Abstract
Translation of the genomes of several positive-sense RNA viruses follows end-independent initiation on an internal ribosomal entry site (IRES) in the viral mRNA. There are four major IRES groups, and despite major differences in the mechanisms that they use, one unifying characteristic is that each mechanism involves essential non-canonical interactions of the IRES with components of the canonical translational apparatus. Thus the ~200nt.-long Type 4 IRESs (epitomized by Cricket paralysis virus) bind directly to the intersubunit space on the ribosomal 40S subunit, followed by joining to a 60S subunit to form active ribosomes by a factor-independent mechanism. The ~300nt.-long type 3 IRESs (epitomized by Hepatitis C virus) binds independently to eukaryotic initiation factor (eIF) 3, and to the solvent-accessible surface and E-site of the 40S subunit: addition of eIF2-GTP/initiator tRNA is sufficient to form a 48S complex that can join a 60S subunit in an eIF5/eIF5B-mediated reaction to form an active ribosome. Recent cryo-electron microscopy and biochemical analyses have revealed a second general characteristic of the mechanisms of initiation on Type 3 and Type 4 IRESs. Both classes of IRES induce similar conformational changes in the ribosome that influence entry, positioning and fixation of mRNA in the ribosomal decoding channel. HCV-like IRESs also stabilize binding of initiator tRNA in the peptidyl (P) site of the 40S subunit, whereas Type 4 IRESs induce changes in the ribosome that likely promote subsequent steps in the translation process, including subunit joining and elongation.
Keywords: Cricket paralysis virus, Classical Swine fever virus, Hepatitis C virus, IRES, ribosome, translation initiation
Introduction
Initiation of translation of a number of eukaryotic viral mRNAs occurs by non-canonical end-independent mechanisms that are mediated by an internal ribosomal entry site (IRES) in the mRNA [1–3]. IRESs are functionally defined RNA elements that are classified into four major structural groups, epitomized by poliovirus (PV; Type 1), encephalomyocarditis virus (EMCV; Type 2), hepatitis C virus (HCV; Type 3) and cricket paralysis virus (CrPV; Type 4). (This nomenclature extends the generally accepted designation for Type 1 and Type 2 IRESs [4]). Although evidence to date suggests that these groups of IRESs mediate initiation by distinct mechanisms, a characteristic of all of these mechanisms is that they involve direct and essential interactions with components of the translational apparatus (i.e. eukaryotic initiation factors (eIFs) and the ribosomal 40S subunit). Recent structural and biochemical studies on Type 3 and Type 4 IRESs have revealed a second important characteristic of IRES-mediated initiation, which is that these IRESs induce conformational changes in the ribosome that include influencing the entry, positioning and fixation of mRNA in the ribosomal decoding channel, stabilizing binding of initiator tRNA in the peptidyl (P) site of the 40S subunit, and priming the resulting initiation complex for subsequent steps in the translation process, including subunit joining and ribosomal translocation during the first elongation cycle.
The canonical mechanism of translation initiation in eukaryotes
Initiation of translation on the majority of mRNAs can be described by the scanning model, and occurs in two stages: 48S complex formation, followed by subunit joining to form active 80S ribosomes [5]. Initiation requires a free pool of ribosomal 40S and 60S subunits, which are generated by recycling of post-termination ribosomes by eIF3, eIF3j (a loosely associated subunit of eIF3), eIF1 and eIF1A [6]. The first step in initiation is assembly of an initiator tRNA (Met-tRNAMeti)/eIF2•GTP ternary complex that, with eIF3, eIF1, eIF1A and a 40S subunit forms a 43S preinitiation complex. Attachment of the 43S complex to the cap-proximal region of mRNA is promoted by eIF4F, a heterotrimer that comprises eIF4E (which binds the mRNA m7GTP ‘cap’), eIF4A (a DEAD-box RNA helicase) and eIF4G (which binds eIF4E, eIF4A, and eIF3). The mechanism of attachment involves coordination of unwinding of the cap-proximal region of mRNA by eIF4F, eIF4A and its cofactor eIF4B to prepare an attachment site, and establishment of interactions between eIF3, eIF4G, mRNA and the 40S subunit. 43S complexes need eIF1 and eIF1A for the subsequent scanning step [7]; scanning on all 5’UTRs is enhanced by the DExH-box protein DHX29, and scanning on highly structured 5’UTRs requires this factor [8]. The 43S complex stops when it recognizes the initiation codon (which is usually the first AUG triplet with a favorable nucleotide context) forming a 48S complex in which the initiation codon is base-paired with the anticodon of Met-tRNAMeti. Initiation codons that deviate from the optimum context AccAUGG, particularly at the −3 and +4 positions (bold) may be bypassed by scanning ribosomal complexes, leading to initiation downstream [9].
Hydrolysis of eIF2-bound GTP/Pi release are key steps in commitment of a scanning ribosomal complex to arrest at an initiation codon, and eIF1 plays an important role in this process, by inhibiting premature hydrolysis of eIF2-bound GTP/Pi release during scanning [10]. Repression is relieved by displacement of eIF1 following establishment of codon-anticodon base-pairing, which is therefore linked by eIF1 to this commitment step [10–12]. eIF1 also contributes to the fidelity of initiation by dissociating 48S complexes assembled on non-AUG codons and AUG codons with poor context [7, 14].
The transition from the scanning conformation of 43S complexes to the arrested conformation of 48S complexes is associated with conformational changes in the 40S subunit, with changes in the relative positions of Met-tRNAMeti and mRNA (see below) and with changes in the interactions and relative positions of initiation factors. eIF1A occupies the ribosomal A-site, but its N- and C-terminal tails (NTT and CTT, respectively) extend into the P-site in 43S complexes [15], near to eIF1, which binds to the interface surface, close to Met-tRNAMeti and the P-site [16]. The eIF1A-NTT is thought to promote the ‘closed’, arrested conformation of the 40S subunit, whereas the CTT promotes the ‘open’ scanning-competent conformation [15, 17]. Start codon recognition leads to dissociation of eIF1 from the 40S subunit [10–12] and ejection of the eIF1A-CTT from the P-site [15, 17], whereas the folded domain of eIF1A binds more tightly in the aminoacyl (A)-site [18]. These changing interactions have been hypothesized to change the position and interactions of eIF5, enabling it to promote hydrolysis of eIF2-bound GTP, and/or triggering release of Pi from the ribosome-associated eIF2/GDP/Pi complex [19, 20]. Cross-linking studies have shown that in 48S complexes, nucleotides at the −3 and +4 positions establish specific interactions with eIF2α and AA(1818–1819) of 18S rRNA, respectively, and biochemical data indicate that the eIF2α/−3nt. interaction is an important determinant of initiation codon selection [21]. In addition to promoting scanning and influencing initiation codon selection, eIF1A also interacts with eIF5B, enhancing its activity in subunit joining [22, 23]. eIF5B•GTP displaces the remaining eIFs from the 40S subunit and promotes joining of a 60S subunit to form an 80S ribosome [10, 24]. The eIF5B C-terminal domain may replace eIF2 in interacting with the acceptor end of Met-tRNAMeti, leading to displacement of eIF2•GDP and likely reorienting Met-tRNAMeti so that it can enter the P-site of the 60S subunit correctly and that subunit joining can proceed. Finally, hydrolysis of the bound GTP leads to release of eIF5B•GDP, yielding a ribosome that can enter the elongation phase of translation [24].
Comparisons between initiation of translation in eukaryotes and prokaryotes
Translation initiation mechanisms in eukaryotes and in prokaryotes are fundamentally similar: both begin with separated ribosomal subunits, involve binding of a unique initiator tRNA in the P-site of the small ribosomal subunit, and proceed via formation of an intermediate complex containing mRNA and this tRNA before the stage of subunit joining. The core of the ribosome is highly conserved, and major differences are restricted to rRNA expansion segments and additional proteins at peripheral locations [25]: accordingly, insights gained from studies of prokaryotic ribosomes can (cautiously) be extrapolated to eukaryotic translation. The small ribosomal subunit consists of the body, which contains major morphological features such as the shoulder and the platform, and is connected via the narrow neck to the head (Figure 1). tRNA is held tightly in the P-site by multiple interactions with the prokaryotic small (30S) subunit so that its anticodon can base-pair accurately with each P-site codon and maintain the reading frame [26, 27]. Prokaryotic mRNA binds in a channel that wraps around the neck of the 30S subunit, passing through two non-covalently closed tunnels, one as it enters between the head and shoulder and the other as it exits between the head and platform [26, 28, 29]. mRNA is anchored onto the platform by base-pairing between the mRNA Shine-Dalgarno (SD) sequence upstream of the initiation codon and the anti-SD sequence at the 3’-end of 16S ribosomal rRNA (rRNA). This sequence is absent from eukaryotic ribosomes, but the path of mRNA on the eukaryotic 40S subunit is in most respects analogous to that in prokaryotes [30].
Figure 1.
Structural features of the 40S ribosomal subunit and conformational changes induced in it by eIF1 and eIF1A. CryoEM reconstructions of (A) apo 40S yeast ribosomal subunits and (B) 40S-eIF1-eIF1A complexes viewed from the solvent side (top) and the showing the intersubunit surface (bottom). The apo 40S model is labeled to show landmarks on the small ribosomal subunit, including the beak (bk), neck (n), shoulder (sh), left foot (lf), right foot (rf) and platform (pf), as well as the latch, and the mRNA entry and exit channels. The 40S-eIF1-eIF1A complex (B, bottom panel) is labeled to show the approximate position of the A-, P- and E-sites in the mRNA-binding channel, and the position of the rRNA helices (h16, h18 and h34) and ribosomal proteins (rpS3, rpS5, rpS14) that are involved in forming the mRNA entry channel (h18-h34), the mRNA exit channel (rpS5-rpS14) and the new eIF1/1A-induced head-shoulder connection (rpS3-h16) (which is also indicated by an asterisk). Adapted from Passmore et al. (2007) Molecular Cell 26, 41–50 (© Elsevier Press) and reprinted with permission. Figures generously provided by L. Passmore.
However, the initiation process in prokaryotes is considerably simpler than in eukaryotes: mRNA and initiator tRNA can bind directly to the 30S subunit in the complete absence of the three single-subunit initiation factors IF1, IF2 and IF3. Their functions are to accelerate the rate and to control the fidelity of initiation. IF2, the bacterial homolog of eIF5B, accelerates binding of initiator tRNA to the 30S subunit, positions its acceptor end for insertion into the 50S subunit during subunit joining and accelerates joining [31, 32]. IF1, which is homologous to the central domain of eIF1A, binds in the ribosomal A-site [33] whereas the C-terminal domain of IF3 (a functional analog of eIF1) binds to the interface surface of the platform near the P-site [34]. IF1 and IF3 promote the accuracy of initiation, accelerating dissociation of mispaired tRNA-mRNA complexes from the subunit, likely by a mechanism that includes factor-induced conformational changes in the 30S subunit [33, 35]. eIF1 binds to a similar location as IF3 and also promotes the accuracy of initiation [14,16]. The observation that eIF1 and IF3 can play these roles in heterologous systems is indicative of a conserved mode of action that may involve similar induced conformational changes in the small ribosomal subunit [36].
Conformational changes in ribosomal complexes during the canonical initiation process
The small ribosomal subunit is a metastable structure that undergoes large and small-scale conformational changes in response to the binding or release of ligands during successive steps in the translation process. Conformational changes in bacterial 30S subunits induced by mRNA, tRNA and factors have been visualized by X-ray crystallography and cryo-electron microscopy (cryoEM) [31, 37]. The neck connecting the head to the body consists of a single helix (h) 28 of 16S rRNA, and it permits hinge-like and rotational movement of the head relative to the body: binding of mRNA and the anticodon stem-loop (ASL) of tRNA in the P-site lead to tilting of the head to close the P-site cleft, and to deeper binding of the ASL in the P-site [27], binding of IF1 induces tilting of the head towards the A-site and changes in the conformation of h44 [33], and binding of cognate tRNA in the A-site alters the conformation of the universally conserved rRNA bases A1492/A1493 and G530 (which line the floor of the A-site) and induces closure of the small subunit around the A-site by movements that include rotation of the head and shoulder towards each other [38, 39]. IF2 alters the conformation and orientation of initiator tRNA on the 30S subunit, anchoring the tRNA acceptor end until subunit joining occurs [31].
The head of the eukaryotic 40S subunit is similarly mobile [40, 41]: several conformational changes can be hypothesized to occur in it during initiation, and experimental data to support them is accumulating. Thus to account for eIF1’s dual roles in promoting scanning and in ensuring the fidelity of initiation codon selection, a model was suggested in which eIF1A and particularly eIF1 are required for preinitiation complexes to adopt an ‘open’ scanning-competent conformation that isomerizes to form a stable ‘closed’ complex following initiation codon recognition [14, 42]. The entry channel of the mRNA-binding cleft is occluded in vacant 40S subunits [43], so that there must be a mechanism that opens it to permit entry of mRNA. Data from biochemical and genetic studies have supported and extended this model [5, 11–19]. Directed hydroxyl radical probing experiments indicated that eIF1 binds to the interface surface of the 40S subunit between the platform and Met-tRNAMeti, and likely influences the conformation of ribosomal complexes [16]. Similar experiments place the IF1-like OB domain of eIF1A in the A-site and are consistent with induced tilting of the head of the 40S subunit towards the shoulder [15], similar to the changes induced by binding of IF1 to the 30S subunit [33]. Consistently, cryoEM analysis showed that conformational changes are induced in yeast 40S subunits by binding of eIF1 and eIF1A that lead to opening of the entry channel ‘‘latch’’ formed between h18 in the body and h34 and ribosomal protein (rp) S3 in the neck and to establishment of a new head/body connection likely mediated by h16 and rpS3 (Figure 1; [44]). These changes are consistent with an open conformation of the preinitiation complex that is able to bind to mRNA (see above). There are indications that DHX29 may induce conformational changes at the entry region of the mRNA-binding channel, partially closing it, which would prevent ribosomal ‘drop-off’ during scanning and thus increase its processivity [8].
Other changes might occur in the 40S subunit as a result of the transition from scanning to stable arrest at the initiation codon. Thus establishment of codon-anticodon base-pairing and of contacts between +4 and −3 context nucleotides and eIF2α/18S rRNA [21] could constrain mRNA by inducing and stabilizing the ‘open’ to ‘closed’ transition of the mRNA-binding channel, and might move the codon-anticodon helix deeper into the P-site binding pocket. This latter hypothetical change would be analogous to the movement to a deeper position of the bacterial P-site codon that is induced by its base-pairing with the anticodon of cognate tRNA [27, 45]. These changes could permit movement of tRNA from an orientation in which it can inspect the 5’UTR to identify the initiation codon during scanning to a rigid position that is compatible with entry of the acceptor end into the 60S subunit to permit subunit joining [46]. Modeling of eIF5B’s position on the ribosome on the basis of directed hydroxyl radical probing data is consistent with it mediating such a change in tRNA’s orientation [47].
Initiation of translation by internal ribosomal entry
Whereas the majority of eukaryotic mRNAs are translated following end-dependent initiation, initiation on a subset of viral mRNAs occurs by end-independent internal ribosomal entry [1–3]. IRESs are functionally defined RNA elements that, when inserted into the intercistronic region of a dicistronic mRNA, mediate initiation of translation of the downstream cistron independently of translation of the upstream cistron. The IRESs of the picornaviruses poliovirus and encephalomyocarditis virus (EMCV) were the first to be identified [48, 49]: both are ~450nt. long and exemplify the major Type 1 and Type 2 classes of IRES, respectively. Biochemical reconstitution experiments [50, 51] revealed that initiation on the EMCV IRES requires the same set of eIFs as the canonical initiation mechanism, except for eIF4E and the N-terminal domain of eIF4G to which eIF4E binds. These experiments also led to the important conceptual advance that initiation mediated by IRESs involves specific non-canonical interactions of IRESs with canonical components of the translation apparatus: eIF4G binds specifically to a domain of the EMCV IRES adjacent to the initiation codon, recruits eIF4A and promotes binding of the 43S complex to the IRES. The same fragment of eIF4G also binds directly and specifically to Type 1 IRESs and recruits eIF4A to them, suggesting that underlying aspects of the mechanism of initiation on these two classes of IRES are similar [52]. Type 3 and Type 4 IRESs, exemplified by Hepatitis C virus (HCV) and Cricket paralysis virus (CrPV), respectively, have sequences and structures that are wholly distinct from one another and from Type 1 and Type 2 IRESs. As described below, initiation on them occurs by different mechanisms, but they both involve direct factor-independent interaction with the 40S subunit.
Initiation of translation on the intergenic region (IGR) IRESs of dicistroviruses
The smallest of the four major classes of viral IRESs occupy the ~190–220nt-long intergenic regions of the genomes of dicistroviruses such as Cricket paralysis virus (CrPV) and Taura shrimp virus (TSV), between the large open reading frames that encode structural and nonstructural proteins [53–69]. These IRESs also use the simplest mechanism of initiation of all IRESs and remarkably do not require initiation factors to assemble elongation-competent 80S ribosomes.
IGR IRESs are divided into two sub-groups, exemplified by CrPV and TSV, respectively, but they all have closely related tertiary structures consisting of three domains (Figure 2), each of which contains a pseudoknot (PK). Domains 1 and 2 form a single rigid structural element, whereas the Domain 3 is attached to this element by a linker and is intrinsically more flexible [56–58](Figure 2C). The TSV-like subgroup differs from CrPV-like IRESs principally by having an additional hairpin in domain 3 (Figure 2B) and an enlarged L1.1 loop in domain 1. There is no evidence for any difference in the mechanism of initiation mediated by these two IRES subgroups. Mutational analyses showed that some helices in the IRES play a structural role (so that loss of activity caused by disruption of base-pairing is restored by compensatory mutations), whereas the loss of function caused by mutation of conserved unpaired elements suggests that they play specific functional roles at different stages in initiation of translation, several of which have been identified [56–62].
Figure 2.
Secondary structure models of (A) the CrPV IGR IRES and (B) of Domain 3 of the Taura shrimp virus (TSV) IRES annotated to show domains 1, 2 and 3, pseudoknots I, II and III, stem-loops IV and V, and loops 1.1. and 1.2. CrPV IRES nucleotides that cryoEM reconstructions indicate are likely to interact with the ribosome [76] are circled, and likely interacting components of the ribosome are indicated in the secondary structure diagram. (C) Combined crystal structures of the ribosome-binding Domain 3 of the PSIV IGR IRES [58] and the P-site domain of the CrPV IGR IRES [57], with structural elements labeled. The relative orientation of the two structures corresponds to their location when docked into cryo-EM reconstructions of CrPV IGR IRES-80S ribosome complexes [57, 75, 76]. Adapted from Kieft (2009) Virus Research 139, 148–56 (© Elsevier Press) and reprinted with permission. Figure generously provided by J. Kieft.
Biochemical studies have established (a) that IGR IRESs bind directly and productively either to 40S subunits (which can then join with 60S subunits independently of eIF5B) or to 80S ribosomes, (b) that binding leads to occupation of the P-site by the IRES to the exclusion of Met-tRNAMeti, and that IGR IRESs mimic the base-paired initiation codon and anticodon of Met-tRNAMeti in this site, (c) that binding sets the correct reading frame for translation by positioning the first triplet to be decoded (usually a GCU, GCA or GCC (alanine) or CAA (glutamine) triplet) in the A-site (in contrast to the conventional initiation process, which begins with positioning of the (AUG) initiation codon in the P-site), (d) that cognate amino-acyl tRNA is recruited to the A-site by eukaryotic elongation factor 1 (eEF1) and binds weakly before translocation to the P-site by eEF2, and (e) that IGR IRESs promote the initial eEF2-mediated translocation event [59, 63–69]. This translocation step is exceptional because it occurs without prior peptide bond formation, and in the absence of deacylated tRNA in the ribosomal exit (E)-site. Factor-mediated ribosomal translocation on mRNA, at least in prokaryotes, but likely also in eukaryotes, is strongly dependent on the interaction of the acceptor stem of deacylated tRNA with the E-site of the large subunit [70]. Accordingly, it is likely that the IRES mimics deacylated E-site tRNA as well as P-site initiator tRNA. Consequently, and quite exceptionally, initiation occurs without the involvement of initiation factors, initiator tRNA or an initiation codon, and translation begins following recruitment of a cognate aminoacyl-tRNA to the A-site and its translocation to the P-site, rather than following delivery of Met-tRNAMeti to the P-site by eIF2.
Studies in mammalian and yeast cells showed that IGR IRES-mediated translation is up-regulated when levels of active initiation factors are reduced [59, 71, 72] and are thus consistent with in vitro findings that initiation on the CrPV IRES is factor-independent and is impaired by factors that promote recruitment of eIF2•GTP/Met-tRNAMeti ternary complex to the 40S subunit [66]. These experimental conditions in cells, in which competition with the ternary complex for access to the P-site is reduced, may mimic circumstances during viral infection when eIF2 is strongly phosphorylated and host protein synthesis is shut-off (but viral capsid protein synthesis is paradoxically at its peak)[73, 74].
Functional interactions of the IGR IRES with the ribosome during initiation
Chemical and enzymatic footprinting experiments, cryoEM analysis of IRES/ribosome complexes and biochemical analyses of mutant IRESs have related individual structural elements in the IRES to the distinct steps in the initiation process described above and have yielded the outlines of a preliminary model for it (Figure 3). Initial attachment of the IRES to the 40S subunit requires only Domain 2: two conserved stemloops (SL-IV and SL-V) in it that are adjacent to each other in the crystal structure make direct contacts with the 40S subunit that are required for stable binding, and alteration of their relative spatial organization by disruption of PKIII greatly reduced binding affinity to the ribosome [57, 60, 61]. Observations that IGR IRESs are active in all eukaryotes, including plants and yeast, and that they bind productively to purified ribosomes from vertebrates and invertebrates [61, 65, 69] suggest that IGR IRESs interact with conserved ribosomal elements. CryoEM analysis of CrPV IRES/40S subunit complexes (Figure 4D, E; [75, 76]) indicated that SL-IV interacts with rpS5 in the E-site and exit channel of the mRNA-binding cleft of the 40S subunit, and that SL-V interacts with rpS5 and an adjacent protein of unknown identity; genetic studies support the importance of the interaction with rpS5 for recruitment of the IRES to 40S subunits [77]. On the other hand, cross-linking studies suggest that SL-IV from the related Plautia stali intestine virus IGR IRES interacts with rpS25 rather than rpS5 [78]. Binding of the IRES to the 40S subunit induces conformational changes in it that include clockwise rotation of the head relative to the body and establishment of a new connection between rpS3 (in the head) and h16 (in the body) that may be associated with opening of the mRNA-binding channel so that the coding region of the viral RNA can enter it (Figure 4E). Significantly, these changes in the mRNA-binding channel appear to be analogous to those induced by eIF1 and eIF1A (Figure 1; [44]), although the current level of resolution of these cryoEM models is not sufficient to conclude unambiguously that these changes lead to identical ribosomal states.
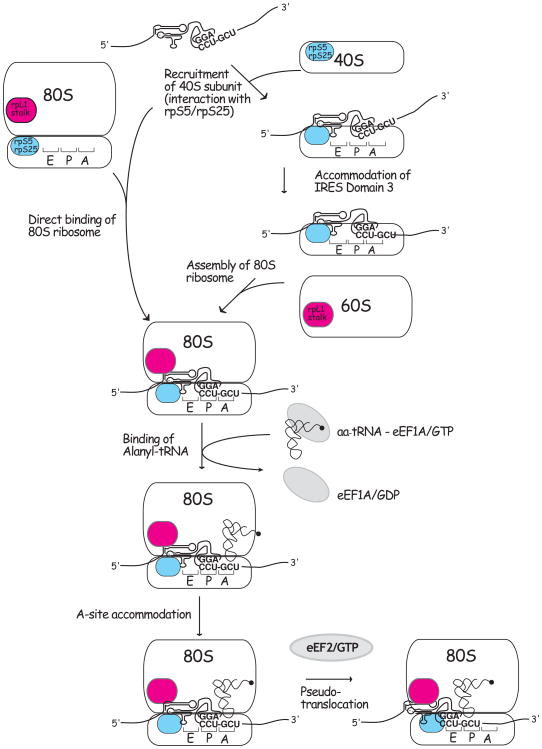
Figure 3.
Model for the mechanism of initiation on the dicistrovirus Type 4 IGR IRES. Assembly of 80S ribosomes can occur either by direct binding of 80S ribosomes to the IRES [66] or by factor-independent binding of the 40S subunit, followed by direct joining of the 60S ribosomal subunit to the 40S/IRES complex [59, 60, 65, 67]. Initial binding of the IRES to the 40S subunit is dependent on interactions of Domain 2 with ribosomal protein (rp)S5 and rp25 (blue oval), whereas Domain 3 does not contribute significantly to affinity to the ribosome [60, 61]. Helix 2 of the Domain 3 pseudoknot takes the place of the conventional base-pairing interaction of the anticodon of Met-tRNAMeti and the P-site AUG initiation codon. Subunit joining is associated with withdrawal of Domain 3 from the A-site, allowing eEF1 to deliver cognate amino-acyl tRNA to it; amino-acyl tRNA then base-pairs with the GCU codon (in CrPV) [65, 67]. Models for binding of aa-tRNA to prokaryotic ribosomes (e.g. [81]) suggest that aa-tRNA likely also binds to the A-site of the eukaryotic ribosome in the pre-accommodated P/T conformation, but could switch to a putative A/P hybrid state, due to the change in orientation of IRES Domain 3 to mimic the P/E hybrid state that occurs during subunit joining [57, 69, 76]. Ribosomal complexes containing the IRES and A-site aminoacyl-tRNA have not yet been observed by cryo-EM. The IRES induces ordering of the P1 stalk region of the 60S subunit [75] (not shown), promoting functional engagement with eEF2, which then mediates ‘pseudo-translocation’ (i.e. in the absence of peptide bond formation and a deacylated E-site tRNA] of the base-paired GCU codon of the IRES and the cognate tRNA anticodon from the A-site to the P-site and the based-paired CCU-GGA triplets of PK1 from the P-site to the E-site [59, 65]. IRES-induced ordering of the rpL1 protuberance (magenta oval) may promote translocation from P- to E-sites. Thereafter, the resulting complex is able to undergo conventional cycles of elongation.
Figure 4.
IRES-induced conformational changes in the ribosomal 40S subunit. Cryo-EM reconstructions of (A–C) HCV IRES/rabbit 40S subunit complexes and (D, E) CrPV IRES/40S complexes, viewed from (A, D) the 60S side, (B) the platform side and (C, E) the solvent side of the 40S subunit. The 40S subunit is shown in yellow, the HCV IRES is purple and the CrPV IRES is pink. (D) Some landmarks of the 40S subunit are indicated, using the same nomenclature as in Figure 1. h16, h18, h30 and h44 indicate the positions of helices 16, 18, 30 and 44, respectively, of 18S rRNA. Some conformational changes in the IRES/40S subunit complexes are indicated with asterisks (C, E). (A–C) From Spahn et al. (2001) Science 291:1959–62 (© American Society for the Advancement of Science) and reprinted with permission. (D, E) From Spahn et al., (2004) Cell 118:4 65–75 (© Elsevier Press) and reprinted with permission. Figures generously provided by J. Frank and C. Spahn.
The next step in initiation, which is dependent on this initial high-affinity interaction, is insertion of Domain 3 into the mRNA-binding cleft, where it overlaps with the sites occupied by P-site and A-site tRNAs [75](Figure 4D), accounting for the competition between the IGR IRES and Met-tRNAMeti for the P-site [66]. This IRES/ribosome interaction is key for the correct placement of the first codon to be decoded into the A-site and of the immediate downstream coding sequence in the mRNA-binding cleft. Mutations that disrupt the structural integrity of Domain 3 abrogate this interaction [59].
In IRES/80S ribosome complexes, IRES Domain 1 interacts with rpL11 of the 60S subunit (via helix PKII) and with rpL1, Helix (H) 76 and H77 (via loop 1.1), structures that normally interact with P- and E-site tRNAs. The suggestion [75] that interaction of the 60S subunit with these elements of the IRES may contribute directly to the process of subunit joining is consistent with the impairment of 80S ribosome assembly caused by mutation of the structurally disordered loop L1.1 [58, 62]. 40S ribosomes bound to an IRES fragment containing only Domain 2 cannot form an 80S ribosome [61], consistent with the hypothesis that subunit joining requires additional contacts of the ribosome with IRES elements outside of Domain 2 and/or the associated conformational changes that they induce.
Several changes occur in both the IRES and the ribosome on subunit joining [75, 76]: Domain 3 withdraws from a position in which it overlaps with the position of A-site tRNA to a position between A- and P-sites so that the A-site becomes accessible for an incoming aminoacyl-tRNA. The IRES-induced rotation of the head of the 40S subunit relative to the body is reversed, but the h18-h34 latch interaction at the entry channel of the mRNA-binding cleft remains open, which would also facilitate binding of aminoacyl-tRNA in the A-site. Third, the IRES induces ordering of the P proteins in the stalk region of the 60S subunit (which is part of the elongation factor binding site on the ribosome [79, 80]) in a manner that appears to enhance the functional interaction of eEF2 with the IRES/80S ribosome complex, thus promoting the first translocation step [69]. CryoEM analyses of IRES/ribosome complexes [76] and modeling of the IGR IRES on the ribosome by fitting crystal structures of Domain 1/Domain 2 and Domain 3 to CryoEM density [57] indicate that Domain 3 occupies a position in 80S ribosomes resembling that of tRNA in the P/E hybrid state, so that after its initial recruitment to the A-site (likely in the A/T conformation; [81]), incoming aminoacyl-tRNA would be able to change its orientation to mimic that of tRNA in the A/P hybrid state: the IRES/80S ribosomal complex consequently resembles the pre-translocation state that is recognized by eEF2, which would thus be able to induce translocation before formation of a peptide bond [69]. The rpL1 stalk in the large ribosomal subunit establishes a connection with deacylated tRNA in the P-site and acts to direct it to the E-site during translocation [82], and the establishment of an interaction of IRES loop 1.1 with rpL1 in the IRES/80S complex may play a similar role.
In summary, the IGR IRES is strongly conserved, and consists of a rigid core and several exposed, frequently mobile elements that interact with distinct elements of the ribosome and manipulate its conformation to promote stages in an exceptional factor-independent translation initiation mechanism, from initial specific binding of the IRES to the 40S subunit to completion of the first elongation cycle.
Initiation of translation on the Hepatitis C virus IRES and on related pestivirus and picornavirus IRESs
The HCV IRES exemplifies a major class of viral IRES that is structurally distinct from IGR IRESs, mediates initiation by a distinct mechanism, but nevertheless also directly induces conformational changes in the ribosome that promote initiation. IRESs that are structurally similar to the HCV IRES occur in the 5’UTRs of GB virus B, of pestiviruses such as classical swine fever virus (CSFV) and bovine viral diarrhea virus (BVDV), and of a growing number of viruses from various picornavirus genera, including porcine teschovirus (PTV) and simian picornavirus type 9 (SPV9) [53, 83–91].
The structure of Type 3 IRESs
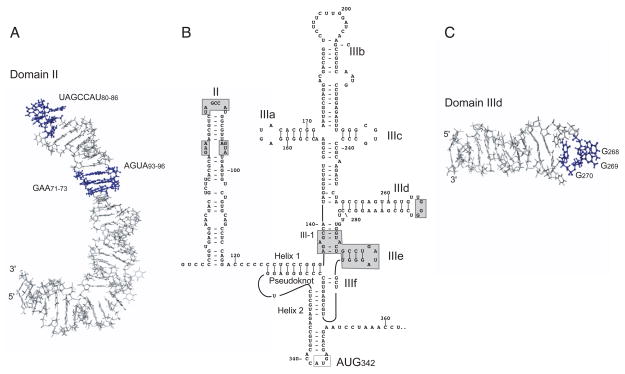
The HCV IRES occupies all but the 5’-proximal ~40nt. of the 341nt.-long 5’UTR and consists of three domains (Figure 5B). Domain II is a long irregular stem-loop structure (Figure 5A; [92–94]), Domain III consists of a complex pseudoknot, hairpin subdomains IIIa–IIIf, and intervening helices, and Domain IV is a short hairpin that sequesters the initiation codon. Structures of many subdomains of the HCV IRES and some elements of GBV-B and CSFV IRESs have been determined by NMR and X-ray crystallography (e.g. Figure 5A, C; see [53, 87, 88, 95] for reviews), whereas predicted structures of other IRESs have mostly not yet been tested by chemical and enzymatic probing, and particularly in picornaviruses should be considered as provisional rather than definitive [84–86, 90, 91]. Nevertheless, HCV-like IRESs all appear to have globally similar but non-identical structures. Their common core comprises a pseudoknot, sub-domain IIIe (which engages in a conserved tertiary interaction with a mis-matched nucleotide in the adjacent helix III1; P. Lukavsky, personal communication) and subdomain IIId (which contains a conserved apical GGG motif; Figure 5C). Other regions of these IRESs are structurally variable: for example, domain IV is often unstructured, additional hairpins are present in GBV-B, pestivirus and likely some picornavirus IRESs, and the equivalents of some peripheral subdomains in the HCV IRES may either be absent or differ significantly in length in picornavirus HCV-like IRESs. The full activity of HCV and related IRESs requires sequences extending from the 5’-border of Domain II to ~40nt. beyond the initiation codon (e.g. 96–98]); further 5’-truncation strongly impairs IRES function, whereas at the 3’-border, coding sequences can be replaced by heterologous sequences if they are relatively unstructured. In common with other classes of viral IRES, helical elements in HCV and related IRESs often play a primarily structural role (so that loss of activity caused by disruption of secondary structure can be overcome by mutations that restore base-pairing) whereas some conserved unpaired nucleotides function by interacting directly with components of the translation apparatus (see below)(reviewed in [53, 87, 88, 95]).
Figure 5.
Structure of Domains II, III and IV of the HCV IRES. (A) Domain II (PDB accession code 1P5P) is annotated to show nucleotides that are conserved in HCV, pestivirus and several picornavirus HCV-like IRESs. (C) Domain IIId (PDB accession code 1F84) is annotated to show the conserved apical GGG motif, in which GG266–267 are exposed to the minor groove and G268 is in the major groove. (B) The nomenclature of domains and helices is as in [53].
Factor requirements for initiation on Type 3 IRESs
Biochemical reconstitution experiments done using the HCV IRES and the related IRESs of pestiviruses (BVDV, CSFV) and picornaviruses (PTV, SPV9) have shown that a 40S subunit, Met-tRNAMeti, and eIF2 are sufficient for 48S complex formation. eIF3 can join at this stage and stabilizes the 48S complex, but is only required during the next stage, subunit joining, with eIF5 and eIF5B [99–104]. Initiation on the HCV IRES occurs by direct assembly of the 48S complex at the initiation codon without scanning [105] and is independent of eIFs 4A, 4B, 4F, 1 and 1A [99]; indeed eIF1 destabilizes 48S complexes assembled on the CSFV IRES (but not on the SPV9 IRES) [7, 104]. A key property of HCV-like IRESs is that they all bind directly and independently to the 40S subunit [43, 99, 100, 102, 104, 106–108] and to eIF3 [99, 104, 110, 111], engaging in multiple interactions with both that would be sufficient to recruit a 43S complex directly to the IRES (reviewed in [53, 87, 95]). This model for initiation complex assembly is consistent with observations that free 40S subunits in the cytoplasm are predominantly already associated with eIF3 [112], likely as a consequence of eIF3’s involvement in recycling of post-termination ribosomes [6]. However, other reports suggest that instead of forming by recruitment of preformed 43S complexes to the IRES, 48S complexes assemble on the HCV IRES by sequential binding of a 40S subunit, eIF3 and then the eIF2•GTP/Met-tRNAMeti ternary complex [e.g. 113]: stable binding of the viral mRNA coding region in the mRNA-binding cleft of the 40S subunit was reported to occur only as a consequence of binding of the ternary complex, but without a requirement for the establishment of codon-anticodon basepairing in the P-site [114]. The eIF2α subunit stabilizes 48S complexes assembled on the CSFV IRES [101] and by the canonical 5’-end-dependent mechanism [21] to similar extents; it is not yet known whether eIF2α interacts with mRNA at the −3 position relative to the CSFV initiation codon in the same way as in conventional 48S complexes. The final step in this initiation mechanism is subunit joining, which parallels the canonical mechanism [24] in requiring eIF5 to induce hydrolysis of eIF2-bound GTP and eIF5B to mediate displacement of factors from the 40S subunit and concurrent subunit joining [101, 103, 104]. eIF3 likely contributes to subunit joining in several ways, by stabilizing binding of the eIF2 ternary complex to the 40S subunit, thereby enhancing 48S complex formation, likely by stabilizing binding of Met-tRNAMeti in the P-site of the 40S subunit after eIF5-mediated hydrolysis of eIF2-bound GTP (see below), and possibly by inducing conformational changes in the 40S subunit that favor subunit joining. eIF3 primarily binds to the solvent-exposed surface of the 40S subunit [111].
eIF2-independent assembly of initiation complexes on Type 3 IRESs
Although the picornavirus SPV9 IRES can initiate translation by the same mechanism as flavivirus HCV-like IRESs, closer analysis [104] revealed unusual characteristics that may illuminate aspects of the ‘conventional’ mechanism of initiation on Type 3 IRESs. Thus the SPV9 IRES promotes efficient and specific factor-independent binding of Met-tRNAMeti to the 40S subunit, which is enhanced by eIF1A and eIF3, but almost completely abrogated by eIF1, even in the presence of eIF1A/eIF3. This destabilizing activity of eIF1 is suppressed by eIF2. eIF1 is thought to function in initiation by inducing conformational changes in the 40S subunit [16, 44], and its destabilization of binding of Met-tRNAMeti to these IRES/40S complexes suggest that it induces conformational changes in them that lead to ejection of Met-tRNAMeti despite the presence of a cognate P-site AUG codon. Primer extension inhibition can be used to characterize IRES/40S subunit interactions, particularly at the site of entry to the mRNA-binding cleft [115], and results from such experiments indicate that eIF1 induces conformational changes in SPV9 IRES/40S complexes that are exacerbated by eIF3 [104]. The mechanism by which the SPV9 IRES, and thus potentially other HCV-like IRESs (see below), promote specific binding of Met-tRNAMeti to the ribosome independently of eIF2 may involve induction of conformational changes that facilitate its access to and stable binding in the P-site. The observation that eIF3 stabilizes direct binding of Met-tRNAMeti to 40S subunits supports the hypothesis [53] that a reason for the requirement for eIF3 in subunit joining to form active 80S ribosomes is that it stabilizes Met-tRNAMeti in the P-site during subunit joining following loss of eIF1 and hydrolysis of eIF2-bound GTP (which weakens eIF2’s affinity to the ribosome [5].
Infection of cells by HCV and other flaviviruses commonly leads to activation of PKR and PERK protein kinases, which phosphorylate eIF2α and ultimately inhibit cap-dependent initiation of translation. Initiation on HCV and CSFV IRESs is more resistant to inhibition by phosphorylation of eIF2α than cap-dependent translation (e.g. [116], and experiments to elucidate the basis of this resistance revealed that in addition to eIF2, eIF5B could also independently promote recruitment of Met-tRNAMeti to 40S subunits bound to CSFV [101] and HCV [117] IRESs. There are thus circumstances under which eIF5B can act like its prokaryotic homolog IF2 in promoting recruitment of Met-tRNAMeti to the small ribosomal subunit. Taking into account the activity of the SPV9 IRES in promoting factor-independent binding of Met-tRNAMeti to 40S subunits (see above) and the fact that eIF5B does not promote recruitment of Met-tRNAMeti to 40S subunits in the absence of CSFV/HCV IRESs, it is likely that this class of IRESs induces conformational changes in the 40S subunit that promote eIF2-independent 48S complex formation. Formation of such 48S complexes by eIF5B was enhanced by eIF3 to a level that was comparable to that of conventional eIF2-mediated binding of Met-tRNAMeti, which, taken together with observations that eIF3 promotes factor-independent binding of Met-tRNAMeti to SPV9 IRES/40S complexes [104] and eIF2-mediated binding of Met-tRNAMeti to 40S subunits in the canonical initiation mechanism, suggests that it plays a general role in promoting association of Met-tRNAMeti with the 40S subunit. However, eIF1 significantly destabilized 48S complexes that had been assembled by the eIF5B/eIF3-mediated mechanism on the CSFV IRES [101] (and, by implication, on the HCV IRES, although this has not been tested), so that this mechanism may thus not contribute significantly to initiation on Type 3 IRESs in stress conditions when levels of the Met-tRNAMeti/eIF2•GTP ternary complex are limiting.
Two alternative mechanisms may contribute more substantially to the residual level of initiation on HCV-like IRESs during viral infection. Elongation-competent 80S ribosomes can assemble on the HCV IRES in the complete absence of eIFs at elevated [118] but not at physiological [117] Mg2+ concentrations, whereas the CSFV IRES supported factor-independent assembly of active 80S ribosomes under physiological conditions, albeit at a ten-fold lower level than by the eIF2-mediated mechanism [101]. It is not known whether Met-tRNAMeti first associates with IRES/40S complexes before subunit joining, or can enter pre-assembled 80S/IRES complexes. Lastly, it is conceivable and even probable (given the apparent influence of the IRES on the affinity of Met-tRNAMeti to 40S subunits), that the Met-tRNAMeti/eIF2•GTP ternary complex has a higher affinity to IRES/40S/eIF3 complexes than to 40S subunits, and that residual ternary complex in virus-infected cells is sufficient to sustain a reduced level of initiation on Type 3 IRESs.
Functional interactions of Type 3 IRESs with eIF3 and the 40S subunit
The first step in the exceptional initiation process on Type 3 IRESs is the direct, factor-independent binding of the IRES to the 40S subunit, which occurs in such a way that the initiation codon is placed in the immediate vicinity of the P-site and that the downstream coding region is fixed in the mRNA-binding cleft [99, 100, 102, 104]. Ribosomal binding involves multiple sites of interaction which have been mapped by chemical and enzymatic footprinting to the apex of HCV domain II, the four-way junction of domains IIIa/IIIb/IIIc, domains IIId and IIIe, the pseudoknot and to mRNA flanking the initiation codon (reviewed in [53, 87, 88, 95]). CryoEM analysis indicated that the HCV IRES binds in an extended conformation to the solvent-exposed surface of the 40S subunit: domain III binds to the platform and body, whereas domain II interacts with the head of the 40S subunit, partially occupying the E-site and extending towards the P-site in the mRNA-binding cleft (Figure 4A, B; [43, 119]). Binding is a multi-stage process, and is likely initiated by interaction of elements of domain III with the platform: both Domain IIIe/helix III1 and the exposed apical GGG motif in domain IIId (Fig. 5C; [109]) are protected from chemical modification by binding to the 40S subunit, with domain IIId interacting particularly extensively [43, 106–109, 119]. Mutations that disrupt the structure of Domain IIIe/helix III1 or alter the apical sequence of Domain IIId strongly impair ribosomal binding and particularly in the case of Domain IIId, lead to almost total abrogation of IRES function [106, 108, 109]. The similar effects of mutations in domains IIId [104] and IIIe [84] on the ribosomal binding and/or initiation activity of picornavirus HCV-like IRESs suggest that they use a similar mechanism for initial engagement with the 40S subunit.
The next stage is entry of mRNA flanking the initiation codon into the mRNA-binding cleft. Domain II and sequences downstream of Helix 1 of the pseudoknot do not contribute to the affinity of the IRES to the ribosome (e.g. [108]) but deletion of domain II or disruption of pseudoknot helix 2 in HCV and CSFV IRESs strongly impairs or even abrogates this entry process [99, 106, 107, 114]. Initiation on Type 3 IRESs can therefore not simply be described in terms of binding to the 40S subunit, but is instead an ordered, multi-step process. Importantly, entry of mRNA into the mRNA-binding cleft involves active manipulation of the 40S subunit by the IRES, which induces significant conformational changes in the 40S subunit that include rotation of the head relative to the body, and opening of the entry latch of the mRNA-binding channel so that mRNA flanking the initiation codon can be accommodated in it (Figure 4A–4C; [43]). Significantly, these changes do not occur in the absence of domain II, indicating that it is responsible for the change induced by binding of the IRES that allows entry and stable binding of mRNA in the cleft: accordingly, the function of pseudoknot helix 2 is likely to be to ensure correct orientation of adjacent downstream sequences so that they enter this cleft. As noted above, domain II binds so that it partially occupies the E-site. IRES function is readily impaired by mutations in the apex of domain I [105, 120, 121], but the interactions of domain II with the E-site have not been determined at high resolution. Domain II may interact with rpS5 [122], and it is accordingly tempting to speculate that the similar interactions of HCV [43, 119] and CrPV [75, 76] IRESs in this region are responsible for the similar changes that they induce in ribosomal conformation.
Analysis of HCV and CSFV IRES deletion mutants lacking domain II revealed that they are defective in promoting initiation, in part at the stage of subunit joining [99, 103, 106, 107, 113]. It was initially thought that domain II or changes induced by it are required for eIF5 to be able to induce hydrolysis of eIF2-bound GTP [103], but the fact that deletion of domain II from the CSFV IRES also impaired subunit-joining following initiation in vitro by the eIF5B/eIF3-mediated mechanism indicates that the defect is more general, and likely reflects an inability of initiation complexes assembled on the mutant IRES to undergo conformational changes required both for induction of hydrolysis of eIF2-bound GTP and subsequent subunit joining. One hypothesis [101] is that establishment of base-pairing between the initiation codon and the anticodon of Met-tRNAMeti in the P-site is followed by a conformational change in the orientation of Met-tRNAMeti that is required both to provide a signal leading to induction of hydrolysis of eIF2-bound GTP and for docking of the 60S subunit onto the IRES/40S/Met-tRNAMeti complex.
Remarkably, the CSFV IRES lacking domain II (ΔII-IRES) was able to promote factor-independent binding of Met-tRNAMeti to the 40S subunit [101], just like the SPV9 IRES [104], which has a domain II that is predicted to differ markedly in size and structure from domain II of the CSFV and HCV IRESs [92, 103], and may thus be unable to induce the same conformational changes in the 40S subunit that they do. As with the SPV9 IRES, 48S complexes assembled on the CSFV ΔII-IRES are resistant to destabilization by eIF1 [101, 104], observations which suggest that eIF1 amplifies conformational changes caused by domain II of the CSFV IRES, leading to loss of Met-tRNAMeti from the P-site.
The overall appearance of the HCV IRES in IRES/40S and IRES/80S complexes is similar, although there is evidence of conformational change in the pseudoknot and in the four-way IIIa/IIIb/IIIc junction [43, 119]. Domain II is still present in the E-site of the 80S ribosome, but must be displaced before or during the first cycle of elongation to allow translocation of deacylated tRNA from P- to E-sites. Domain II also interacts with the L1 stalk in 80S ribosomes, and as in prokaryotic ribosomes (in which rpL1 promotes transfer of deacylated tRNA to the E-site and ejection from it) and in IGR IRES/80S complexes (in which rpL1 has been hypothesized to promote ejection of the IRES from the E-site (see above)), this interaction of Domain II may promote its ejection from elongating ribosomes. The mRNA exit channel is closed in IRES/80S complexes just as in IRES/40S complexes, possibly because of Domain II’s continued presence in the E-site of initiation complexes at this stage.
SUMMARY
Type 3 (HCV-like) and Type 4 (CrPV IGR-like) IRESs are structurally unrelated and mediate initiation by distinct mechanisms. However, both classes of IRES engage directly with the ribosome, and their structural organization enables them to interact with the ribosome at multiple sites and to induce conformational changes in it that affect multiple stages in the initiation process. Thus the Type 4 IGR IRES has a compact and relatively rigid core that provides a platform that orients elements that are responsible for the specificity and affinity of the IRES’ factor-independent association with the 40S subunit, for promoting subunit joining and likely for promoting elongation-factor mediated ribosomal translocation. The IRES alters its conformation during the different stages of this initiation process, and importantly, also induces a series of conformational changes in small and large ribosomal subunits that are likely to promote these stages, and in some instances have been experimentally found to do so. The Type 3 HCV-like IRESs are larger and less compact than Type 4 IRESs, but also contain distinct elements that play discrete functions at different stages during the initiation process, including specific binding to eIF3 and importantly, factor-independent association of the IRES with the 40S subunit, inducing conformational changes in it that promote entry and fixation of the initiation codon region in the mRNA-binding cleft, stabilizing binding of Met-tRNAMeti in the P-site and promoting subunit joining. As in the case of the IGR IRES, Type 3 IRESs likely also undergo conformational transitions during these different stages.
Acknowledgments
I thank Joachim Frank, Jeffrey Kieft, Lori Passmore and Christian Spahn for figures. Work in the author’s laboratory is funded by NIH/NIAID Award AI-51340 and by American Heart Association (Heritage Affiliate) Award 0755900T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hellen CUT, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 3.Doudna JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JBW, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 2007. pp. 297–318. [Google Scholar]
- 4.Hellen CUT, Wimmer E. Translation of encephalomyocarditis virus RNA by internal ribosomal entry. Curr Top Microbiol Immunol. 1995;203:31–63. doi: 10.1007/978-3-642-79663-0_2. [DOI] [PubMed] [Google Scholar]
- 5.Pestova TV, Lorsch JR, Hellen CUT. The Mechanism of Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, Hershey JBW, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 2007. pp. 87–128. [Google Scholar]
- 6.Pisarev AV, Hellen CUT, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 8.Pisareva VP, Pisarev AV, Komar AA, Hellen CUT, Pestova TV. Translation initiation on mammalian mRNAs with structured 5'UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 10.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 12.Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2222. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hog P, Wagner G, Hellen CUT, Pestova TV. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Research. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maag D, Algire MA, Lorsch JR. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J Mol Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell SF, Lorsch JR. Should I stay or should I go? Eukaryotic translation initiation factors 1 and 1A control start codon recognition. J Biol Chem. 2008;283:27345–27349. doi: 10.1074/jbc.R800031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conte MR, Kelly G, Babon J, Sanfelice D, Youell J, Smerdon SJ, Proud CG. Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry. 2006;45:4550–4558. doi: 10.1021/bi052387u. [DOI] [PubMed] [Google Scholar]
- 21.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CUT, Pestova TV. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marintchev A, Kolupaeva VG, Pestova TV, Wagner G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proc Natl Acad Sci USA. 2003;100:1535–1540. doi: 10.1073/pnas.0437845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acker MG, Shin BS, Dever TE, Lorsch JR. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J Biol Chem. 2006;281:8469–8475. doi: 10.1074/jbc.M600210200. [DOI] [PubMed] [Google Scholar]
- 24.Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 25.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 26.Selmer M, Dunham CM, Murphy FV, IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 27.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 29.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 30.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CUT, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 32.Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol. 2009;21:435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 33.Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 34.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 35.Milon P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Mol Cell. 2008;30:712–720. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 36.Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 39.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 40.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jørgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestova TV, Hellen CU. The structure and function of initiation factors in eukaryotic protein synthesis. Cell Mol Life Sci. 2000;57:651–674. doi: 10.1007/PL00000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 44.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 45.Schäfer MA, Tastan AO, Patzke S, Blaha G, Spahn CM, Wilson DN, Nierhaus NH. Codon-anticodon interaction at the P site is a prerequisite for tRNA interaction with the small ribosomal subunit. J Biol Chem. 2002;277:19095–19105. doi: 10.1074/jbc.M108902200. [DOI] [PubMed] [Google Scholar]
- 46.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 47.Unbehaun A, Marintchev A, Lomakin IB, Didenko T, Wagner G, Hellen CU, Pestova TV. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J. 2007;26:3109–3123. doi: 10.1038/sj.emboj.7601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 49.Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pestova TV, Hellen CUT, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pestova TV, Shatsky IN, Hellen CUT. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CUT. Direct Functional interaction of initiation factor eIF4G with the Type 1 internal ribosomal entry site of enteroviruses. Proc Natl Acad Sci USA. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pisarev AV, Shirokikh NE, Hellen CUT. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C R Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Nakashima N, Uchiumi T. Functional analysis of structural motifs in dicistroviruses. Virus Res. 2009;139:137–147. doi: 10.1016/j.virusres.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 55.Kieft JS. Comparing the three-dimensional structures of Dicistroviridae IGR IRES RNAs with other viral RNA structures. Virus Res. 2009;139:148–156. doi: 10.1016/j.virusres.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanamori Y, Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–74. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 60.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 61.Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jang CJ, Lo MCY, Jan E. Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J Mol Biol. 2009;387:42–58. doi: 10.1016/j.jmb.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 63.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci USA. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pestova TV, Hellen CUT. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pestova TV, Lomakin IB, Hellen CUT. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci USA. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cevallos RC, Sarnow P. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J Virol. 2005;79:677–683. doi: 10.1128/JVI.79.2.677-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J Biol Chem. 2007;282:7770–7776. doi: 10.1074/jbc.M610887200. [DOI] [PubMed] [Google Scholar]
- 70.Shoji S, Walker SE, Fredrick K. Ribosomal Translocation: One Step Closer to the Molecular Mechanism. ACS Chem Biol. 2009 Jan 27; doi: 10.1021/cb8002946. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc Natl Acad Sci USA. 2001;98:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deniz N, Lenarcic EM, Landry DM, Thompson SR. Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA. 2009;15:932–946. doi: 10.1261/rna.1315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bushell M, Sarnow P. Hijacking the translation apparatus by RNA viruses. J Cell Biol. 2002;158:395–399. doi: 10.1083/jcb.200205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moore NF, Kearns A, Pullin JS. Characterization of Cricket paralysis virus-induced polypeptides in Drosophila cells. J Virol. 1980;33:1–9. doi: 10.1128/jvi.33.1.1-9.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 76.Schüler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 77.Galkin O, Bentley AA, Gupta S, Compton BA, Mazumder B, Kinzy TG, Merrick WC, Hatzoglou M, Pestova TV, Hellen CU, Komar AA. Roles of the negatively charged N-terminal extension of Saccharomyces cerevisiae ribosomal protein S5 revealed by characterization of a yeast strain containing human ribosomal protein S5. RNA. 2007;13:2116–2128. doi: 10.1261/rna.688207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35:1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uchiumi T, Hori K, Nomura T, Hachimori A. Replacement of L7/L12.L10 protein complex in Escherichia coli ribosomes with the eukaryotic counterpart changes the specificity of elongation factor binding. J Biol Chem. 1999;274:27578–27582. doi: 10.1074/jbc.274.39.27578. [DOI] [PubMed] [Google Scholar]
- 80.Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, Frank J. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 A resolution. EMBO J. 2000;19:2710–2718. doi: 10.1093/emboj/19.11.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schuette JC, Murphy FV, 4th, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CM. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 83.Rijnbrand R, Abell G, Lemon SM. Mutational analysis of the GB virus B internal ribosome entry site. J Virol. 2000;74:773–783. doi: 10.1128/jvi.74.2.773-783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chard LS, Kaku Y, Jones B, Nayak A, Belsham GJ. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and a picornavirus, porcine teschovirus-1 Talfan. J Virol. 2006;80:1271–1279. doi: 10.1128/JVI.80.3.1271-1279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chard LS, Bordeleau ME, Pelletier J, Tanaka J, Belsham GJ. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J Gen Virol. 2006;87:927–936. doi: 10.1099/vir.0.81546-0. [DOI] [PubMed] [Google Scholar]
- 86.Hellen CUT, de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse Picornavirus genera: Evidence for modular exchange of functional noncoding RNA elements by recombination. J Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 88.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Belsham GJ. Divergent picornavirus IRES elements. Virus Res. 2009;139:183–192. doi: 10.1016/j.virusres.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 90.Bakhshesh M, Groppelli E, Willcocks MM, Royall E, Belsham GJ, Roberts LO. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J Virol. 2008;82:1993–2003. doi: 10.1128/JVI.01957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kapoor A, Victoria J, Simmonds P, Wang C, Shafer RW, Nims R, Nielsen O, Delwart E. A highly divergent picornavirus in a marine mammal. J Virol. 2008;82:311–320. doi: 10.1128/JVI.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 93.Dibrov SM, Johnston-Cox H, Weng YH, Hermann T. Functional architecture of HCV IRES domain II stabilized by divalent metal ions in the crystal and in solution. Angew Chem Int Ed Engl. 2007;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- 94.Zhao Q, Han Q, Kissinger CR, Hermann T, Thompson PA. Structure of hepatitis C virus IRES subdomain IIa. Acta Crystallogr D Biol Crystallogr. 2008;64:436–443. doi: 10.1107/S0907444908002011. [DOI] [PubMed] [Google Scholar]
- 95.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fletcher SP, Jackson RJ. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5' untranslated region important for IRES function. J Virol. 2002;76:5024–5033. doi: 10.1128/JVI.76.10.5024-5033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fletcher SP, Ali IK, Kaminski A, Digard JP, Jackson RJ. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA. 2002;8:1558–1571. [PMC free article] [PubMed] [Google Scholar]
- 99.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pestova TV, Hellen CUT. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology. 1999;258:249–256. doi: 10.1006/viro.1999.9741. [DOI] [PubMed] [Google Scholar]
- 101.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CUT. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Breyne S, Yu Y, Pestova TV, Hellen CU. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA. 2008;14:367–380. doi: 10.1261/rna.696508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 106.Kolupaeva VG, Pestova TV, Hellen CU. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 110.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CUT. Specific interaction of eukaryotic translation initiation factor 3 with the 5' nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 112.Sundkvist IC, Staehelin T. Structure and function of free 40S ribosome subunits: Characterization of initiation factors. J Mol Biol. 1975;99:401–418. doi: 10.1016/s0022-2836(75)80135-5. [DOI] [PubMed] [Google Scholar]
- 113.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fraser CS, Hershey JW, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat Struct Mol Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kolupaeva VG, de Breyne S, Pestova TV, Hellen CU. In vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry. Methods Enzymol. 2007;430:409–39. doi: 10.1016/S0076-6879(07)30016-5. [DOI] [PubMed] [Google Scholar]
- 116.Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR, Pelletier J. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol Biol Cell. 2006;17:4632–4644. doi: 10.1091/mbc.E06-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 118.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 120.Odreman-Macchioli F, Baralle FE, Buratti E. Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J Biol Chem. 2001;276:41648–41655. doi: 10.1074/jbc.M104128200. [DOI] [PubMed] [Google Scholar]
- 121.Kalliampakou KI, Psaridi-Linardaki L, Mavromara P. Mutational analysis of the apical region of domain II of the HCV IRES. FEBS Lett. 2002;511:79–84. doi: 10.1016/s0014-5793(01)03300-2. [DOI] [PubMed] [Google Scholar]
- 122.Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]