let-7 miRNAs have emerged as major regulators of ES cell fate, and exhibit opposing effects on ES cell self-renewal, pluripotency, and differentiation. ESCC miRNAs belonging to the miR-290 cluster are responsible for ensuring proper ES cell proliferation, as well as proper ES cell differentiation when subjected to appropriate differentiation cues. let-7 family miRNAs are involved in a negative feedback regulatory loop with Lin28 and c-Myc, both of which have important functions in ES cells and iPS cells. Lin28, apart from preventing the generation of mature let-7, inhibits the processing of miR-107, miR-143, and miR-200c in ES cells. Furthermore, Lin28 facilitates the translation of Oct4 in ES cells. c-Myc is involved in a positive feedback regulatory loop with members of the ESCC miRNAs. Additionally, c-Myc induces the expression of miR-141, miR-200, and miR-429, which attenuate mouse ES cell differentiation upon LIF withdrawal. c-Myc has also been shown to activate the transcription of the miR-17-92 cluster in tumor cells.
Note: data were compiled from studies involving mouse and human cells
It is becoming evident that the trio of Sox2, Oct4, and Nanog utilize miRNAs to fine tune the expression of their target genes in ES cells. This is exemplified by the involvement of Sox2, Oct4, and Nanog in incoherent and coherent feed-forward regulatory networks, by activating expression of specific miRNAs, to modulate the levels of proteins, such as Lefty1 and DNA methyltransferases 3a and 3b (Dnmt 3a, Dnmt 3b), in ES cells (Marson, et al. 2008). Lefty1 participates in pluripotency regulating transcription networks (Nakatake, et al. 2006), whereas Dnmt 3a and Dnmt 3b are necessary for proper differentiation of ES cells when they are subjected to appropriate differentiation cues (Benetti, et al. 2008; Sinkkonen, et al. 2008). Recently, two independent studies have investigated the mechanism responsible for the inability of Dicer null ES cells to silence the self-renewal program when subjected to conditions that normally promote differentiation. These studies determined that the miR-290 cluster of miRNAs is required for expression of de novo DNA methyltransferases in ES cells, and that differentiation defects observed in Dicer null ES cells are due, at least in part, to incomplete and reversible silencing of Oct4 expression, resulting from improper promoter methylation (Benetti, et al. 2008; Sinkkonen, et al. 2008).
In addition to the association of Sox2, Oct4, and Nanog with promoters of miRNAs that are preferentially expressed in ES cells, Sox2, Oct4, and Nanog also associate with the promoters of silenced tissue-specific miRNAs (Marson, et al. 2008). It has been suggested that binding of Sox2, Oct4, and Nanog to these tissue-specific miRNAs primes them for expression upon differentiation of ES cells; however, they are kept silent in ES cells due to the association of these miRNA genes with inhibitory polycomb repressor proteins (Marson, et al. 2008). In this regard, several tissue-specific miRNAs are involved in regulating the expression of critical pluripotency factors, and they induce differentiation when expressed in ES cells. For example, miR-296, miR-470, and miR-134, whose expression is up-regulated upon retinoic acid induced differentiation of mouse ES cells, inhibit the expression of Sox2, Oct4, and Nanog in various combinations (Tay, et al. 2008a; Tay, et al. 2008b). In a related study, miR-145 has been shown to repress pluripotency in human ES cells by down-regulating the expression of Sox2, Oct4, and Klf4 (Xu, et al. 2009b). These findings demonstrate that miRNAs are an integral part of the core ES cell transcriptional regulatory network and play important roles in regulating ES cell fate.
Role of miRNAs in ES cell specific cell cycle structure
As discussed earlier, DGCR8 null mouse ES cells exhibit cell cycle defects. To extend these findings, Wang and co-workers screened a library of 266 mouse miRNAs for their ability to rescue the cell cycle defects of DGCR8 null ES cells (Wang, et al. 2008b). They determined that a subset of miRNAs, referred to as ES cell-specific cell cycle-regulating miRNAs (ESCC miRNAs) (miR-291a-3p, miR-291b-3p, miR-294, miR-295, and miR-302), can rescue the ES cell cycle defect. This occurs, at least in part, by promoting the G1-S transition. These workers also demonstrated that ESCC miRNAs exert their effect by suppressing the expression of multiple cyclin E-Cdk2 inhibitors, such as p21, Rbl2, and Lats2 (Wang, et al. 2008b).
To fully understand the roles played by miRNAs in ES cells, it is important to stress that mouse and human ES cells represent different stages of mammalian development (Brons, et al. 2007; Tesar, et al. 2007). Consequently, it is likely that there are differences in the mechanisms that control the self-renewal of mouse and human ES cells. For instance, of the miRNAs belonging to the miR-290 and miR-302 clusters, miR-302 cluster miRNAs are expressed in both human and mouse ES cells; whereas, miR-290 cluster is only expressed in mouse ES cells (Kim, et al. 2009c). Furthermore, work with several miRNAs has only been conducted in human ES cells. For example, miR-520 cluster miRNAs, whose seed sequence is similar to miR-302 cluster miRNAs, have been shown to regulate important cellular functions in human ES, including cell proliferation and chromatin structure (Ren, et al. 2009). Another miRNA whose function has only been studied in human ES cells is miR-92b, which promotes the G1-S transition by repressing the Cdk inhibitor p57 (Sengupta, et al. 2009). Interestingly, genome-wide transcription factor binding assays have demonstrated that pluripotency factors Sox2 and Oct4 associate with the promoter of miR-92b in mouse ES cells, suggesting direct regulation of miR-92b expression by Sox2 and Oct4 (Marson, et al. 2008), and also likely involvement of miR-92b in establishing the mouse ES cell phenotype.
Opposing roles of ESCC and let-7 miRNAs in the control of ES cell fate
ESCC and let-7 family miRNAs have begun to emerge as important regulators of ES cell self-renewal, pluripotency, and differentiation (Melton, et al. 2010). ESCC and let-7 miRNAs represent major miRNAs expressed in ES cells and somatic cells, respectively (Marson, et al. 2008). Athough mature let-7 is not expressed in ES cells, the let-7 promoter is bound by Sox2, Oct4, and Nanog (Marson, et al. 2008). In accordance with this finding, pri-let-7 transcripts are present at high levels in ES cells and depletion of Oct4 decreases the levels of pri-let-7 transcripts (Marson, et al. 2008). However, processing of pri-let-7 into mature let-7 is prevented by RNA binding protein Lin28. Lin28 inhibits both Drosha-mediated (Newman, et al. 2008; Viswanathan, et al. 2008) and Dicer-mediated (Heo, et al. 2008; Rybak, et al. 2008) processing of pri-let-7 into mature let-7. Mature let-7, in turn, inhibits the expression of Lin28 (Rybak, et al. 2008). Thus, the negative feedback loop between Lin28 and let-7 has a major influence over ES cell fate. Lin28, apart from its role in preventing the generation of mature let-7, is necessary for proper ES cell proliferation, and also for efficient translation of Oct4 (Xu, et al. 2009a; Qiu, et al. 2010).
c-Myc, which is required for maintenance of ES cell self-renewal (Cartwright, et al. 2005), is also involved in a negative feedback regulatory loop with let-7. c-Myc, through an indirect mechanism involving transcriptional activation of Lin28, inhibits the biogenesis of mature let-7 (Chang, et al. 2009). Additionally, in lymphoma cells where expression of c-Myc leads to downregulation of let-7 expression, c-Myc has been shown to bind let-7 promoter, which suggest that c-Myc directly inhibits let-7 expression (Chang, et al. 2008). Mature let-7, in turn, directly inhibits the expression of c-Myc (Kumar, et al. 2007; Melton, et al. 2010). Using DGCR8 null ES cells, described above, Melton and co-workers demonstrated that ESCC and let-7 miRNAs play opposing roles in regulating the ES cell phenotype (Melton, et al. 2010). Specifically, they demonstrated that introduction of ESCC miRNAs into DGCR8 null ES cells rescued the cell cycle defect, whereas introduction of let-7 miRNAs induced DGCR8 null ES cells to differentiate.
The effects of let-7 in ES cells can be explained by its inhibitory effect on the expression of Lin28, c-Myc, Sall4 and downstream target genes of pluripotency factors, in particular Sox2, Oct4, and Nanog (Melton, et al. 2010). Additionally, let-7 also represses positive regulators of cell cycle, such as CDK6, CDC25A, and Cyclin D, in human cancer cells (Johnson, et al. 2007). Introduction of let-7 into wild-type ES cells failed to induce ES cells to differentiate, which suggests that ESCC miRNAs antagonize the effects of let-7 in ES cells (Melton, et al. 2010). The importance of ESCC miRNAs in regulating ES cell fate is further reinforced by the presence of a positive feed-back loop between ESCC miRNAs and Myc. In this regard, ChIP-seq data have shown that c-Myc and n-Myc bind to the promoter of the miR-290 cluster, which expresses several ESCC miRNAs, suggesting direct activation of expression of these miRNAs by Myc (Chen, et al. 2008). c-Myc has also been shown to induce, by an indirect mechanism, the expression of the miR-302 cluster, which codes for an ESCC miRNA (Lin, et al. 2009). Moreover, the ESCC miRNA, miR-294, has been shown to indirectly activate the expression of c-Myc (Melton, et al. 2010).
Recently, c-Myc has been shown to bind to promoter regions of miR-141, miR-200, and miR-429 and to induce their expression in mouse ES cells (Lin, et al. 2009). Introduction of these c-Myc induced miRNAs into ES cells attenuated the differentiation of these cells in response to LIF withdrawal. The miR-17-92 cluster of miRNAs, whose expression is elevated in many cancers, are also enriched in ES cells, and have similar seed sequences to those of ESCC miRNAs (Laurent, et al. 2008; Marson, et al. 2008; Mendell. 2008; Wang, et al. 2008b; Judson, et al. 2009). Additionally, c-Myc has been shown to induce the expression of the miR-17-92 cluster in tumor cells (O'Donnell, et al. 2005). However, the function of the miR-17-92 cluster in ES cells has not been investigated. The fact that miRNAs from the miR-17-92 cluster have seed sequences similar to those of ESCC miRNAs, and that c-Myc induces expression of both miR-17-92 and ESCC miRNAs, highlights the impact of c-Myc-regulated miRNAs on the self-renewal of stem cells (He, et al. 2005; O'Donnell, et al. 2005; Chen, et al. 2008).
Regulatory mechanisms involved in establishing the ES cell-specific miRNA profile
Although it is clear that miRNAs play critical roles in regulating the self-renewal and pluripotency of ES cells, much less is known about the molecular mechanisms that regulate miRNA expression in pluripotent stem cells. This will necessitate systematic identification and study of both functional core promoter/enhancer elements, and specific cis-regulatory elements involved in the regulation of miRNA expression in ES cells and differentiated cells. A recent study involving large scale identification of miRNA promoters in both human and mouse cells is an excellent starting point for characterizing individual miRNA promoters (Marson, et al. 2008). Recently, the miR-302 cluster promoter has been characterized and functionally validated in human ES cells (Barroso-delJesus, et al. 2008; Card, et al. 2008). Specifically, it has been demonstrated that Sox2 and Oct4 bind the miR-302 promoter and are essential for expression of miR-302 miRNAs in human ES cells (Card, et al. 2008). Similar studies are warranted for miRNAs, such as the miR-290 cluster, miR-92b, miR-145, miR-134 and the let-7 cluster, that are known to play important functions in ES cell self-renewal and differentiation.
Biogenesis of miRNAs, which interfere with ES cell self-renewal and pluripotency, appears to be tightly regulated at multiple levels in ES cells. This is exemplified for let-7 miRNAs, whose biogenesis is controlled at both transcriptional and post-transcriptional levels. As noted above, c-Myc has been shown to transcriptionally repress let-7 expression in lymphoma cells (Chang, et al. 2008), whereas Lin28 prevents processing of pri-let-7 to mature let-7 in ES cells (Heo, et al. 2008; Newman, et al. 2008; Rybak, et al. 2008; Viswanathan, et al. 2008). Additionally, mouse Lin41 has been shown to suppress let-7 activity, at least in part, by antagonizing Argonaute 2 (Rybak, et al. 2009). Considering the importance of the control of miRNA biogenesis, identification of proteins that regulate biogenesis of other lineage-specific miRNAs, such as miR-134, miR-145, miR-296, in ES cells is expected to provide greater insight into ES cell biology. Additionally, a recent study demonstrated that the inhibitory effect of Lin28 on miRNA processing is not limited to let-7. Processing of several other miRNAs, including miR-107, miR-143, and miR-200c, is also inhibited by Lin28 (Heo, et al. 2009). In accordance with this finding, miR-107, miR-143, and miR-200c are more abundantly expressed in differentiated cells compared to undifferentiated cells (Heo, et al. 2009). Therefore, additional studies to investigate the effects of these miRNAs on the fate of ES cells are warranted.
Roles of miRNAs in the reprogramming of somatic cells into iPS cells
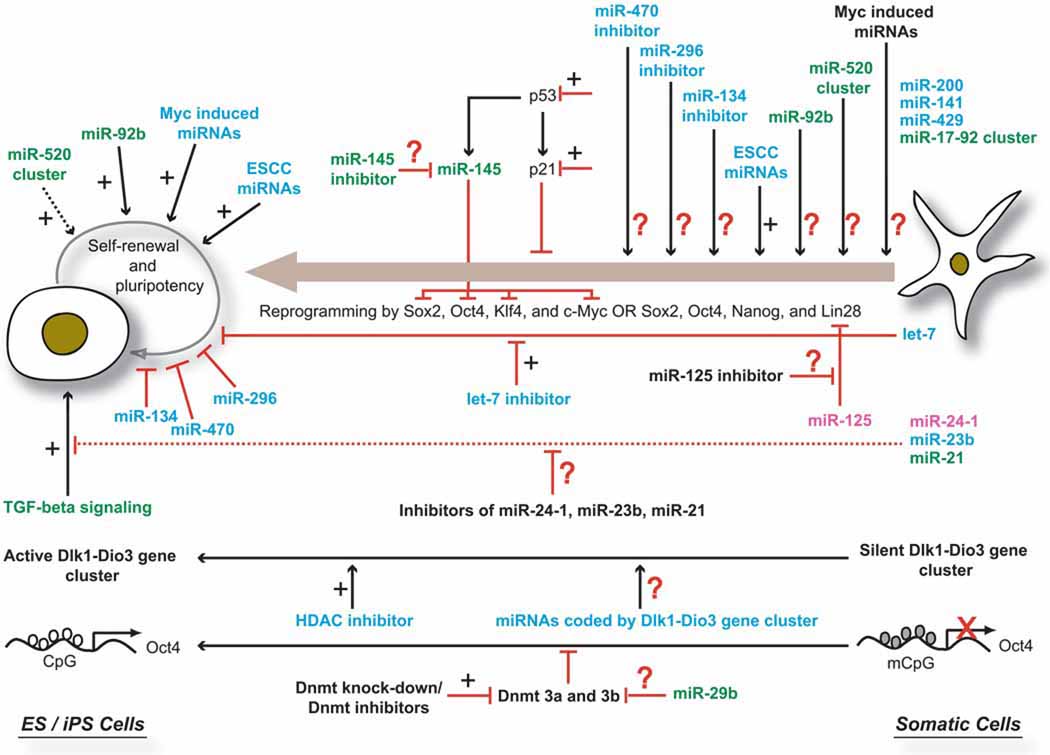
Only four years ago, it was discovered that somatic cells could be reprogrammed into iPS cells (Takahashi, Yamanaka. 2006). This brought about a true paradigm shift in the field of stem cell biology. Over the past four years, the original combination of reprogramming factors [Sox2, Oct4, Klf4, and c-Myc (SOKM)] has been used to generate iPS cells from a wide-range of cell types (Maherali, Hochedlinger. 2008; Cox, Rizzino. 2010). Other combinations, such as Sox2, Oct4, Lin28 and Nanog, have also been used to generate iPS cells (Yu, et al. 2007). However, relatively little is known about the roles played by miRNAs in the reprogramming of somatic cells into iPS cells. The known effects of miRNAs during reprogramming and several examples of miRNAs with potential roles in modulating reprogramming are summarized in Figure 3.
Figure 3. Known and predicted roles of miRNAs in reprogramming of somatic cells into iPS cells.
Plus mark: positive influence on ES cell self-renewal and / or somatic cell reprogramming
Black dashed arrow: indirect evidence for positive influence on ES cell self-renewal
Red line with vertical stub: inhibition of ES cell self-renewal and / or somatic cell reprogramming
Red dashed line with vertical stub: indirect evidence for inhibition of TGF-β signaling in human ES cells
Question mark: has the potential to promote reprogramming, but effects have not yet been investigated
Green colored fonts: indicated function demonstrated in human cells
Blue colored fonts: indicated function demonstrated in mouse cells
Pink colored fonts: indicated function demonstrated in both human and mouse cells
Several miRNAs, including ESCC miRNAs, Myc-induced miRNAs, miR-92b, and the miR-520 cluster, have been shown to positively regulate the self-renewal and pluripotency of ES cells. Among these, only ESCC miRNAs have been tested for their ability to promote reprogramming. Additionally, a number of tissue-specific miRNAs, such as let-7, miR-134, miR-470, miR-296, and miR-145, have been shown to interfere with the self-renewal and pluripotency of ES cells. However, with the exception of let-7, the effects of inhibiting the activity of these miRNAs on reprogramming are not known. Recent study by Melton and co-workers demonstrated that inhibition of let-7 activity promotes reprogramming (Melton, et al. 2010). miR-125, which inhibits the expression of Lin28, is also expected to positively influence reprogramming (Wu, Belasco. 2005). Additionally, miRNAs that target specific signaling pathways (e.g TFG-beta signaling) and epigenetic processes (e.g. DNA methylation) can also be tested for their ability to promote reprogramming. miRNAs encoded by Dlk1-Dio3 gene cluster are also attractive candidates for promoting reprogramming because activation of imprinted Dlk1-Dio3 gene cluster is essential for generating fully reprogrammed iPS cells, which are functionally equivalent to ES cells.
Note: data were compiled from studies involving mouse and human cells
Expression of ES cell-specific miRNAs promotes reprogramming
As discussed earlier, transient transfection of ESCC miRNAs into DGCR8 knockout mouse ES cells rescued their proliferation defect (Wang, et al. 2008b). Interestingly, Judson and co-workers investigated the effects of ESCC miRNAs on reprogramming of somatic cells into iPS cells (Judson, et al. 2009). For this purpose, mouse embryonic fibroblasts were infected with retroviruses that express Sox2, Oct4, and Klf4, and miRNA mimics were introduced into the cells by transient transfection. They determined that ESCC miRNAs increase the generation of mouse iPS cells induced by the combination of Sox2, Oct4, and Klf4. Among different ESCC miRNAs, miR-294 exhibited the greatest effect on reprogramming and increased efficiency of iPS cell generation from 0.01–0.05% to 0.4–0.7%. Additionally, miR-294 increased the kinetics of Sox2, Oct4, and Klf4 mediated reprogramming. However, when miR-294 was introduced with Sox2, Oct4, Klf4, and c-Myc, it had no effect on reprogramming. Therefore, ESCC miRNAs appear to promote Sox2, Oct4, and Klf4 mediated reprogramming by substituting for c-Myc. Importantly, iPS cells generated without c-Myc are likely to be safer for future use in cell-based clinical therapies. As discussed below, miRNAs from the miR-302 cluster have also been shown to promote reprogramming.
Inhibition of tissue-specific miRNAs promotes the formation of iPS cells
The pro-differentiation effect of let-7 on ES cells prompted Melton and co-workers to test the effect of inhibiting the activity of let-7 miRNA on the reprogramming of somatic cells into iPS cells (Marson, et al. 2008; Rybak, et al. 2008; Melton, et al. 2010). For this purpose, they introduced let-7 antisense inhibitor into mouse embryonic fibroblasts by transient transfection and studied its effects on reprogramming mediated by Sox2, Oct4, and Klf4, in the presence or absence of c-Myc. They determined that inhibition of let-7 activity increased Sox2, Oct4, and Klf4 mediated reprogramming 4.3 fold, whereas Sox2, Oct4, Klf4, and c-Myc-mediated reprogramming increased only 1.75 fold. These data argue that increased reprogramming in response to let-7 inhibition is mediated by let-7 target genes, such as c-Myc and Lin28 (Figure 2). Interestingly, recent studies have shown that Lin28 is also repressed by miR-125, which is abundantly expressed in differentiated cells (Wu, Belasco. 2005; Wilson, et al. 2009). This raises the possibility that inhibiting the activity of both miR-125 and let-7 miRNAs may result in additional beneficial effects during reprogramming, due to robust activation of Lin28 expression. However, elevating Lin28 levels beyond a critical level could have deleterious effects (Darr, Benvenisty. 2009). Collectively, these results illustrate the important roles played by miRNAs in reprogramming somatic cells into iPS cells.
Mechanisms for modulating the activities and levels of miRNAs
Multiple methods are available to modulate the activities and levels of miRNAs. Anti-miRNA oligonucleotides, known as antagomirs, are routinely used to inhibit miRNA activity (Liu, et al. 2008). Antagomirs bind to mature miRNAs, mediated by Watson-Crick base pairing, and interfere with their target recognition. The affinity, stability, safety and delivery of antagomirs have been improved through chemical modifications. Four chemical modifications are commonly used: replacement of the 2’-OH in the ribose moiety with 2’-O-methyl or 2’-O-methoxyethyl, addition of an extra methylene bridge to the ribose moiety, and replacement of a non-bridging oxygen with a sulfur atom in the phosphate backbone (Wahlestedt, et al. 2000; Meister, et al. 2004; Davis, et al. 2006; Orom, et al. 2006; Liu, et al. 2008). Antagomirs are delivered to cells by transient transfection. Therefore, achieving efficient delivery and stable expression of these oligonucleotides is not possible. This short-coming of antagomirs can be overcome by using viral vectors that code for miRNA sponges, which can provide efficient delivery and stable expression of anti-miRNAs (Gentner, et al. 2009). miRNA sponges are RNA molecules that contain multiple miRNA binding sites (Ebert, et al. 2007), and function by sequestering corresponding miRNAs. miRNA sponges often inhibit the activity of closely related miRNAs within the same family (Ebert, et al. 2007). In addition to inhibiting miRNA activity by various methods, one can exogenously express miRNAs by transient transfection of either miRNA mimics or pre-miRNAs (Tay, et al. 2008a; Judson, et al. 2009; Sengupta, et al. 2009; Xu, et al. 2009b; Melton, et al. 2010). Alternatively, exogenous miRNA expression can be achieved by viral vector-mediated delivery of pre-miRNA molecules, which provide both efficient delivery and stable expression of miRNAs (Lin, et al. 2008; Xu, et al. 2009b).
Recent studies investigating the roles of miRNAs during reprogramming employed transient transfection of cells with miRNA mimics and miRNA inhibitors (Judson, et al. 2009; Melton, et al. 2010). It is possible that longer expression of miRNA mimics and miRNA inhibitors, for example by using viral vectors, may be necessary to maximize their effects on reprogramming. However, stable expression of miRNA mimics and miRNA inhibitors that are helpful during somatic cell reprogramming may interfere with subsequent lineage-specific differentiation of the iPS cells. This problem can be circumvented by using drug-inducible viral vectors. Moving forward, it would also be desirable to employ methods, such as adenoviral vectors (Stadtfeld, et al. 2008) or non-integrating episomal vectors (Yu, et al. 2009), that deliver miRNA mimics and miRNA inhibitors without directly altering chromosomal integrity.
Do miRNAs mediate the effects of p53 on reprogramming?
Considering the burgeoning role of miRNAs in regulating ES cell self-renewal and differentiation, it is conceivable that miRNAs have much wider and more important roles in reprogramming than is currently recognized. Recent studies have shown that p53 poses a barrier to reprogramming, and deletion of p53 significantly increases the efficiency of generating iPS cells (Banito, et al. 2009; Hong, et al. 2009; Kawamura, et al. 2009; Li, et al. 2009; Marion, et al. 2009). Moreover, the effects of p53 on reprogramming appear to be mediated, at least in part, by p21. Accordingly, knockdown of p21 in cells containing wild type p53 also increases the efficiency of generating iPS cells (Kawamura, et al. 2009; Li, et al. 2009). However, inhibition of the p53 pathway results in iPS cell populations containing a high percentage of cells with DNA damage (Marion, et al. 2009). To overcome the p53-mediated barrier to reprogramming without compromising the genomic integrity of iPS cells, it is necessary to understand the mechanisms by which p53 antagonizes reprogramming. A probable role for miRNAs in the p53-mediated barrier to reprogramming is suggested by the finding that p53 enhances the processing and maturation of several miRNAs in human fibroblasts, including miR-145 (Suzuki, et al. 2009). Additionally, p53 has been shown to bind to the miR-145 promoter and activate its expression (Sachdeva, et al. 2009).
As mentioned earlier, miR-145 induces ES cell differentiation by inhibiting the expression of key pluripotency/reprogramming factors, such as Sox2, Oct4, Klf4, and c-Myc (Sachdeva, et al. 2009; Xu, et al. 2009b). Hence, it is reasonable to speculate that the p53-mediated barrier to reprogramming may be due, at least in part, to miR-145. If this is the case, inhibiting the activity of miR-145 will promote the reprogramming of human somatic cells into iPS cells by enabling the early activation of endogenous reprogramming factors. Furthermore, p53 positively regulates the expression of several miRNAs, in addition to miR-145 (Suzuki, et al. 2009). Therefore, it is possible that the effects of p53 on reprogramming are mediated through multiple miRNAs. Further study into the possible roles of p53-regulated miRNAs may identify still other roles for these non-coding RNAs.
Possible role for miRNAs in promoting epigenetic modifications that favor reprogramming
During generation of iPS cells a significant portion of the cells are trapped in partially reprogrammed states characterized by incomplete epigenetic remodeling involving persistent DNA hypermethylation at the promoters of pluripotency-associated genes (Mikkelsen, et al. 2008). One way to improve reprogramming efficiency could be to coax partially reprogrammed cells to undergo complete reprogramming. In support of this argument, inhibition or knock-down of DNA methyltransferase enhanced iPS cell generation by inducing promoter demethylation of pluripotency-associated genes (Mikkelsen, et al. 2008). Additionally, Dnmt inhibitors are also used to generate iPS cells with only two factors (Oct4 and Klf4) or three factors (Sox2, Oct4, and Klf4) (Huangfu, et al. 2008; Shi, et al. 2008). Recently, miR-29b has been shown to induce global DNA hypomethylation and re-expression of p15INK4b, a tumor suppressor gene, in human acute myeloid leukemia (AML) cells by targeting Dnmt 1, 3a, and 3b (Garzon, et al. 2009). Therefore, co-expression of miR-29b and reprogramming factors is expected to induce complete reprogramming of partially reprogrammed cells by promoting demethylation of promoter regions of pluripotency-associated genes, such as Oct4 and Nanog. It will be interesting to compare the effects of miR-29b and Dnmt knock-down (or Dnmt inhibitors) on reprogramming.
Recently, two independent studies have demonstrated that expression of the imprinted Dlk1-Dio3 gene cluster, which codes for ~50 conserved miRNAs, is often silenced in iPS cells (Liu, et al. 2010; Stadtfeld, et al. 2010). Moreover, treatment of iPS cells with HDAC inhibitors led to activation of the Dlk1-Dio3 miRNA cluster and the generation of iPS cells whose developmental potential appears to be equivalent to that of ES cells (Stadtfeld, et al. 2010). These findings highlight the importance of achieving the appropriate epigenetic status of cells during reprogramming, and further reinforce the critical roles played by miRNAs in the establishment and maintenance of pluripotency. Investigating the effects of exogenous expression of individual miRNAs encoded by the Dlk1-Dio3 cluster during reprogramming will provide further insight into the specific roles of this miRNA cluster.
miRNAs as modulators of pluripotency-promoting signaling pathways during reprogramming
Signaling pathways involved in regulation of a multitude of cellular functions play essential roles in relaying external cues to cells. Among the different pluripotency-sustaining signaling pathways, significant progress has been made towards understanding the roles of TGF-β/Activin signaling in human ES cells (Xu, et al. 2008; Vallier, et al. 2009). Inhibition of TGF-β/Activin signaling using a chemical inhibitor induces human ES cells to differentiate (James, et al. 2005). TGF-β signaling is activated when Activin binds to the ALK4 receptor leading to phosphorylation of SMAD 2/3. Phosphorylated SMAD 2/3 binds to SMAD4 and the resulting complex associates with the promoters of its target genes to activate their expression (Xu, et al. 2008). Recent work has shown that SMAD 2/3 complex binds to SMAD binding elements (SBE) in the Nanog promoter and activates its expression in human ES cells (Xu, et al. 2008; Vallier, et al. 2009).
miRNAs that inhibit TGF-β signaling have been identified in different cell types. In mouse liver stem cells, miR-23b and miR-24-1 inhibit TGF-β signaling by downregulating SMAD3 (Rogler, et al. 2009). In human haematopoietic progenitor cells, miR-24-1 inhibits TGF-β/Activin signaling by targeting the ALK4 receptor (Wang, et al. 2008a). miR-21 induces adipogenic differentiation of human adipose tissue derived mesenchymal stem cells by downregulating the expression of the type II TGF-β receptor (Kim, et al. 2009). Additionally, miRNA profiling experiments have shown that miR-24-1, miR-23b, and miR-21 are expressed at high levels in human IMR90 fibroblasts, whereas their expression is significantly lower in human ES cells and iPS cells generated from IMR90 fibroblasts (Wilson, et al. 2009). This suggests that inhibiting the activity of miR-24-1, miR-23b, and miR-21 may promote reprogramming of human somatic cells into human iPS cells by activating TGF-β/Activin signaling. Apart from its role in regulating TGF-β signaling, miR-24-1 also inhibits cell proliferation by targeting important cell cycle regulators, such as c-Myc, and E2F2 (Lal, et al. 2009). By inhibiting the activity of miR-24-1, it may be possible to generate iPS cells without c-Myc and Klf4. In this regard, c-Myc and Klf4 appear to promote reprogramming, at least in part, by increasing cellular proliferation (Yamanaka. 2007). Apart from the miRNAs discussed above, it will be important to identify and study miRNAs that modulate other signaling pathways, such as Wnt and Mek/Erk, in the process of reprogramming. Again, it is important to recognize that different signaling pathways control the self-renewal and pluripotency of human and mouse ES cells (Yu, Thomson. 2008).
c-Myc induced miRNAs and miRNAs involved in regulation of ES cell self-renewal and cell cycle progression as promoters of reprogramming
As mentioned earlier, c-Myc-regulated miRNAs, miR-141, miR-200, and miR-429, have been shown to attenuate differentiation of mouse ES cells upon LIF withdrawal (Lin, et al. 2009). Additionally, the pro-tumorigenic miR-17-92 cluster, which is transcriptionally activated by c-Myc in tumor cell models, is enriched in both human ES cells and human iPS cells (O'Donnell, et al. 2005; Wilson, et al. 2009). Hence, it would be interesting to determine whether these c-Myc-regulated miRNAs exert positive effects on somatic cell reprogramming. Similarly, miR-92b and miRNAs belonging to the miR-520 cluster should be tested for their effects on reprogramming given their established or predicted roles in promoting ES cell proliferation (Ren, et al. 2009; Sengupta, et al. 2009). In addition, inhibitors of miR-134, miR-296 and miR-470 miRNAs appear to be good candidates for influencing reprogramming, given that these miRNAs have been shown to interfere with the self-renewal of ES cells (Tay, et al. 2008a).
Finally, various miRNA mimics and miRNA inhibitors, when used in the optimal combination with one another, are expected to improve both the efficiency of producing iPS cells and the quality of the iPS cells produced. Interestingly, it has been reported that reprogramming of tumor cells can be achieved using only miRNAs, specifically by exogenous expression of the miR-302 cluster (Lin, et al. 2008). Furthermore, it was claimed that the miR-302 cluster can also reprogram primary cultured somatic cells, but no data was presented (Lin, et al. 2008). Thus, the potential clinical use of human iPS cells generated solely with miRNAs remains to be determined. Furthermore, the effectiveness of miRNA-based reprogramming strategies may be cell-type dependent. From an experimental standpoint, it would be interesting to test the ability of the miRNAs, including the miR-302 cluster, to reprogram neural stem cells, which only require Oct4 for reprogramming (Kim, et al. 2009a; Kim, et al. 2009b).
Conclusion
Research over the past decade has contributed substantially towards understanding the molecular mechanisms that control self-renewal and pluripotency of ES cells. The identification of miRNAs and their varied effects on ES cells has provided a far better understanding of the molecular mechanisms that fine tune the complex gene regulatory networks which control the proliferation and the differentiation of pluripotent stem cells. Specific miRNAs, both ES cell- and tissue-specific, have been shown to regulate the expression of critical transcription factors, cell cycle proteins, epigenetic modifiers, and other regulatory proteins, to confer either ES cell or differentiated cell phenotypes. Notwithstanding the immense progress made recently towards understanding the contribution of miRNAs in maintaining the pluripotent state, much remains to be done. Recent work by several groups have demonstrated that iPS cells and ES cells can be distinguished by gene expression signatures, including expression of miRNAs (Chin, et al. 2009; Liu, et al. 2010; Stadtfeld, et al. 2010). These findings suggest that iPS cells are very similar to ES cells, but there are important differences between them. Finally, as discussed in this review, our understanding of the roles of miRNAs in somatic cell reprogramming is relatively limited. Therefore, future studies that modulate the expression of specific miRNAs during the generation of iPS cells are expected to both improve reprogramming itself and provide greater insights into the mechanistic details surrounding the generation of iPS cells. Equally important, extensive characterization of the miRNA status of human iPS cells is likely to have significant impact on the potential clinical use of these cells in cell-based therapies.
Acknowledgments
Timothy McKeithan, Jesse Cox and Heather Rizzino are thanked for reading this manuscript. Stem cell research in this laboratory is supported by a grant from the National Institutes of Health (GM 080751) and the Nebraska Department of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: Authors declare no potential conflict of interest.
References
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, Vallier L, Gil J. Senescence Impairs Successful Reprogramming to Pluripotent Stem Cells. Genes Dev. 2009;18:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, Ruzzo WL, Ware C, Radich JP, Gentleman R, Ruohola-Baker H, Tewari M. MicroRNA Discovery and Profiling in Human Embryonic Stem Cells by Deep Sequencing of Small RNA Libraries. Stem Cells. 2008;10:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-delJesus A, Romero-Lopez C, Lucena-Aguilar G, Melen GJ, Sanchez L, Ligero G, Berzal-Herranz A, Menendez P. Embryonic Stem Cell-Specific miR302-367 Cluster: Human Gene Structure and Functional Characterization of its Core Promoter. Mol. Cell. Biol. 2008;21:6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;2:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA. A Mammalian microRNA Cluster Controls DNA Methylation and Telomere Recombination Via Rbl2-Dependent Regulation of DNA Methyltransferases. Nat. Struct. Mol. Biol. 2008;3:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of Pluripotent Epiblast Stem Cells from Mammalian Embryos. Nature. 2007;7150:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-Regulated miR-302 Targets Cyclin D1 in Human Embryonic Stem Cells. Mol. Cell. Biol. 2008;20:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 Controls ES Cell Self-Renewal and Pluripotency by a Myc-Dependent Mechanism. Development. 2005;5:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, Thomas-Tikhonenko A, Mendell JT. Lin-28B Transactivation is Necessary for Myc-Mediated Let-7 Repression and Proliferation. Proc. Natl. Acad. Sci. U. S. A. 2009;9:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA Repression by Myc Contributes to Tumorigenesis. Nat. Genet. 2008;1:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell. 2008;6:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP Recruits the Dicer Complex to Ago2 for microRNA Processing and Gene Silencing. Nature. 2005;7051:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Induced Pluripotent Stem Cells and Embryonic Stem Cells are Distinguished by Gene Expression Signatures. Cell Stem Cell. 2009;1:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes KR, Srivastava D. MicroRNA Regulation of Cardiovascular Development. Circ. Res. 2009;6:724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JL, Rizzino A. Induced Pluripotent Stem Cells: What Lies Beyond the Paradigm Shift. Exp. Biol. Med. (Maywood) 2010;2:148–158. doi: 10.1258/ebm.2009.009267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr H, Benvenisty N. Genetic Analysis of the Role of the Reprogramming Gene LIN-28 in Human Embryonic Stem Cells. Stem Cells. 2009;27:352–362. doi: 10.1634/stemcells.2008-0720. [DOI] [PubMed] [Google Scholar]
- Davis S, Lollo B, Freier S, Esau C. Improved Targeting of miRNA with Antisense Oligonucleotides. Nucleic Acids Res. 2006;8:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of Primary microRNAs by the Microprocessor Complex. Nature. 2004;7014:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA Sponges: Competitive Inhibitors of Small RNAs in Mammalian Cells. Nat. Methods. 2007;9:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM, Jones PA. MicroRNAs: Critical Mediators of Differentiation, Development and Disease. Swiss Med. Wkly. 2009;33–34:466–472. doi: 10.4414/smw.2009.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: Key Regulators of Stem Cells. Nat. Rev. Mol. Cell Biol. 2009;2:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b Induces Global DNA Hypomethylation and Tumor Suppressor Gene Reexpression in Acute Myeloid Leukemia by Targeting Directly DNMT3A and 3B and Indirectly DNMT1. Blood. 2009;25:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L. Stable Knockdown of microRNA in Vivo by Lentiviral Vectors. Nat. Methods. 2009;1:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor Complex Mediates the Genesis of microRNAs. Nature. 2004;7014:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a Regulator of Cellular PKR and HIV-1 Virus Expression, Interacts with Dicer and Functions in RNA Silencing. EMBO Rep. 2005;10:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 Complex in Primary microRNA Processing. Genes Dev. 2004;24:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA Polycistron as a Potential Human Oncogene. Nature. 2005;7043:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in Concert with Lin28 Suppresses microRNA Biogenesis through Pre-microRNA Uridylation. Cell. 2009;4:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 Mediates the Terminal Uridylation of Let-7 Precursor MicroRNA. Mol. Cell. 2008;2:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of Induced Pluripotent Stem Cell Generation by the p53-p21 Pathway. Nature. 2009;7259:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic Stem Cell-Specific MicroRNAs. Dev. Cell. 2003;2:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of Pluripotent Stem Cells by Defined Factors is Greatly Improved by Small-Molecule Compounds. Nat. Biotechnol. 2008;7:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal Signaling is Necessary for the Maintenance of Pluripotency in Human Embryonic Stem Cells. Development. 2005;6:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The Let-7 microRNA Represses Cell Proliferation Pathways in Human Cells. Cancer Res. 2007;16:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic Stem Cell-Specific microRNAs Promote Induced Pluripotency. Nat. Biotechnol. 2009;5:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-Deficient Mouse Embryonic Stem Cells are Defective in Differentiation and Centromeric Silencing. Genes Dev. 2005;4:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Slack FJ. MicroRNAs: Small Molecules with Big Roles - C. Elegans to Human Cancer. Biol. Cell. 2008;2:71–81. doi: 10.1042/BC20070078. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 Tumour Suppressor Pathway to Somatic Cell Reprogramming. Nature. 2009;7259:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR. Direct Reprogramming of Human Neural Stem Cells by OCT4. Nature. 2009a;7264 doi: 10.1038/nature08436. 649-643. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Scholer HR. Oct4-Induced Pluripotency in Adult Neural Stem Cells. Cell. 2009b;3:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kim KS, Kim JS, Lee MR, Jeong HS, Kim J. A Study of microRNAs in Silico and in Vivo: Emerging Regulators of Embryonic Stem Cells. FEBS J. 2009c;8:2140–2149. doi: 10.1111/j.1742-4658.2009.06932.x. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA Biogenesis: Coordinated Cropping and Dicing. Nat. Rev. Mol. Cell Biol. 2005;5:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Hwang SJ, Bae YC, Jung JS. MiR-21 Regulates Adipogenic Differentiation through the Modulation of TGF-Beta Signaling in Mesenchymal Stem Cells Derived from Human Adipose Tissue. Stem Cells. 2009;12:3093–3102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA Processing Enhances Cellular Transformation and Tumorigenesis. Nat. Genet. 2007;5:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of Novel Genes Coding for Small Expressed RNAs. Science. 2001;5543:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U, Hart RP. Concise Review: MicroRNA Expression in Multipotent Mesenchymal Stromal Cells. Stem Cells. 2008;2:356–363. doi: 10.1634/stemcells.2007-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. MiR-24 Inhibits Cell Proliferation by Targeting E2F2, MYC, and Other Cell-Cycle Genes Via Binding to "Seedless" 3'UTR microRNA Recognition Elements. Mol. Cell. 2009;5:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The Human DiGeorge Syndrome Critical Region Gene 8 and its D. Melanogaster Homolog are Required for miRNA Biogenesis. Curr. Biol. 2004;23:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis Elegans. Science. 2001;5543:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, Fan JB, Loring JF. Comprehensive microRNA Profiling Reveals a Unique Human Embryonic Stem Cell Signature Dominated by a Single Seed Sequence. Stem Cells. 2008;6:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An Extensive Class of Small RNAs in Caenorhabditis Elegans. Science. 2001;5543:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell. 1993;5:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The Nuclear RNase III Drosha Initiates microRNA Processing. Nature. 2003;6956:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA Maturation: Stepwise Processing and Subcellular Localization. EMBO J. 2002;17:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, often Flanked by Adenosines, Indicates that Thousands of Human Genes are microRNA Targets. Cell. 2005;1:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf Locus is a Barrier for iPS Cell Reprogramming. Nature. 2009;7259:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Jin P. Roles of Small Regulatory RNAs in Determining Neuronal Identity. Nat. Rev. Neurosci. 2010;5:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-Regulated microRNAs Attenuate Embryonic Stem Cell Differentiation. EMBO J. 2009;20:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY. Mir-302 Reprograms Human Skin Cancer Cells into a Pluripotent ES-Cell-Like State. RNA. 2008;10:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Luo GZ, Yang W, Zhao X, Zheng Q, Lv Z, Li W, Wu HJ, Wang L, Wang XJ, Zhou Q. Activation of the Imprinted Dlk1-Dio3 Region Correlates with Pluripotency Levels of Mouse Stem Cells. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sall A, Yang D. MicroRNA: An Emerging Therapeutic Target and Intervention Tool. Int. J. Mol. Sci. 2008;6:978–999. doi: 10.3390/ijms9060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear Export of microRNA Precursors. Science. 2004;5654:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are Repressed as Efficiently by microRNA-Binding Sites in the 5' UTR as in the 3' UTR. Proc. Natl. Acad. Sci. U. S. A. 2007;23:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Guidelines and Techniques for the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell. 2008;6:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A Human, ATP-Independent, RISC Assembly Machine Fueled by Pre-miRNA. Genes Dev. 2005;24:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-Mediated DNA Damage Response Limits Reprogramming to Ensure iPS Cell Genomic Integrity. Nature. 2009;7259:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA Genes to the Core Transcriptional Regulatory Circuitry of Embryonic Stem Cells. Cell. 2008;3:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-Specific Inhibition of microRNA- and siRNA-Induced RNA Silencing. RNA. 2004;3:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA Families Regulate Self-Renewal in Mouse Embryonic Stem Cells. Nature. 2010;7281:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT. MiRiad Roles for the miR-17-92 Cluster in Development and Disease. Cell. 2008;2:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting Direct Reprogramming through Integrative Genomic Analysis. Nature. 2008;7200:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-Deficient Murine Embryonic Stem Cells. Proc. Natl. Acad. Sci. U. S. A. 2005;34:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, Miyazaki J, Matoba R, Ko MS, Niwa H. Klf4 Cooperates with Oct3/4 and Sox2 to Activate the Lefty1 Core Promoter in Embryonic Stem Cells. Mol. Cell. Biol. 2006;20:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 Interaction with the Let-7 Precursor Loop Mediates Regulated microRNA Processing. RNA. 2008;8:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. C-Myc-Regulated microRNAs Modulate E2F1 Expression. Nature. 2005;7043:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Orom UA, Kauppinen S, Lund AH. LNA-Modified Oligonucleotides Mediate Specific Inhibition of microRNA Function. Gene. 2006:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the Sequence and Temporal Expression of Let-7 Heterochronic Regulatory RNA. Nature. 2000;6808:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-Mediated Post-Transcriptional Regulation of Oct4 Expression in Human Embryonic Stem Cells. Nucleic Acids Res. 2010;4:1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-Nucleotide Let-7 RNA Regulates Developmental Timing in Caenorhabditis Elegans. Nature. 2000;6772:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and Gene Expression Patterns in the Differentiation of Human Embryonic Stem Cells. J. Transl. Med. 2009:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of Mammalian microRNA Host Genes and Transcription Units. Genome Res. 2004;10A:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, Rogler LE. MicroRNA-23b Cluster microRNAs Regulate Transforming Growth Factor-beta/bone Morphogenetic Protein Signaling and Liver Stem Cell Differentiation by Targeting Smads. Hepatology. 2009;2:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The Let-7 Target Gene Mouse Lin-41 is a Stem Cell Specific E3 Ubiquitin Ligase for the miRNA Pathway Protein Ago2. Nat. Cell Biol. 2009;12:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A Feedback Loop Comprising Lin-28 and Let-7 Controls Pre-Let-7 Maturation during Neural Stem-Cell Commitment. Nat. Cell Biol. 2008;8:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. P53 Represses c-Myc through Induction of the Tumor Suppressor miR-145. Proc. Natl. Acad. Sci. U. S. A. 2009;9:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Nie J, Wagner RJ, Yang C, Stewart R, Thomson JA. MicroRNA 92b Controls the G1/S Checkpoint Gene p57 in Human Embryonic Stem Cells. Stem Cells. 2009;7:1524–1528. doi: 10.1002/stem.84. [DOI] [PubMed] [Google Scholar]
- Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of Pluripotent Stem Cells from Mouse Embryonic Fibroblasts by Oct4 and Klf4 with Small-Molecule Compounds. Cell Stem Cell. 2008;5:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs Control De Novo DNA Methylation through Regulation of Transcriptional Repressors in Mouse Embryonic Stem Cells. Nat. Struct. Mol. Biol. 2008;3:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant Silencing of Imprinted Genes on Chromosome 12qF1 in Mouse Induced Pluripotent Stem Cells. Nature. 2010 doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced Pluripotent Stem Cells Generated without Viral Integration. Science. 2008;5903:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small Non-Coding RNAs in Animal Development. Nat. Rev. Mol. Cell Biol. 2008;3:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA Processing by p53. Nature. 2009;7254:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;4:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 Coding Regions Modulate Embryonic Stem Cell Differentiation. Nature. 2008a;7216:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, Wang W, Liu R, George J, Ng HH, Perera RJ, Lufkin T, Rigoutsos I, Thomson AM, Lim B. MicroRNA-134 Modulates the Differentiation of Mouse Embryonic Stem Cells, Where it Causes Post-Transcriptional Attenuation of Nanog and LRH1. Stem Cells. 2008b;1:17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New Cell Lines from Mouse Epiblast Share Defining Features with Human Embryonic Stem Cells. Nature. 2007;7150:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, Brons G, Pedersen RA. Activin/Nodal Signalling Maintains Pluripotency by Controlling Nanog Expression. Development. 2009;8:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective Blockade of microRNA Processing by Lin28. Science. 2008;5872:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, Broberger C, Porreca F, Lai J, Ren K, Ossipov M, Koshkin A, Jakobsen N, Skouv J, Oerum H, Jacobsen MH, Wengel J. Potent and Nontoxic Antisense Oligonucleotides Containing Locked Nucleic Acids. Proc. Natl. Acad. Sci. U. S. A. 2000;10:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Huang Z, Xue H, Jin C, Ju XL, Han JD, Chen YG. MicroRNA miR-24 Inhibits Erythropoiesis by Targeting Activin Type I Receptor ALK4. Blood. 2008a;2:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic Stem Cell-Specific microRNAs Regulate the G1-S Transition and Promote Rapid Proliferation. Nat. Genet. 2008b;12:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is Essential for microRNA Biogenesis and Silencing of Embryonic Stem Cell Self-Renewal. Nat. Genet. 2007;3:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA Profiling of Human-Induced Pluripotent Stem Cells. Stem Cells Dev. 2009;5:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Micro-RNA Regulation of the Mammalian Lin-28 Gene during Neuronal Differentiation of Embryonal Carcinoma Cells. Mol. Cell. Biol. 2005;21:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Zhang K, Huang Y. Lin28 Modulates Cell Growth and Associates with a Subset of Cell Cycle Regulator mRNAs in Mouse Embryonic Stem Cells. RNA. 2009a;3:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 Regulates OCT4, SOX2, and KLF4 and Represses Pluripotency in Human Embryonic Stem Cells. Cell. 2009b;4:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, Thomson JA. NANOG is a Direct Target of TGFbeta/activin-Mediated SMAD Signaling in Human ESCs. Cell Stem Cell. 2008;2:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and New Developments in the Generation of Patient-Specific Pluripotent Stem Cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 Mediates the Nuclear Export of Pre-microRNAs and Short Hairpin RNAs. Genes Dev. 2003;24:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science. 2009;5928:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Thomson JA. Pluripotent Stem Cell Lines. Genes Dev. 2008;15:1987–1997. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;5858:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Sequence Requirements for Micro RNA Processing and Function in Human Cells. RNA. 2003;1:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Srivastava D. A Developmental View of microRNA Function. Trends Biochem. Sci. 2007;4:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]