When compared with their E. coli homologue, two X-ray crystal structures of T. maritima endonuclease IV differing in the composition of the trinuclear metal site point to the importance of the trinuclear site and its modulation among species to the function of this enzyme in the AP endonuclease IV family.
Keywords: apurinic/apyrimidinic endonucleases, endonuclease IV, DNA-repair proteins, Thermotoga maritima
Abstract
The most frequent lesion in DNA is at apurinic/apyrimidinic (AP) sites resulting from DNA-base losses. These AP-site lesions can stall DNA replication and lead to genome instability if left unrepaired. The AP endonucleases are an important class of enzymes that are involved in the repair of AP-site intermediates during damage-general DNA base-excision repair pathways. These enzymes hydrolytically cleave the 5′-phosphodiester bond at an AP site to generate a free 3′-hydroxyl group and a 5′-terminal sugar phosphate using their AP nuclease activity. Specifically, Thermotoga maritima endonuclease IV is a member of the second conserved AP endonuclease family that includes Escherichia coli endonuclease IV, which is the archetype of the AP endonuclease superfamily. In order to more fully characterize the AP endonuclease family of enzymes, two X-ray crystal structures of the T. maritima endonuclease IV homologue were determined in the presence of divalent metal ions bound in the active-site region. These structures of the T. maritima endonuclease IV homologue further revealed the use of the TIM-barrel fold and the trinuclear metal binding site as important highly conserved structural elements that are involved in DNA-binding and AP-site repair processes in the AP endonuclease superfamily.
1. Introduction
DNA replication and repair processes are essential for maintaining the fidelity and genomic stability required for life. Abasic or apurinic/apyrimidinic (AP) sites are the most common DNA lesions that occur as a result of DNA-base losses. AP sites are generated by the action of damage-specific DNA glycosylases that remove mismatched and modified bases from the damaged DNA (Krokan et al., 1997 ▶; Cunningham, 1997 ▶; Mol et al., 2000 ▶). AP sites can also be generated by direct interactions between DNA and reactive oxygen species, spontaneous hydrolysis, chemical toxins and radiation (Hutchinson, 1985 ▶; Lindahl et al., 1997 ▶; Huffman et al., 2005 ▶). These AP-site lesions can stall DNA replication and if left unrepaired can be mutagenic, leading to genome instability (Loeb & Preston, 1986 ▶; Sobol et al., 2003 ▶). The repair of AP-site intermediates is initiated by a class of enzymes that are referred to as AP endonucleases (Doetsch & Cunningham, 1990 ▶). During damage-general DNA base-excision repair (BER), these enzymes hydrolytically cleave the 5′-phosphodiester bond at an AP site to generate a free 3′-hydroxyl group and a 5′-terminal sugar phosphate (Bailly & Verly, 1989 ▶; Levin & Demple, 1990 ▶). Following cleavage of the DNA backbone, a DNA deoxyribophosphodiesterase enzyme removes the terminal 5′ sugar phosphate, resulting in a single-nucleotide gap that can be processed by a DNA polymerase and a DNA ligase to complete the repair process (Barzilay & Hickson, 1995 ▶; Levin et al., 1988 ▶).
The AP endonuclease superfamily is comprised of two conserved families of enzymes that catalyze the first damage-general step of BER by cleaving the DNA backbone immediately 5′ to an AP-site (Aravind et al., 1999 ▶). The first enzyme family is typified by exonuclease III (ExoIII; Doetsch & Cunningham, 1990 ▶) from Escherichia coli and the homologous APE-1 (also known as APEX1; Demple et al., 1991 ▶) enzyme in humans. The second conserved AP endonuclease family is typified by the functionally related but structurally distinct endonuclease IV (EndoIV; Ramotar, 1997 ▶; Hosfield et al., 1999 ▶) from E. coli, which is considered to be the archetype for the AP endonuclease superfamily. The thermally stable endonuclease IV homologue from the thermophilic bacterium Thermotoga maritima (Haas et al., 1999 ▶; Hughes et al., 2009 ▶) described here is a member of the second conserved AP endonuclease family. Based on functional homology within the AP endonuclease superfamily, the first damage-general step of BER in which AP-site intermediates are processed is highly conserved from bacteria to humans and archaea (Mol et al., 2000 ▶; Huffman et al., 2005 ▶; Hitomi et al., 2007 ▶). Thus, the endonuclease IV enzymes play a major role in cellular DNA-repair pathways by processing AP-site intermediates.
In addition to AP endonuclease activity, the endonuclease IV enzymes also possess additional 3′-processing activities (Demple et al., 1986 ▶), act as 3′-to-5′ exonucleases on nicked substrates (Kerins et al., 2003 ▶; Ishchenko et al., 2005 ▶) and catalyze nucleotide-incision repair (Ischenko & Saparbaev, 2002 ▶; Ishchenko et al., 2003 ▶), bypassing base removal by glycosylases. Specifically, the T. maritima endonuclease IV homologue has been shown to have AP endonuclease activity and also possesses both phosphomonoesterase and phosphodiesterase repair activities on 3′ DNA-blocking groups such as 3′-phosphates, 3′-phosphoglycolates and 3′-trans-4-hydroxy-2-pentenal-5-phosphates (Haas et al., 1999 ▶) at DNA-strand breaks that result from oxidative DNA damage (Demple et al., 1986 ▶). The T. maritima endonuclease IV exhibits enzymatic activity at both low and high temperatures and is more thermally stable than the E. coli endonuclease IV, with denaturation temperatures approaching 363 K for the T. maritima enzyme (Haas et al., 1999 ▶).
Previous structural studies of the E. coli endonuclease IV enzyme have revealed the use of the widely occurring TIM-barrel fold for recognition and cleavage of DNA at AP sites. In particular, ultrahigh-resolution structures of the DNA-free E. coli endonuclease IV [PDB codes 1qtw (Hosfield et al., 1999 ▶) and 2nqh (Garcin et al., 2008 ▶)] have shown that the active-site region contains three bound zinc ions that are ligated by conserved amino-acid residues. Structures of E. coli endonuclease IV complexed with AP-DNA substrate (PDB code 2nqj; Garcin et al., 2008 ▶) and AP-DNA product [PDB codes 1qum (Hosfield et al., 1999 ▶) and 2nq9 (Garcin et al., 2008 ▶)] have revealed the active-site geometry in the presence of trinuclear zinc before and after catalysis. These structures further characterized the DNA-binding specificity of the endonuclease IV family of enzymes, leading to a more complete understanding of the catalytic mechanism (Mol et al., 2000 ▶; Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶).
As a result of efforts to clone, overexpress and crystallize multiple endonuclease IV gene-product homologues, we obtained crystals of the endonuclease IV homologue from T. maritima that were suitable for X-ray diffraction experiments (Hughes et al., 2009 ▶). Here, we report two X-ray crystal structures of the T. maritima endonuclease IV homologue in the presence of divalent metal ions bound in the active-site region. Our interest in determining these structures of T. maritima endonuclease IV bound to divalent metal ions was to obtain X-ray structural models for future neutron diffraction studies of the enzyme once larger crystal volumes could be obtained. One structure of the T. maritima endonuclease IV, which was solved to 2.30 Å resolution, contained three zinc ions bound in the active-site region. The second structure, which was solved to 2.36 Å resolution, contained a possible mixed-metal configuration of two cadmium ions and one zinc ion bound in the active-site region. These structures of the T. maritima endonuclease IV enzyme have further established the binding of trinuclear metal ions as a necessary motif for the AP endonuclease family of enzymes. A structural analysis of the T. maritima endonuclease IV active-site regions in the presence of the respective divalent metal ions is presented here.
2. Materials and methods
2.1. Protein expression and purification
A synthetic gene encoding the full-length endonuclease IV homologue (287 amino acids) from T. maritima (Q9WYJ7) was designed with E. coli codon preference by iXpressGenes Inc. and inserted into a pET3a expression plasmid (Novagen) by recombining the BamHI- and NdeI-restricted linear expression vector and the PCR product using in vivo homologous recombination in DH5α competent E. coli as described by Marsic et al. (2008 ▶). Recombinant endonuclease IV from T. maritima was expressed in E. coli and purified as described previously by Hughes et al. (2009 ▶). SDS–PAGE was used to confirm the homogeneity of the purified protein. The purified protein was then concentrated by centrifugation (Millipore Amicon Ultra, 10 000 molecular-weight cutoff) and the protein concentration was determined by UV absorbance (A 280nm = 0.95 for 1 mg ml−1). Prior to crystallization, all concentrated protein samples were filtered using 0.45 µm Ultra-free MC spin filters (Millipore, USA).
2.2. Crystallization
Following purification, the enzyme was prepared in a buffer solution consisting of 0.05 M MOPS pH 8.0 and 0.05 M NaCl and the protein solution was concentrated to 10 mg ml−1 for crystallization. Crystals of endonuclease IV from T. maritima with cadmium and zinc were obtained using the sitting-drop vapor-diffusion method at 295 K in a 96-well Intelli-Plate (Art Robbins) as previously described by Hughes et al. (2009 ▶). In the setup, the sitting-drop well contained 3 µl protein solution mixed with 3 µl reservoir solution [0.1 M Bicine pH 9.0, 20%(w/v) polyethylene glycol MME 550] and 1 µl of a micro-crystalline seeding stock. The volume of the reservoir solution was 100 µl. The seeding stock used for crystallization was the well solution from Crystal Screen HT (Hampton Research) condition G10 (0.05 M cadmium sulfate hydrate, 0.1 M HEPES pH 7.5, 1.0 M sodium acetate trihydrate). After one week, well formed rod-shaped crystals were observed (Hughes et al., 2009 ▶). While zinc was not intentionally included during crystallization, we hypothesize that unknown trace amounts of divalent zinc throughout sample preparation may account for the presence of zinc in the crystal structure of the mixed-metal configuration (see §3). Prior to data collection, the crystals of endonuclease IV from T. maritima with cadmium and zinc were placed momentarily in a reservoir solution containing a cryoprotectant (30% glycerol) and flash-frozen in liquid nitrogen. Crystals of endonuclease IV from T. maritima with zinc were obtained as described above followed by soaking the crystals for 10 min in a reservoir solution containing 10 mM zinc sulfate and 30% glycerol prior to flash-freezing in liquid nitrogen for data collection.
2.3. X-ray diffraction data collection and processing
For data collection, single crystals of endonuclease IV from T. maritima in the presence of the respective divalent metal ions were placed momentarily in a reservoir solution containing cryoprotectant (30% glycerol) and flash-frozen in liquid nitrogen. Diffraction data for T. maritima endonuclease IV with zinc were collected over a range of 70° (1° steps) using a MAR Research MX-300 detector on the SER-CAT 22-BM beamline (λ = 1.00 Å) at the Advanced Photon Source (Chicago, USA). The data were processed to 2.30 Å resolution using MOSFLM (Leslie, 1992 ▶); SCALA (Collaborative Computational Project, Number 4, 1994 ▶) was used to scale and merge the observed data. The crystal of T. maritima endonuclease IV with zinc belonged to the hexagonal space group P61, with unit-cell parameters a = 123.37, b = 123.37, c = 35.30 Å. Diffraction data for T. maritima endonuclease IV with cadmium and zinc were collected over a range of 70° (1° steps) using a MAR Research MX-300 detector on the SER-CAT 22-BM beamline (λ = 1.00 Å) at the Advanced Photon Source (Chicago, USA). The data were processed to 2.36 Å resolution using HKL-2000; SCALEPACK (Otwinowski & Minor, 1997 ▶) was used to scale and merge the observed data. The crystal of T. maritima endonuclease IV with cadmium belonged to the hexagonal space group P61, with unit-cell parameters a = 123.19, b = 123.19, c = 35.34 Å. See Table 1 ▶ for a summary of all X-ray crystallographic data-collection statistics.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| With Zn (PDB code 2x7v) | With Cd and Zn (PDB code 2x7w) | |
|---|---|---|
| Unit-cell parameters (Å) | a = b = 123.37, c = 35.30 | a = b = 123.19, c = 35.34 |
| Space group | P61 | P61 |
| No. of unique reflections | 13865 | 12749 |
| Resolution range (Å) | 35.61–2.30 (2.42–2.30) | 40.32–2.36 (2.44–2.36) |
| Multiplicity | 4.2 (4.1) | 4.3 (4.1) |
| I/σ(I) | 4.5 (2.1) | 13.1 (6.2) |
| Rmerge† (%) | 12.3 (36.0) | 9.5 (17.4) |
| Data completeness (%) | 99.4 (99.9) | 99.3 (95.6) |
| R factor‡ (%) | 14.6 | 15.6 |
| Rfree‡ (%) | 20.0 | 19.6 |
| R.m.s.d. bonds§ (Å) | 0.009 | 0.023 |
| R.m.s.d. angles§ (°) | 1.103 | 0.794 |
| Atoms (non-H) | 2446 | 2501 |
| Solvent molecules | 165 | 209 |
| Ramachandran plot¶ | ||
| Outliers (%) | 0.0 | 0.0 |
| Favored (%) | 98.2 | 98.6 |
| Rotamer outliers (%) | 3.3 | 1.6 |
R
merge = 100 × 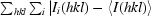
 , where Ii(hkl) is the intensity of reflection hkl and 〈I(hkl)〉 is the mean value of i multiple measurements of reflection hkl.
, where Ii(hkl) is the intensity of reflection hkl and 〈I(hkl)〉 is the mean value of i multiple measurements of reflection hkl.
R factor = 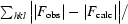
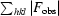 ; R
free is the R factor computed using a 5% subset of the data randomly excluded from structure determination (where F
calc includes all scale factors and corrections for bulk solvent).
; R
free is the R factor computed using a 5% subset of the data randomly excluded from structure determination (where F
calc includes all scale factors and corrections for bulk solvent).
Root-mean-square deviations of bond lengths in angstroms and bond angles in degrees calculated with phenix.refine in the PHENIX program suite.
Ramachandran plot quality assessment using MolProbity.
2.4. Structure determination and refinement
The structures of T. maritima endonuclease IV in the presence of either three zinc ions or the likely mixed-metal configuration of two cadmium ions and one bound zinc ion were both solved by molecular replacement using the Phaser (McCoy et al., 2007 ▶) program from the PHENIX (Adams et al., 2002 ▶) program suite with the known structure of E. coli endonuclease IV (PDB code 1qtw; Hosfield et al., 1999 ▶) as the search model (32% sequence homology to T. maritima endonuclease IV) as described by Hughes et al. (2009 ▶). The molecular-replacement solution for each respective data set was refined using the phenix.refine (Afonine et al., 2005 ▶) program in PHENIX (v.1.6_289). The final model for each structure was obtained after several rounds of maximum-likelihood-based refinement of individual coordinates, atomic displacement parameters (ADP) and occupancies using the phenix.refine program and model building into σA-weighted F o − F c and 2F o − F c electron-density maps using Coot (Emsley & Cowtan, 2004 ▶). In each structure, a total of 286 amino-acid residues were included in the final model; only the C-terminal residue 287 was disordered in the electron-density maps and could not be modeled. The final refinement statistics from phenix.refine for each structure are summarized in Table 1 ▶. MolProbity (Chen et al., 2010 ▶) was used to analyze the stereochemistry of the final models (see Table 1 ▶). Figures of the models were created and rendered with the program PyMOL (DeLano, 2008 ▶).
3. Results
3.1. Overall structure of T. maritima endonuclease IV
Two X-ray crystal structures of the T. maritima endonuclease IV homologue with divalent metal ions bound in the active site were determined by molecular replacement. Final models with good stereochemistry were obtained by completing multiple rounds of model building and maximum-likelihood-based individual coordinate, atomic displacement parameter (ADP) and occupancy refinement against X-ray diffraction data (see Table 1 ▶). One structure of T. maritima endonuclease IV solved to 2.30 Å resolution (R factor = 14.6%; R free = 20.0%) contained three zinc ions bound in the active-site region. The second structure, which was solved to 2.36 Å resolution (R factor = 15.6%; R free = 19.6%), contained a possible mixed-metal configuration of two cadmium ions and one zinc ion bound in the active-site region along with a Bicine buffer molecule. Both structures of T. maritima endonuclease IV in the presence of divalent metal ions are essentially identical (r.m.s.d. = 0.12 Å for the main chain), with the only differences being the radii of the respective metal ions and the presence of a Bicine ligand in the structure containing cadmium; therefore, the structure in the presence of three zinc ions is used to describe the overall three-dimensional structure of the enzyme.
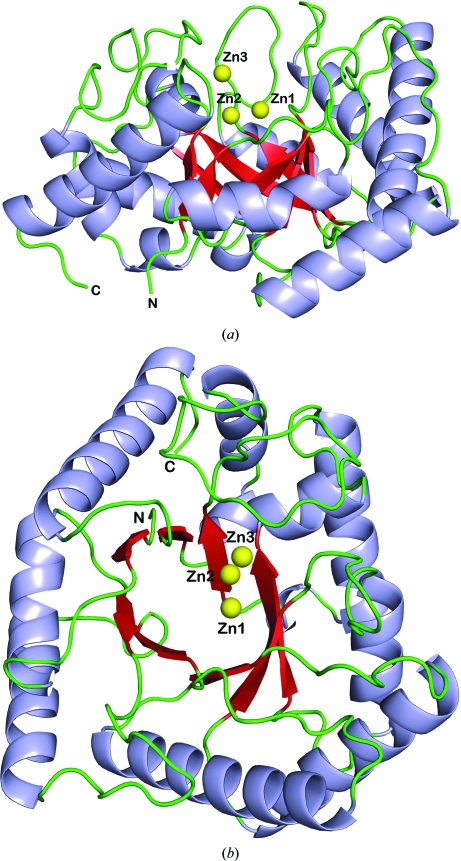
T. maritima endonuclease IV is a single-domain αβ protein in which the secondary-structure elements are arranged as a barrel consisting of eight parallel β-strands that are surrounded by eight peripheral α-helices (see Figs. 1 ▶ a and 1 ▶ b). This eight-stranded αβ-barrel fold was first observed in triose phosphate isomerase (TIM barrel; Banner et al., 1975 ▶) and is a major folding superfamily that occurs in a number of enzymes with diverse functions (Farber & Petsko, 1990 ▶; Reardon & Farber, 1995 ▶). Like other TIM-barrel proteins, all eight peripheral α-helices in the T. maritima endonuclease IV structure are oriented with their N-termini pointing towards the C-terminal face of the β-barrel. A large crescent-shaped pocket is formed from five protruding loops that connect the α-helices and β-sheets at the C-terminal face of the β-barrel. These five loops or R-loops have been shown to be involved in the binding of AP-DNA substrate by binding to the phosphate backbone and providing DNA-base interactions at the AP site (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶). The base of the large pocket forms the active-site region of the enzyme where three zinc ions are bound to absolutely conserved amino-acid residues (see Figs. 1 ▶ a and 1 ▶ b). Previous structures of E. coli endonuclease IV demonstrated how structural features of the TIM-barrel fold, such as the positive helix dipoles pointing toward the C-terminal face of the enzyme and the active-site pocket containing the trinuclear ‘mainly’ zinc binding site, are optimized for DNA binding (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶).
Figure 1.
(a) Ribbon diagram of T. maritima endonuclease IV in the presence of three bound zinc ions (Zn1, Zn2 and Zn3; shown in yellow) as viewed from the side of the αβ-barrel fold, which consists of eight parallel β-strands (shown in red) that are surrounded by eight peripheral α-helices (shown in light blue) and are connected by loop regions (shown in green). (b) Ribbon diagram of T. maritima endonuclease IV in the presence of three bound zinc ions (Zn1, Zn2 and Zn3) as viewed from the top or the C-terminal face of the αβ-barrel fold (colors are the same as noted in a).
3.2. Metal-ion arrangement in T. maritima endonuclease IV
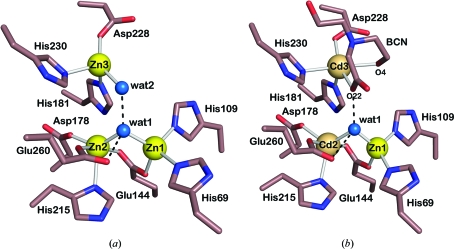
The T. maritima endonuclease IV active site, which is located at the base of the pocket formed by the β-barrel, contains three metal ions that are bound to absolutely conserved amino-acid residues. The active-site regions of T. maritima endonuclease IV bound to three zinc ions and the likely mixed-metal configuration of two cadmium ions, one zinc ion and a Bicine buffer molecule are shown in Figs. 2 ▶(a) and 2 ▶(b), respectively. The locations of all active-site divalent metal ions for each structure were determined based on a thorough analysis of σA-weighted F o − F c electron-density maps calculated using phenix.refine (Afonine et al., 2005 ▶). While ambiguous, our interpretation of the electron density for the mixed-metal configuration as containing two cadmium ions and one zinc ion is in agreement with the refined occupancy and B-factor values for these respective metal sites as discussed below. Strong positive electron-density peaks associated with the positions of divalent zinc ions were observed up to contour levels of 21σ for both Zn1 and Zn3 and of 17σ for Zn2. In the cadmium- and zinc-containing structure strong positive electron-density peaks associated with the positions of divalent metal ions were observed up to contour levels of 15σ for both Cd2 and Cd3 and of 8σ for Zn1. The zinc and cadmium divalent metal ions were introduced into the crystals either through soaking experiments (zinc-containing structure) or through crystallization screening and seeding experiments (cadmium- and zinc-containing structure) as described above in §2. In the structure of T. maritima endonuclease IV, the bound zinc ions refined to occupancy values of 82, 72 and 92% for Zn1, Zn2 and Zn3, respectively, with refined B-factor values of 24.6, 30.4 and 30.5 Å2, respectively. In the structure bound to cadmium and zinc, the cadmium ions refined to occupancy values of 100 and 92% for Cd2 and Cd3, respectively, with refined B-factor values of 10.5 and 10.7 Å2, respectively, and the Zn1 zinc ion refined to an occupancy value of 44%, with a refined B-factor value of 13.5 Å2. The Bicine buffer molecule bound adjacent to the Cd3 metal site refined to an occupancy value of 97%. As the crystals used for determining the cadmium and zinc structure were grown by seeding techniques in the presence of cadmium as previously described in our crystallization report (Hughes et al., 2009 ▶), the metal ion bound in site 1 could also be a partially occupied cadmium ion. The refined occupancy value for a cadmium ion (Cd1) bound in metal site 1 was 35% (data not shown). Additional refinements were also completed with zinc ions in both metal sites 2 and 3 in order to confirm the presence of the heavier cadmium ions. An analysis of σA-weighted F o − F c electron-density maps along with refined ADP and occupancy values confirmed that metal sites 2 and 3 are most likely to be occupied by cadmium ions (data not shown). In particular, when a zinc ion was refined in metal site 2 additional F o − F c electron density was present around the position of the refined zinc, providing further evidence that metal site 2 is likely to be occupied by the heavier cadmium ion (data not shown). While zinc was not intentionally included in the cell-culture media, purification buffers or crystallization screening reagents, the presence of unknown trace amounts of divalent zinc throughout sample preparation may account for the presence of a partially bound zinc ion in metal site 1.
Figure 2.
(a) Active-site region of T. maritima endonuclease IV in the presence of three bound zinc ions (Zn1, Zn2 and Zn3) and two water molecules (wat1 and wat2). Hydrogen-bonding interactions are shown by dashed lines. (b) Active-site region of T. maritima endonuclease IV in the presence of two bound cadmium ions (Cd2 and Cd3), one bound zinc ion (Zn1) and two water molecules (wat1 and wat2). The active-site region also contains a Bicine buffer molecule (BCN) that is bound adjacent to Cd3. Hydrogen-bonding interactions are shown as dashed lines.
In the structure of T. maritima endonuclease IV bound to three zinc ions, both Zn1 and Zn2 are located at the base of the pocket formed by the β-barrel and are separated by 3.37 Å. The third bound zinc ion, Zn3, is displaced away from the base of the pocket and from both Zn1 and Zn2, with a Zn1–Zn3 distance of 5.94 Å and a Zn2–Zn3 distance of 4.85 Å (see Fig. 2 ▶ a). Based on its location in the active-site pocket, Zn3 is more solvent-accessible than both Zn1 and Zn2. The Zn1 ion is coordinated by the side chains of His69, His109, Glu144 and water molecule 1 (wat1) in a tetrahedral arrangement. Both the side chain of Glu144 and wat1 bridge between the bound Zn1 and Zn2 ions. It has been hypothesized that the bridging water molecule (wat1) is likely to be the nucleophile in the phosphodiester-hydrolysis reaction once it has been deprotonated (Garcin et al., 2008 ▶; Ivanov et al., 2007 ▶). The Zn2 ion is coordinated by the side chains of Glu144, Asp178, Glu260, His215 and wat1 in a trigonal bipyramidal arrangement in which Glu144 and Glu260 are axial ligands and Asp178, Glu260 and wat1 are equatorial ligands. The displaced Zn3 ion is coordinated in a distorted tetrahedral arrangement to the side chains of His181, Asp228, His230 and water molecule 2 (wat2) as shown in Fig. 2 ▶(a). A summary of the bond distances in the active-site region of T. maritima endonuclease IV bound to zinc is given in Table 2 ▶.
Table 2. Bond distances for the trinuclear metal sites in the active sites of T. maritima endonuclease IV and E. coli endonuclease IV (PDB code 1qtw; Hosfield et al., 1999 ▶).
(a).
T. maritima endonuclease IV.
| Structure with Zn | Structure with Cd and Zn | |
|---|---|---|
| Metal site 1 | ||
| Zn1—His109 N∊2 | 2.19 | 2.05 |
| Zn1—His69 N∊2 | 2.26 | 2.08 |
| Zn1—Glu144 O∊1 | 2.23 | 2.08 |
| Zn1—wat1 | 2.26 | 2.05 |
| Zn1 geometry | Tetrahedral | Tetrahedral |
| Metal site 2 | ||
| Zn2/Cd2—Glu144 O∊2 | 2.26 | 2.11 |
| Zn2/Cd2—Glu260 O∊1 | 2.18 | 2.28 |
| Zn2/Cd2—His215 Nδ1 | 2.37 | 2.36 |
| Zn2/Cd2—Asp178 Oδ1 | 2.09 | 2.44 |
| Zn2/Cd2—wat1 | 2.22 | 2.45 |
| Zn2/Cd2 geometry | Trigonal bipyramidal | Trigonal bipyramidal |
| Metal site 3 | ||
| Zn3/Cd3—Asp228 Oδ2 | 2.38 | 2.33 |
| Zn3/Cd3—His230 N∊2 | 2.21 | 2.51 |
| Zn3/Cd3—His181 N∊2 | 2.16 | 2.31 |
| Zn3—wat2 | 2.41 | — |
| Cd3—Bicine O22 | — | 2.42 |
| Cd3—Bicine O4 | — | 2.54 |
| Zn3/Cd3 geometry | Distorted tetrahedral | Distorted trigonal bipyramidal |
(b).
E. coli endonuclease IV.
| Structure with Zn (PDB code 1qtw; Hosfield et al., 1999 ▶) | |
|---|---|
| Metal site 1 | |
| Zn1—His109 N∊2 | 1.98 |
| Zn1—His69 N∊2 | 2.01 |
| Zn1—Glu145 O∊2 | 2.07 |
| Zn1—wat1 | 1.92 |
| Zn1 geometry | Tetrahedral |
| Metal site 2 | |
| Zn2—Glu145 O∊1 | 2.11 |
| Zn2—Glu261 O∊1 | 2.21 |
| Zn2—His216 Nδ1 | 2.07 |
| Zn2—Asp179 Oδ2 | 1.98 |
| Zn2—wat1 | 2.00 |
| Zn2 geometry | Trigonal bipyramidal |
| Metal site 3 | |
| Zn3—Asp229 Oδ1 | 2.19 |
| Zn3—Asp229 Oδ2 | 2.34 |
| Zn3—His231 N∊2 | 1.98 |
| Zn3—His182 N∊2 | 2.00 |
| Zn3—wat2 | 2.01 |
| Zn3—wat3 | 2.39 |
| Zn3 geometry | Distorted trigonal bipyramidal |
The overall active-site structure of T. maritima endonuclease IV bound to cadmium and zinc is quite similar to the structure bound to zinc: both Zn1 and Cd2 are also located at the base of the β-barrel and are separated by 3.25 Å. Similarly, the third bound ion, Cd3, is also displaced away from the base of the pocket and from both Zn1 and Cd2, with a Zn1—Cd3 distance of 5.85 Å and a Cd2—Cd3 distance of 5.14 Å (see Fig. 2 ▶ b). The coordination of amino-acid side chains around metal sites 1 and 2 in the presence of zinc and cadmium, respectively, is identical to that of the zinc-bound structure; Zn1 is coordinated in a tetrahedral arrangement and Cd2 is coordinated in a trigonal bipyramidal arrangement. The coordination of Cd3 in the third metal site is different from the zinc-bound structure owing to the displacement of wat2 by the Bicine buffer molecule. The Cd3 ion is coordinated in a distorted trigonal bipyramidal arrangement to the side chains of His181, Asp228 and His230 and to the anionic Bicine buffer molecule, which forms a bidentate ligand to Cd3 (see Fig. 2 ▶ b) through Bicine O4 and O22. The complexation of divalent transition-metal ions with zwitterionic buffers such as Bicine is well known (Corfu & Bin Song, 1992 ▶). A summary of the bond distances in the active-site region of T. maritima endonuclease IV bound to two cadmium ions and one zinc ion is given in Table 2 ▶.
3.3. Structure comparison of T. maritima endonuclease IV and E. coli endonuclease IV
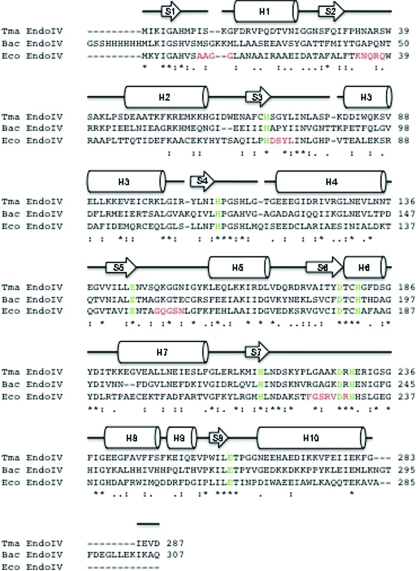
Endonuclease IV from E. coli is considered to be the archetype for the second conserved AP endonuclease superfamily of enzymes (Ramotar, 1997 ▶; Hosfield et al., 1999 ▶). It has been shown that T. maritima endonuclease IV is a member of the second conserved AP endonuclease family as a result of its possessing enzymatic activities that are characteristic of the endonuclease IV family of DNA-repair enzymes, including AP endonuclease activity and repair activities on 3′-phosphates, 3′-phosphoglycolates and 3′-trans-4-hydroxy-2-pentenal-5-phosphates (Haas et al., 1999 ▶). Based on a primary sequence alignment, T. maritima endonuclease IV has only 32% sequence identity to E. coli endonuclease IV (see Fig. 3 ▶). A superposition of the ribbon structures of both T. maritima endonuclease IV and E. coli endonuclease IV (PDB code 1qtw; Hosfield et al., 1999 ▶) bound to trinuclear zinc is shown in Fig. 4 ▶(a). The secondary-structure matching program within Coot was used for all structure superpositions (Krissinel & Henrick, 2004 ▶). Even with such a low sequence identity, the overall structural folds of these enzymes are remarkably similar, with an r.m.s.d. of 1.65 Å for main-chain atoms. Both enzymes have the characteristic TIM-barrel fold (Marsic et al., 2008 ▶) consisting of eight parallel β-strands that are surrounded by eight peripheral α-helices. The active-site pocket formed from the central β-barrel that contains the trinuclear zinc site in each enzyme is also structurally conserved in both T. maritima endonuclease IV and E. coli endonuclease IV.
Figure 3.
Amino-acid sequence alignment of T. maritima endonuclease IV (287 residues), E. coli endonuclease IV (285 residues) and Bacillus anthracis endonuclease IV (307 residues). The amino-acid sequences of T. maritima endonuclease IV (Tma EndoIV; Q9WYJ7), E. coli endonuclease IV (Eco EndoIV; P0A6C1) and B. anthracis endonuclease IV (Bac EndoIV; Q81LV1) from PDB code 1xp3 (M. J. Fogg, V. M. Levdikov, E. V. Blagova, J. A. Brannigan, A. J. Wilkinson & K. S. Wilson, unpublished work) were aligned with the program ClustalW (Thompson et al., 1994 ▶). The secondary-structure elements of T. maritima endonuclease IV bound to zinc are shown aligned with its primary sequence, where H and S represent α-helices and β-sheets, respectively. The absolutely conserved amino-acid residues in the active-site regions of Tma EndoIV, Eco EndoIV and Bac EndoIV that are involved in the binding of divalent metal ions are colored green. The five R-loop regions (residues 10–13, 34–38, 70–73, 149–153 and 224–230) in E. coli endonuclease IV that have been shown to be involved in DNA binding are colored in red below the corresponding residues in both T. maritima endonuclease IV and B. anthracis endonuclease IV. The ‘*’ symbol indicates identical residues in the sequence alignment, the ‘:’ symbol indicates conserved substitutions in the sequence alignment and the ‘.’ symbol indicates semi-conserved substitutions in the sequence alignment.
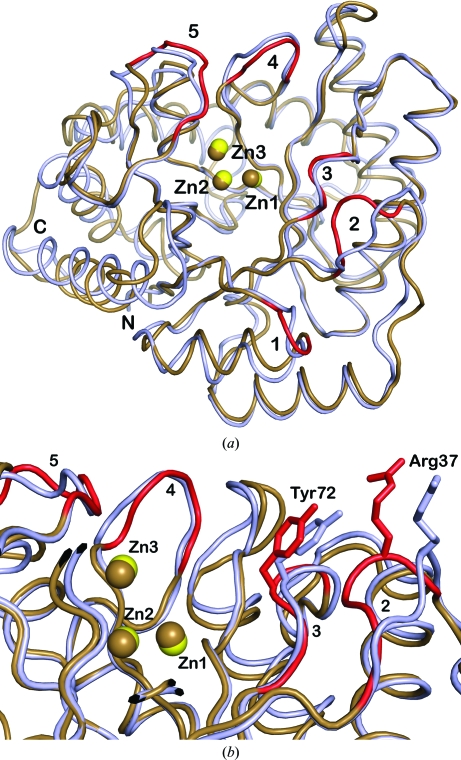
Figure 4.
(a) Backbone superpostion of the T. maritima endonuclease IV and E. coli endonuclease IV (PDB code 1qtw; Hosfield et al., 1999 ▶) structures both in the presence of three bound zinc ions (Zn1, Zn2 and Zn3). The T. maritima endonuclease IV structure is shown in light blue, with the three bound zinc ions colored yellow. The E. coli endonuclease IV structure is shown in brown, with the three bound zinc ions also colored brown. The five R-loop regions (residues 10–13, 34–38, 70–73, 149–153 and 224–230) in the E. coli endonuclease IV structure are colored red and labeled 1–5, respectively. These five loop regions connect the α-helices and β-sheets at the C-terminal face of the β-barrel and have also been shown to be involved in the binding to AP-DNA substrate (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶). Although it only has 32% sequence identity to E. coli endonuclease IV, the structure of T. maritima endonuclease IV has the same overall αβ-barrel fold consisting of eight parallel β-strands that are surrounded by eight peripheral α-helices. (b) Backbone superposition of the T. maritima endonuclease IV and E. coli endonuclease IV structures, showing a comparison of the conformations of residues Arg37 and Tyr72. The colors of the endonuclease IV enzymes and bound zinc ions are the same as described in (a). Four R-loop regions (residues 34–38, 70–73, 149–153 and 224–230) in the E. coli endonuclease IV structure are colored red and labeled 2–5, respectively. The positions of Arg37 and Tyr72 in E. coli endonuclease IV are colored red, corresponding to their respective locations in R-loop 2 and R-loop 3. The positions of Arg37 and Tyr72 in the T. maritima endonuclease IV structure are colored light blue.
Structural differences between these enzymes are present within the five protruding loops (R-loops) that connect the α-helices and β-sheets at the C-terminal face of the β-barrel. The orientations of these loop regions in T. maritima endonuclease IV are shifted slightly with respect to their positions in E. coli endonuclease IV, with the largest shift (approximately 5 Å) being present in R-loop 2. Smaller main-chain R-loop conformational differences are present between the two enzymes in R-loop 3 (1.05 Å shift), R-loop 4 (1.33 Å shift) and R-loop 4 (1.36 Å shift). It is likely that primary sequence differences within these R-loop regions (see Fig. 3 ▶) and crystal lattice contacts involving these R-loop regions account for these structural differences between T. maritima endonuclease IV and E. coli endonuclease IV. In E. coli endonuclease IV, conformational changes in these loop regions have been shown to be important for binding AP-DNA substrate. Specifically, residues Arg37 (R-loop 2) and Tyr72 (R-loop 3) in E. coli endonuclease IV are involved in important minor-groove-penetrating interactions with AP-DNA substrate that result from conformational changes in these respective R-loops (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶). In the T. maritima endonuclease IV structure, the conserved Arg37 and Tyr72 residues are also located within R-loop 2 and R-loop 3, respectively. In the T. maritima endonuclease IV structure the Arg37 side chain is rotated by approximately 30° and is located 4.7 Å away from the center of the β-barrel with respect to the location of Arg37 in E. coli endonuclease IV. The position of Tyr72 in the T. maritima endonuclease IV structure is in a similar location to that of Tyr72 in E. coli endonuclease IV. A comparison of the conformations of residues Arg37 (R-loop 2) and Tyr72 (R-loop 3) in both the T. maritima endonuclease IV and E. coli endonuclease IV structures is shown in Fig. 4 ▶(b). In addition to the differences in the R-loop regions, structural differences between the two enzymes are also present in the α-helical regions in their C-terminal regions. The C-terminal α-helical regions (residues 241–249 and 267–282) of T. maritima endonuclease IV are shifted outward and away from the central β-barrel region by approximately 2–5 Å in comparison to the positions of these C-terminal regions in E. coli endonuclease IV.
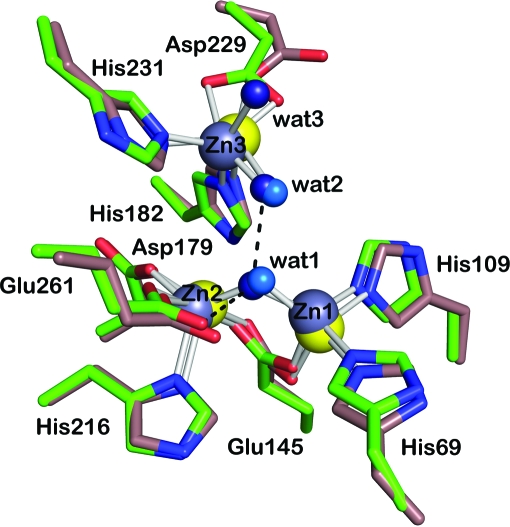
Interestingly, even with such a low sequence identity the residues involved in forming the trinuclear metal binding site are absolutely conserved in both T. maritima endonuclease IV and E. coli endonuclease IV (see Fig. 3 ▶). A highly conserved region (residues 171–189) within the second family of AP endonucleases is also present in T. maritima endonuclease IV. Analogous to the structure of E. coli endonuclease IV (Hosfield et al., 1999 ▶), this conserved region in T. maritima endonuclease IV also provides important packing and hydrogen-bonding interactions within the central β-barrel that serve to stabilize the relative positions of Zn2 and Zn3 within the trinuclear metal site. In the E. coli endonuclease IV structure, Zn1 and Zn2 are located at the base of the β-barrel and are separated by 3.40 Å. The third metal site, Zn3, is displaced from the first two metal centers, with a Zn1–Zn3 distance of 5.43 Å and a Zn2–Zn3 distance of 4.67 Å. The active-site region of E. coli endonuclease IV is shown in Fig. 5 ▶. A summary of the bond distances in the active-site region of E. coli endonuclease IV (Hosfield et al., 1999 ▶) bound to trinuclear zinc is shown in Table 2 ▶. The overall active-site structure and coordination arrangement around the Zn1 and Zn2 metal ions are nearly identical in both E. coli endonuclease IV and T. maritima endonuclease IV. The only distinct difference within the active-site region between the enzymes is the coordination and arrangement around Zn3. In the E. coli endonuclease IV active site the Zn3 ion is coordinated to the side chains of His182, His231 and Asp229 and water molecule 2 (wat2) in a distorted trigonal bipyramidal arrangement. The two side-chain carboxyl O atoms of Asp229 form a unibidentate ligand to Zn3, in contrast to the active site of T. maritima endonuclease IV, where only one of the Asp228 carboxyl O atoms can coordinate to Zn3. In the T. maritima endonuclease IV structure a conformational change in R-loop 5 (as discussed above and shown in Fig. 4 ▶) repositions the side chain of Asp228 so that only one of the carboxyl O atoms can coordinate to Zn3. Also, there is an additional water molecule (wat3) located 2.4 Å away from Zn3 in E. coli endonuclease IV that is not present in the T. maritima endonuclease IV structure (see Fig. 5 ▶). In the structure of the T. maritima endonuclease IV structure bound to cadmium and zinc the Cd3 metal ion is also coordinated to the side chain of Asp228 through one of the carboxyl O atoms (see Fig. 2 ▶ b). The bound Bicine molecule coordinates the Cd3 metal ion and displaces the conserved water molecule (wat2) observed in both the T. maritima and E. coli endonuclease IV structures along with a second water molecule (wat3) that is only observed in the E. coli endonuclease IV structure. No other significant structural differences in the active-site region were observed between T. maritima endonuclease IV bound to cadmium and zinc and E. coli endonuclease IV bound to zinc.
Figure 5.
Active-site region of E. coli endonuclease IV with C atoms shown in green (PDB code 1qtw; Hosfield et al., 1999 ▶) in the presence of three bound zinc ions (Zn1, Zn2 and Zn3, shown in gray) and three water molecules (wat1, wat2 and wat3, shown in dark blue). Hydrogen-bonding interactions are shown by dashed lines. All labels correspond to the E. coli endonuclease IV structure. The active-site region of T. maritima endonuclease IV from Fig. 2 ▶(a) in the presence of three bound zinc ions (Zn1, Zn2 and Zn3 shown in yellow) and two water molecules (wat1 and wat2 shown in light blue) is shown superimposed with C atoms in brown.
4. Discussion
The X-ray crystal structures of the T. maritima endonuclease IV homologue reveal divalent metal ions bound in the active-site region. Consequently, observation of the structural interplay between the protein and selective metal coordination further characterizes the AP endonuclease family of enzymes. One structure of T. maritima endonuclease IV contains three zinc ions bound in the active-site region. The second structure is likely to contain a mixed-metal configuration of two cadmium ions and one zinc ion bound in the active-site region. These structures of T. maritima endonuclease IV further revealed the use of the eight-stranded αβ-barrel fold (TIM barrel; Banner et al., 1975 ▶) for DNA binding and catalysis in the AP endonuclease superfamily. Furthermore, the presence of a trinuclear metal binding site in the active site of the respective T. maritima endonuclease IV structures further establishes the binding of trinuclear metal ions as a necessary motif for the AP endonuclease family of enzymes. Based on the previously characterized AP endonuclease activity (Haas et al., 1999 ▶) and the overall structural similarity of the T. maritima endonuclease IV and E. coli endonuclease IV enzymes, it is likely that T. maritima endonuclease IV binds AP-DNA substrate in a similar manner to that previously described for E. coli endonuclease IV (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶). DNA binding to T. maritima endonuclease IV is likely to be mediated by interactions between the R-loop regions that emanate from the C-terminal end of the TIM-barrel fold and the negatively charged DNA phosphate backbone. The conserved TIM-barrel fold of T. maritima endonuclease IV positions the five R-loop regions to form an interface that resembles the known AP-DNA-binding interface for E. coli endonuclease IV (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶). In T. maritima endonuclease IV the conserved residues Arg37 (R-loop 2) and Tyr72 (R-loop 3) may interact with AP-DNA substrate in a similar manner to that described for E. coli endonuclease IV, where R-loop conformational changes provide specific interactions with DNA to promote DNA bending. These residues have been shown to interact specifically with the minor groove of the AP-DNA substrate, in which residue Tyr72 base-stacks with the base pair 5′ to the AP site to occupy the gap left by the extrahelical AP site and residue Arg37 forms a base-stacking interaction with the base pair 3′ to the AP site (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶). Thus, the key structural features that have been shown to be responsible for AP-RNA recognition and binding of the scissile phosphate to the trinuclear metal site in the active-site region in E. coli endonuclease IV (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶) are also present in T. maritima endonuclease IV. In the absence of an AP-DNA substrate bound to T. maritima endonuclease IV, further structural studies of complexes with substrate will be needed to fully characterize the specific DNA-binding interactions related to the recognition of AP-DNA.
Previous mutational (Ishchenko et al., 2006 ▶; Yang et al., 1999 ▶), structural (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶) and molecular-dynamics simulation (Ivanov et al., 2007 ▶) studies on the E. coli endonuclease IV enzyme have shown that all three zinc ions participate directly in a three-metal-ion mechanism in which an adjacent hydroxide bridging Zn1 and Zn2 is positioned to be the attacking nucleophile for cleavage of the scissile phosphate bond of AP-DNA. It has been proposed that residue Glu261 in E. coli endonuclease IV (conserved residue Glu260 in T. maritima endonuclease IV) participates in activating the hydroxide nucleophile (Garcin et al., 2008 ▶; Ivanov et al., 2007 ▶). Even with such a low sequence identity the residues that are involved in forming the trinuclear metal binding site are absolutely conserved in both the T. maritima endonuclease IV and E. coli endonuclease IV structures (see Figs. 2 ▶ a, 2 ▶ b and 5 ▶).
A structural analysis of the three active-site regions of T. maritima endonuclease IV bound to either trinuclear zinc or the possible mixed-metal configuration of two cadmium ions and one zinc ion and E. coli endonuclease IV bound to trinuclear zinc revealed that the active-site structure and coordination to metal sites 1 and 2 is conserved in all three structures (see Figs. 2 ▶ a, 2 ▶ b and 5 ▶). However, occupancy refinements of the respective metal ions in the T. maritima endonuclease IV structures show that metal site 1 may not be fully occupied when divalent cadmium is present in the active site in both metal sites 2 and 3. In the structure bound to cadmium and zinc the refined occupancy value for metal site 1 (Zn1, 44%) suggests that a zinc ion (or possibly a cadmium ion) does not completely occupy this site, whereas in the structure of T. maritima endonuclease IV bound to trinuclear zinc the Zn1 metal ion refined to a higher occupancy value (Zn1, 82%), suggesting that metal site 1 is nearly fully occupied by zinc. It has been shown that cadmium can compete with divalent zinc binding to proteins. Specifically, cadmium has a preference for cysteine, glutamate, aspartate and histidine ligands in tetrahedral geometries (McMurray & Tainer, 2003 ▶). It has been suggested that heavy-metal-ion toxicity may disrupt the metal-site geometry as cadmium ions can replace zinc ions to become an environmental mutagen (Garcin et al., 2008 ▶; McMurray & Tainer, 2003 ▶). In the T. maritima endonuclease IV active site, metal site 1 coordinates to the side chains of His69, His109 and Glu144 and water molecule 1 (wat1) in a tetrahedral arrangement and is in an ideal geometry for either zinc or cadmium binding. The presence of a partially bound zinc or cadmium ion in metal site 1 does not disrupt the coordination geometry of the partially occupied metal site 1 in comparison to either the structure of T. maritima endonuclease IV or E. coli endonuclease IV with zinc bound in site 1. Further studies involving the effects of heavy-metal-ion binding to T. maritima endonuclease IV are necessary to understand the impact of metal binding with respect to catalytic activity. The observation that metal site 1 is also partially inaccessible to solvent cannot be ruled out as a possible explanation for the low-occupancy binding of zinc to metal site 1 in the mixed-metal configuration of T. maritima endonuclease IV.
As initially shown in the structure of wild-type E. coli endonuclease IV (Hosfield et al., 1999 ▶), the third metal site (Zn3) is mostly solvent-accessible and is coordinated in a distorted trigonal bipyramidal arrangement (see Fig. 5 ▶). Additional structures of phosphate-bound (E261Q mutant; distorted trigonal bipyramidal Zn3 geometry) and both AP-DNA substrate (E261Q mutant; trigonal Zn3 geometry) and product complexes (both wild-type and Y72A mutant; trigonal bipyramidal Zn3 geometry) of E. coli endonuclease IV have suggested that the coordination geometry around metal site 3 varies extensively throughout catalysis (Hosfield et al., 1999 ▶; Garcin et al., 2008 ▶). In the structures of T. maritima endonuclease IV, metal site 3 is coordinated in a distorted tetrahedral arrangement when bound to zinc, whereas metal site 3 is coordinated in a distorted trigonal bipyramidal arrangement when bound to cadmium and Bicine (see Figs. 2 ▶ a and 2 ▶ b). The coordination around metal site 3 in the zinc-bound structure closely resembles the structure of the E. coli endonuclease IV E261Q mutant bound to phosphate. The phosphate O atom coordinating Zn3 in the E. coli endonuclease IV structure is located in nearly the same position as water molecule 2 in T. maritima endonuclease IV. Additionally, the coordination around metal site 3 in the mixed-metal configuration of the T. maritima endonuclease IV structure bound to cadmium and Bicine resembles the structure of the E. coli endonuclease IV E261Q mutant–DNA substrate complex. In both structures the Zn3 coordination sphere is incomplete as the coordinating side-chain residues are arranged in a trigonal geometry in which only one of the carboxyl atoms in the side chains of Asp229 in the E. coli structure and Asp228 in the T. maritima structure coordinate to Zn3. In the structure of the E. coli endonuclease IV E261Q mutant–DNA substrate complex both the 3′-O atom of the intact phosphodiester bond and the unesterified phosphate O atom are constrained, which prevents completion of the Zn3 coordination sphere. In the T. maritima endonuclease IV structure bound to cadmium and Bicine, the Zn3 coordination sphere is completed by a bidentate interaction with Bicine through O4 and O22. Further structural studies of T. maritima endonuclease IV will be needed in order to assess the significance of these structural observations in relation to catalysis. However, these results provide further evidence that the coordination geometry around metal site 3 is quite variable and are in agreement with previous structural results on E. coli endonuclease IV that suggest that the Zn3 coordination sphere is variable throughout the catalytic mechanism in order for these enzymes to possess such remarkable substrate recognition and specificity toward AP-site DNA. Thus, these structural studies of the T. maritima endonuclease IV homologue further reveal the use of the TIM-barrel fold and the trinuclear metal binding site as key structural elements involved in AP-site repair processes that are highly conserved from bacteria to humans and archaea. Future activity and structural studies of T. maritima endonuclease IV in complex with both AP-DNA and catalytic divalent metal ions, including toxic heavy-metal ions such as cadmium, will be important for further characterization of the role of the AP endonuclease family of enzymes in the BER pathway.
Supplementary Material
PDB reference: T. maritima endonuclease IV, 2x7v
PDB reference: 2x7w
Acknowledgments
We thank Dr Jenny P. Glusker for a critical review of the manuscript and valuable comments. This research was sponsored by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory (ORNL), managed by UT-Battelle LLC for the US Department of Energy under Contract No. DE-AC05-00OR22725. The research at Oak Ridge National Laboratory’s Center for Structural Molecular Biology (CSMB) was supported by the Office of Biological and Environmental Research, using facilities supported by the US Department of Energy, managed by UT-Battelle LLC under contract No. DE-AC05-00OR22725.
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Afonine, P. V., Grosse-Kunstleve, R. W. & Adams, P. D. (2005). CCP4 Newsl.42, contribution 8.
- Aravind, L., Walker, D. R. & Koonin, E. V. (1999). Nucleic Acids Res.27, 1223–1242. [DOI] [PMC free article] [PubMed]
- Bailly, V. & Verly, W. G. (1989). Nucleic Acids Res.17, 3617–3618. [DOI] [PMC free article] [PubMed]
- Banner, D. W., Bloomer, A. C., Petsko, G. A., Phillips, D. C., Pogson, C. I., Wilson, I. A., Coran, P. H., Furth, A. J., Milman, J. D. & Offord, R. E. (1975). Nature (London), 255, 609–614. [DOI] [PubMed]
- Barzilay, G. & Hickson, I. D. (1995). Bioessays, 17, 713–719. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Corfu, N. A. & Bin Song, L. J. (1992). Inorg. Chim. Acta, 192, 243–251.
- Cunningham, R. P. (1997). Mutat. Res.383, 189–196. [DOI] [PubMed]
- DeLano, W. L. (2008). PyMOL Molecular Viewer. DeLano Scientific LLC, Palo Alto, California, USA. http://www.pymol.org.
- Demple, B., Herman, T. & Chen, D. S. (1991). Proc. Natl Acad. Sci. USA, 88, 11450–11454. [DOI] [PMC free article] [PubMed]
- Demple, B., Johnson, A. & Fung, D. (1986). Proc. Natl Acad. Sci. USA, 83, 7731–7735. [DOI] [PMC free article] [PubMed]
- Doetsch, P. W. & Cunningham, R. P. (1990). Mutat. Res.236, 173–201. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Farber, G. K. & Petsko, G. A. (1990). Trends Biochem. Sci.15, 228–234. [DOI] [PubMed]
- Garcin, E. D., Hosfield, D. J., Desai, S. A., Haas, B. J., Björas, M., Cunningham, R. P. & Tainer, J. A. (2008). Nature Struct. Mol. Biol.15, 515–522. [DOI] [PubMed]
- Haas, B. J., Sandigursky, M., Tainer, J. A., Franklin, W. A. & Cunningham, R. P. (1999). J. Bacteriol.181, 2834–2839. [DOI] [PMC free article] [PubMed]
- Hitomi, K., Iwai, S. & Tainer, J. A. (2007). DNA Repair, 6, 410–428. [DOI] [PubMed]
- Hosfield, D. J., Guan, Y., Haas, B. J., Cunningham, R. P. & Tainer, J. A. (1999). Cell, 98, 397–408. [DOI] [PubMed]
- Hughes, R. C., Tomanicek, S. J., Ng, J. D. & Coates, L. (2009). Acta Cryst. F65, 1317–1319. [DOI] [PMC free article] [PubMed]
- Huffman, J. L., Sundheim, O. & Tainer, J. A. (2005). Mutat. Res.577, 55–76. [DOI] [PubMed]
- Hutchinson, F. (1985). Prog. Nucleic Acid Res. Mol. Biol.32, 115–154. [DOI] [PubMed]
- Ishchenko, A. A., Deprez, E., Maksimenko, A., Brochon, J.-C., Tauc, P. & Saparbaev, M. K. (2006). Proc. Natl Acad. Sci. USA, 103, 2564–2569. [DOI] [PMC free article] [PubMed]
- Ishchenko, A. A., Sanz, G., Priveventzev, C. V., Maksimenko, A. V. & Saparbaev, M. (2003). Nucleic Acids Res.31, 6344–6353. [DOI] [PMC free article] [PubMed]
- Ischenko, A. A. & Saparbaev, M. K. (2002). Nature (London), 415, 183–187. [DOI] [PubMed]
- Ishchenko, A. A., Yang, X., Ramotar, D. & Saparbaev, M. (2005). Mol. Cell. Biol.25, 6380–6390. [DOI] [PMC free article] [PubMed]
- Ivanov, I., Tainer, J. A. & McCammon, J. A. (2007). Proc. Natl Acad. Sci. USA, 104, 1465–1470. [DOI] [PMC free article] [PubMed]
- Kerins, S. M., Collins, R. & McCarthy, T. V. (2003). J. Biol. Chem.278, 3048–3054. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Krokan, H. E., Standal, R. & Slupphaug, R. (1997). Biochem. J.325, 1–16. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Levin, J. D. & Demple, B. (1990). Nucleic Acids Res.18, 5069–5075. [DOI] [PMC free article] [PubMed]
- Levin, J. D., Johnson, A. W. & Demple, B. (1988). J. Biol. Chem.263, 8066–8071. [PubMed]
- Lindahl, T., Karran, P. & Wood, R. D. (1997). Curr. Opin. Genet. Dev.7, 158–169. [DOI] [PubMed]
- Loeb, L. A. & Preston, B. D. (1986). Annu. Rev. Genet.20, 201–230. [DOI] [PubMed]
- Marsic, D., Hughes, R. C., Byrne-Steele, M. L. & Ng, J. D. (2008). BMC Biotechnol.8, 44. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- McMurray, C. T. & Tainer, J. A. (2003). Nature Genet.34, 239–241. [DOI] [PubMed]
- Mol, C. D., Hosfield, D. J. & Tainer, J. A. (2000). Mutat. Res.460, 211–229. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Ramotar, D. (1997). Biochem. Cell Biol.75, 327–336. [PubMed]
- Reardon, D. & Farber, G. K. (1995). FASEB J.9, 497–503. [DOI] [PubMed]
- Sobol, R. W., Kartalou, M., Almeida, K. H., Joyce, D. F., Engelward, B. P., Horton, J. K., Prasad, R., Samson, L. D. & Wilson, S. H. (2003). J. Biol. Chem.278, 39951–39959. [DOI] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Yang, X., Tellier, P., Masson, J. Y., Vu, T. & Ramotar, D. (1999). Biochemistry, 38, 3615–3623. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: T. maritima endonuclease IV, 2x7v
PDB reference: 2x7w