Tom70 family members may exhibit structural plasticity when complexed with Hsp70/Hsp90. This structural plasticity enables Tom70 to accommodate various precursor substrates for mitochondrial translocation.
Keywords: Tom71, mitochondrial precursors, structural plasticity
Abstract
Mitochondrial precursors are transported through the translocase of the outer membrane (TOM) complex. Tom70/Tom71 is a major surface receptor of the TOM complex for mitochondrial precursors and facilitates Hsp70/Hsp90-escorted precursor translocation into the mitochondrion. Previous structural studies of Tom71 have revealed that it contains an N-terminal and a C-terminal domain and that the two domains may remain in an open conformation when binding to Hsp70/Hsp90. In a newly obtained crystal form of a complex of Tom71 and the Hsp70 C-terminus, the N-terminal domain was found to have rotated about 12° towards the C-terminal domain compared with the previous determined crystal structure of Tom71 in the open conformation. This newly solved structure is defined as the ‘intermediate conformation’. The domain rearrangements in Tom71 significantly change the surface hydrophobicity and the volume of the precursor-binding pocket. This work suggests that Tom70/Tom71-family members may exhibit structural plasticity from the intermediate conformation to the fully open conformation when complexed with Hsp70/Hsp90. This structural plasticity enables the precursor receptors to accommodate different precursor substrates for mitochondrial translocation.
1. Introduction
The mitochondrion contains a large number of proteins. However, it can only synthesize a few of these proteins by itself (Sickmann et al., 2003 ▶; Gray et al., 1999 ▶). The majority of the proteins are translated in the cytosol and are transported to the mitochondrion by multi-protein complexes: the translocase of the outer membrane complex (TOM) and the translocase of the inner membrane complex (TIM) (Neupert & Herrmann, 2007 ▶). The TOM complex has two types of surface receptor: Tom20 and Tom70 (Neupert & Herrmann, 2007 ▶; Hines et al., 1990 ▶; Sollner et al., 1989 ▶, 1990 ▶; Steger et al., 1990 ▶; Endo & Yamano, 2009 ▶). Tom20 recognizes the N-terminal mitochondrial targeting signals of precursor proteins in cooperation with Tom22 (Yamano et al., 2008 ▶; Brix et al., 1997 ▶), while Tom70/Tom71 specializes in binding internal targeting sequences (Neupert & Herrmann, 2007 ▶; Chan et al., 2006 ▶). Yeast Tom71 and Tom70 have significant protein sequence identity (53%) and overlapping cellular functions (Bomer et al., 1996 ▶; Koh et al., 2001 ▶; Kondo-Okamoto et al., 2008 ▶) and it is reasonable to propose that they function in mitochondrial precursor translocations using a similar mechanism. The partially folded precursors are chaperoned to Tom70/Tom71 by Hsp70/Hsp90 (Young et al., 2003 ▶).
The structure of the complex between the cytosolic domain of yeast Tom71 and the Hsp70 Ssa1 C-terminal EEVD motif showed that Tom71 contains two domains which consist of 11 TPR motifs (Li et al., 2009 ▶; Wu & Sha, 2006 ▶). The two domains can be arranged into a closed conformation when free of Hsp70/Hsp90 or an open conformation when complexed with Hsp70/Hsp90 (Li et al., 2009 ▶; Wu & Sha, 2006 ▶). The N-terminal domain rotates by about 180° when Tom70/Tom71 switches from the closed conformation to the open conformation. The N-terminal domain of Tom71 interacts with the Hsp70/Hsp90 C-terminus, like Hop and CHIP (Li et al., 2009 ▶). A hydrophobic precursor-binding pocket is formed by the Tom71 C-terminal domain and part of the N-terminal domain (Li et al., 2009 ▶; Wu & Sha, 2006 ▶). The binding of Hsp70/Hsp90 to Tom71 may keep Tom71 in the open conformation for precursor loading (Li et al., 2009 ▶). In the open conformation, the hydrophobic precursor-binding pocket of Tom71 has estimated dimensions of about 25 × 35 × 20 Å, which is large enough to accommodate several α-helices (Li et al., 2009 ▶).
Tom70/Tom71 can bind and receive a broad range of precursor substrates, including linearized polypeptides and folding intermediates with secondary structure (Brix et al., 2000 ▶; Yamamoto et al., 2009 ▶). Using peptide-scanning methods, two peptide fragments of 13 amino acids in length from the yeast protein PiC (residues 181–193 and 211–223) have been identified as substrates of the Tom70 peptide (Brix et al., 2000 ▶). The binding affinity (K d) between Tom70 and PiC (181–193) is about 68 µM as measured by fluorescence anisotropy (Mills et al., 2009 ▶). In addition, Tom70 also interacts with precursors with N-terminal mitochondrial targeting peptides and protects them from aggregation (Yamamoto et al., 2009 ▶). In order to accommodate different precursor substrates, it is possible that Tom70/Tom71 may exhibit conformational flexibility at the precursor-binding pocket. Biophysical analysis revealed that Tom70 is in a relatively unstable conformation under physiological conditions. It has been suggested that this instability might be important for Tom70 to recognize the wide range of precursor substrates and might be vital for protein translocation (Beddoe et al., 2004 ▶). However, the mechanism by which Tom70/Tom71 provides conformation flexibility is not clear at the atomic level.
2. Materials and methods
Saccharomyces cerevisiae Tom71 was expressed and purified as described in Li et al. (2009 ▶). Tom71 was mixed with the synthetic S. cerevisiae Hsp70 Ssa1 C-terminal peptide (PEAEGPTVEEVD; residues 651–662; Genscript) in a 1:2 molar ratio in 20 mM Tris pH 7.5, 50 mM NaCl. A crystal was grown using the hanging-drop vapor-diffusion method with 100 mM Tris pH 7.5, 20% PEG 6000, 20% ethylene glycol. The complex crystals diffracted X-rays to 2.19 Å resolution on the SER-CAT synchrotron beamline at APS. The atomic coordinates of the N-terminal domain and C-terminal domain of yeast Tom71 complexed with the Ssa1 C-terminus (PDB code 3fp4; Li et al., 2009 ▶) were used individually as the search model in the molecular-replacement method using the program Phaser (McCoy et al., 2007 ▶). The model was manually built by Coot (Emsley & Cowtan, 2004 ▶). The coordinates and structure factors have been deposited in the PDB with accession code 3lca.
3. Results and discussion
In this study, we describe a crystal structure of yeast Tom71 complexed with the Hsp70 C-terminus that differs significantly from that which we reported previously (Li et al., 2009 ▶). The structural data strongly support the idea that the Tom70/Tom71 precursor-binding pocket may exhibit significant structural flexibility to recognize and bind various precursor substrates for mitochondrial biogenesis. The results are in good accordance with the previous biophysical and SAXS analyses (Mills et al., 2009 ▶; Beddoe et al., 2004 ▶) and provide insight into the conformation plasticity of Tom70/Tom71 at the molecular level.
The structure of the complex of the S. cerevisiae Tom71 cytosolic domain (residues 107–639) and the Hsp70 Ssa1 C-terminus in this study is named Tom71_new. The complex structure of the Tom71 cytosolic domain with the Ssa1 C-terminus in the open conformation described previously is termed Tom71_old (Li et al., 2009 ▶). The structure of Tom71_new was determined to 2.19 Å resolution by the molecular-replacement method using Tom71_old as the search model (Table 1 ▶). The final model contains residues 124–637 of yeast Tom71 and residues 659–662 of the yeast Hsp70 Ssa1 C-terminal fragment. In the structure, the electron-density map is not clear for residues 107–123, 225–231, 254–260, 331–338, 390–421, 535–540 and 638–639 of Tom71 and residues 650–658 of Ssa1, indicating that these regions may be disordered in Tom71_new. In Tom71_new the Tom71 structure contains 27 α-helices (A1–A27) and no β-strands. The 27 α-helices A1–A27 form 11 TPR motifs (TPR1–11) in the structure. The N-terminal domain of Tom71 in Tom71_new covers A1–A7 (TPR1–3) and the C-terminal domain consists of A8–A27. The whole complex molecule exhibits an elongated monomer that is consistent with the characterization of Tom70 by analytical ultracentrifugation and solution small-angle X-ray scattering (SAXS; Mills et al., 2009 ▶).
Table 1. Data collection and structure determination of Tom71 complexed with the Ssa1 C-terminus.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 51.148, b = 97.971, c = 129.102, α = β = γ = 90 |
| Wavelength (Å) | 0.9500 |
| Resolution (Å) | 2.19 (2.27–2.19) |
| Rsym or Rmerge | 0.089 (0.43) |
| I/σ(I) | 26.6 (2.7) |
| Completeness (%) | 98.6 (89.5) |
| Redundancy | 5.7 (3.1) |
| Refinement | |
| Resolution (Å) | 2.19 |
| No. of reflections | 31246 |
| Rwork/Rfree (%) | 23.0 (26.7)/27.4 (30.0) |
| No. of atoms | |
| Protein | 4133 |
| Water | 232 |
| B factors (Å2) | |
| Protein | 33.94 |
| Water | 47.3 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.904 |
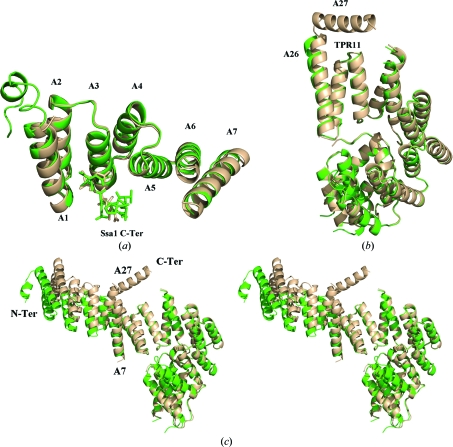
When Tom71_new is compared with Tom71_old, the individual N-terminal and C-terminal domain structures are very similar. The structure of the N-terminal domain in Tom71_new can be superimposed very well with its counterpart in Tom71_old with a root-mean-square deviation (r.m.s.d.) of 0.368 Å (Fig. 1 ▶ a). Meanwhile, the r.m.s.d. for the C-terminal domains is 0.320 Å when the C-terminal domain from Tom71_new is superimposed with that from Tom71_old (Fig. 1 ▶ b). Moreover, the Ssa1 C-terminal peptide in Tom71_new is located in a groove in the N-terminal domain and stays in a very similar conformation as that in Tom71_old.
Figure 1.
The crystal structure of Tom71 complexed with the Hsp70 Ssa1 C-terminus. (a) The N-terminal domain of Tom71 complexed with the Ssa1 C-terminal tail. The Tom71 structures are shown as ribbon drawings and the bound Ssa1 C-termini are shown in stick mode. A superimposition of the N-terminal domain of Tom71_old (in green) and Tom71_new (in wheat) revealed that they share similar structures. (b) The C-terminal domains of Tom71. Superimposition of the C-terminal domain of Tom71_new (in wheat) and Tom71_old (in green) shows they have similar structures. (c) Ribbon drawings of full-length Tom71 structures. The C-terminal domain of Tom71_new (in wheat) is superimposed on that of Tom71_old (in green). In Tom71_new the N-terminal domain rotates towards the C-terminal domain by approximately 12° along helix A7 from its position in Tom71_old.
However, the N-terminal and C-terminal domains of Tom71 in the Tom71_new structure are arranged in a significantly different fashion compared with those in the Tom71_old structure. In Tom71_new the N-terminal domain rotates towards the C-terminal domain by approximately 12° along helix A7 from its position in Tom71_old (Fig. 1 ▶ c). To distinguish Tom71_new from the previously determined Tom71_old which is in the open conformation, we define this newly solved structure (Tom71_new) as the ‘intermediate conformation’. It is likely that Tom70/Tom71 could utilize helix A7 as a hinge to rotate the N-terminal domain by 12° to achieve the intermediate conformation (Tom71_new) from the position in the open conformation (Tom71_old).
The Tom71–Ssa1 complex forms a monomer in both the Tom71_new and Tom71_old structures when Tom71 is complexed with the Hsp70/Hsp90 C-terminus. We reported that in the absence of Hsp70/Hsp90 yeast Tom70 formed a dimer in the closed conformation (Wu & Sha, 2006 ▶). It is likely that Tom70/Tom71 forms a monomer when complexed with Hsp70/Hsp90 but remains as a dimer in the absence of Hsp70/Hsp90 (Li et al., 2009 ▶; Wu & Sha, 2006 ▶).
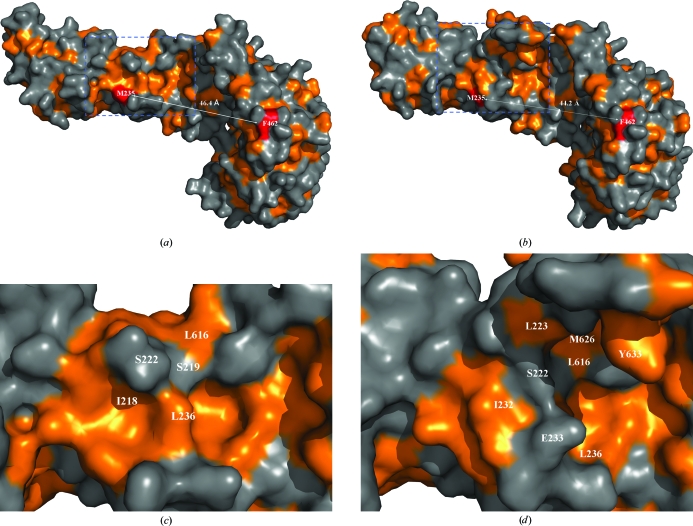
When compared with Tom71_old in the open conformation, Tom71_new in the intermediate conformation contains significant alterations within the precursor-binding pocket of Tom71. Firstly, the volume of the precursor binding site is decreased. Since helices A6 and A7 are involved in forming one side of the precursor-binding pocket, the rotation of the two helices in the N-terminal domain towards the C-terminal domain closes up the pocket. For example, residues Met235 and Phe462 are located on opposite sides of the precursor-binding pocket of Tom71. The distance between the Cα atoms of Met234 and Phe462 is 46.4 and 44.2 Å in Tom71_old and Tom71_new, respectively (Figs. 2 ▶ a and 2 ▶ b). A conformational change is also observed in helix A26. Compared with Tom71_old, helix A26 in Tom71_new has moved toward the precursor-binding pocket by ∼2 Å. This movement further decreases the size of the binding pocket in Tom71_new. This observation is nicely consistent with the proposal that N-terminal rotation is in concert with TPR11 (Beddoe et al., 2004 ▶).
Figure 2.
Hydrophobicity drawings of Tom71 structures. (a) Hydrophobicity drawing of Tom71_old. The orange color denotes the hydrophobic patches. The distance between Met235 and Phe462 is shown as a dotted line. The exposed surfaces of Met235 and Phe263 are shown in red. (b) Hydrophobicity drawing of Tom71_new. The distance between Met235 and Phe462 is shown as a dotted line. (c) A magnified version of the area within the blue box in (a). (d) A magnified version of the area within the blue box in (b). Some residues around Ser222 and Glu233 are labeled.
Secondly, the hydrophobic properties of the precursor-binding pocket have changed as a result of the conformational changes. In the Tom71_old structure a large continuous hydrophobic patch exists along the hinge region between the N-terminal and the C-terminal domains (Fig. 2 ▶ c). After the rotation of the N-terminal domain, this hydrophobic patch was disrupted by exposure of the polar side chains of Ser222 and Glu223 in Tom71_new (Fig. 2 ▶ d). We proposed that the hydrophobic regions within the precursor-binding pocket may play important roles in recognizing the internal targeting sequences of the mitochondrial precursors (Li et al., 2009 ▶). The differences in the hydrophobicity within the precursor-binding pocket in the Tom71_new and Tom71_old structures indicate that Tom71 may recognize various precursor substrates. The data strongly suggest that the Tom71 precursor-binding pocket exhibits significant structural plasticity when Tom71 is complexed with Hsp70/Hsp90, which might be essential for Tom71 to accommodate and receive a broad range of precursor substrates with different hydrophobicities.
In the Tom71_new structure the C-terminal helix A27 of Tom71 is clearly visible all the way to the C-terminus of Tom71 in the electron-density map, while in previously determined Tom70/Tom71 structures only a few turns of helix A27 could be located or it was totally missing from the electron density (Li et al., 2009 ▶; Wu & Sha, 2006 ▶). Helix A27 forms the upper side wall of the precursor-binding pocket in the Tom71_new structure (Figs. 1 ▶ c and 2 ▶). Furthermore, several hydrophobic residues such as Met626, Leu630 and Tyr633 from helix A27 face towards the precursor-binding pocket and might be involved in binding the hydrophobic precursor substrates.
Based on previous structural data, we proposed that binding of Hsp70/Hsp90 to Tom71 may stabilize Tom71 in the open conformation for precursor loading (Li et al., 2009 ▶). In this study, we provide crystallographic data to show that the Tom71 precursor-binding pocket might exhibit significant structural flexibility even when Tom71 is bound to Hsp70/Hsp90. When Hsp70/Hsp90 is bound to Tom71, the N-terminal and C-terminal domains of Tom71 may rotate between the intermediate conformation and the open conformation. This will provide Tom71 with the flexibility to accommodate a broad range of substrates, which could be essential for the function of Tom70/Tom71 in mitochondrial biogenesis. This hypothesis is also supported by other biochemical and biophysical studies. Firstly, Tom70/Tom71 has various precursor substrates and is involved in the translocation of all precursors with internal mitochondrial targeting signals and N-terminal pre-sequences (Yamamoto et al., 2009 ▶; Brix et al., 2000 ▶). Secondly, biophysical studies have indicated that Tom70 may exhibit significant structural flexibility and that several structural intermediates may exist (Mills et al., 2009 ▶; Beddoe et al., 2004 ▶). Thirdly, the buried surface between the N-terminal and C-terminal domains of Tom71_new is 1464 Å2, which accounts for only 5.7% of the molecular surface of Tom71. The small interaction surface might provide intrinsic flexibility between the N-terminal and C-terminal domains within Tom71.
Supplementary Material
PDB reference: Tom71 complexed with Hsp C-terminus, 3lca
Acknowledgments
We are grateful to the staff scientists at the APS SER-CAT and GM-CAT beamlines for their help in data collection. This work was supported by grants from NIH (R01 DK56203 and R01 GM080261) and the Army Research Office (51894LS) to BS.
References
- Beddoe, T., Bushell, S. R., Perugini, M. A., Lithgow, T., Mulhern, T. D., Bottomley, S. P. & Rossjohn, J. (2004). J. Biol. Chem.279, 46448–46454. [DOI] [PubMed]
- Bomer, U., Pfanner, N. & Dietmeier, K. (1996). FEBS Lett.382, 153–158. [DOI] [PubMed]
- Brix, J., Dietmeier, K. & Pfanner, N. (1997). J. Biol. Chem.272, 20730–20735. [DOI] [PubMed]
- Brix, J., Ziegler, G. A., Dietmeier, K., Schneider-Mergener, J., Schulz, G. E. & Pfanner, N. (2000). J. Mol. Biol.303, 479–488. [DOI] [PubMed]
- Chan, N. C., Likić, V. A., Waller, R. F., Mulhern, T. D. & Lithgow, T. (2006). J. Mol. Biol.358, 1010–1022. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Endo, T. & Yamano, K. (2009). Biol. Chem.390, 723–730. [DOI] [PubMed]
- Gray, M. W., Burger, G. & Lang, B. F. (1999). Science, 283, 1476–1481. [DOI] [PubMed]
- Hines, V., Brandt, A., Griffiths, G., Horstmann, H., Brutsch, H. & Schatz, G. (1990). EMBO J.9, 3191–3200. [DOI] [PMC free article] [PubMed]
- Koh, J. Y., Hájek, P. & Bedwell, D. M. (2001). Mol. Cell. Biol.21, 7576–7586. [DOI] [PMC free article] [PubMed]
- Kondo-Okamoto, N., Shaw, J. M. & Okamoto, K. (2008). EMBO Rep.9, 63–69. [DOI] [PMC free article] [PubMed]
- Li, J., Qian, X., Hu, J. & Sha, B. (2009). J. Biol. Chem.284, 23852–23859. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Mills, R. D., Trewhella, J., Qiu, T. W., Welte, T., Ryan, T. M., Hanley, T., Knott, R. B., Lithgow, T. & Mulhern, T. D. (2009). J. Mol. Biol.388, 1043–1058. [DOI] [PubMed]
- Neupert, W. & Herrmann, J. M. (2007). Annu. Rev. Biochem.76, 723–749. [DOI] [PubMed]
- Sickmann, A., Reinders, J., Wagner, Y., Joppich, C., Zahedi, R., Meyer, H. E., Schonfisch, B., Perschil, I., Chacinska, A., Guiard, B., Rehling, P., Pfanner, N. & Meisinger, C. (2003). Proc. Natl Acad. Sci. USA, 100, 13207–13212. [DOI] [PMC free article] [PubMed]
- Sollner, T., Griffiths, G., Pfaller, R., Pfanner, N. & Neupert, W. (1989). Cell, 59, 1061–1070. [DOI] [PubMed]
- Sollner, T., Pfaller, R., Griffiths, G., Pfanner, N. & Neupert, W. (1990). Cell, 62, 107–115. [DOI] [PubMed]
- Steger, H. F., Sollner, T., Kiebler, M., Dietmeier, K. A., Pfaller, R., Trulzsch, K. S., Tropschug, M., Neupert, W. & Pfanner, N. (1990). J. Cell Biol.111, 2353–2363. [DOI] [PMC free article] [PubMed]
- Wu, Y. & Sha, B. (2006). Nature Struct. Mol. Biol.13, 589–593. [DOI] [PubMed]
- Yamamoto, H., Fukui, K., Takahashi, H., Kitamura, S., Shiota, T., Terao, K., Uchida, M., Esaki, M., Nishikawa, S., Yoshihisa, T., Yamano, K. & Endo, T. (2009). J. Biol. Chem.284, 31635–31646. [DOI] [PMC free article] [PubMed]
- Yamano, K., Yatsukawa, Y. I., Esaki, M., Hobbs, A. E. A., Jensen, R. E. & Endo, T. (2008). J. Biol. Chem.283, 3799–3807. [DOI] [PubMed]
- Young, J. C., Hoogenraad, N. J. & Hartl, F. U. (2003). Cell, 112, 41–50. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Tom71 complexed with Hsp C-terminus, 3lca