The N-terminal PAS domains from the eukaryotic EAG potassium channels are thought to have a regulatory function. Here the expression, purification, crystallization and preliminary crystallographic characterization of two of these domains are described.
Keywords: PAS domains, EAG, potassium channels
Abstract
Per–Arnt–Sim (PAS) domains are ubiquitous in nature; they are ∼130-amino-acid protein domains that adopt a fairly conserved three-dimensional structure despite their low degree of sequence homology. These domains constitute the N-terminus or, less frequently, the C-terminus of a number of proteins, where they exert regulatory functions. PAS-containing proteins generally display two or more copies of this motif. In this work, the crystallization and preliminary analysis of the PAS domains of two eukaryotic potassium channels from the ether-à-go-go (EAG) family are reported.
1. Introduction
Per–Arnt–Sim (PAS) domains function as sensory and regulatory domains in many different protein systems (Taylor & Zhulin, 1999 ▶). In archaea and bacteria they are essentially associated with two-component regulatory systems, while in eukarya they have mostly been found to be associated with kinases, transcription factors and voltage-gated potassium channels (McIntosh et al., 2010 ▶). Potassium channels mediate the selective movement of potassium ions across the membrane and are essential for the generation and propagation of electrical activity in organisms; these membrane proteins are therefore at the basis of several phenomena such as vision and muscle contraction. EAG potassium channels are voltage-gated potassium channels (Morais Cabral et al., 1998 ▶) that have an N-terminal PAS domain (Bauer & Schwarz, 2001 ▶). It has previously been demonstrated that deletion of this PAS domain does not affect the assembly process of the channel but modifies the kinetics of channel function; little else is known about the functional role of these domains in EAG channels. However, in analogy with PAS domains from other protein systems, it is thought that they function as channel regulators. In an effort to clarify the function and mechanism of action of EAG PAS domains, we have crystallized and solved the structures of PAS domains from two members of the EAG potassium-channel family: the mouse EAG channel and the fruit-fly ELK channel.
2. Materials and methods
2.1. Purification of recombinant mouse EAG PAS domain
The open reading frame encoding residues 27–136 of the mouse EAG potassium channel (mEAG; UniGene Mm.4489) was amplified by PCR with Pfu Taq polymerase (Fermentas) according to the manufacturer’s specifications. The PCR product was cloned into pET-15b (Novagen) between NdeI and BamHI restriction sites. The resulting construct (pET-15b-mEAG PAS) encodes a 6×His N-terminal tag and a thrombin cleavage site (corresponding to the amino-acid sequence MGSSHHHHHHSSGLVPRGSHM) followed by residues 27–136 of the mEAG channel, as confirmed by DNA sequencing. Escherichia coli BL21 (DE3) cells harbouring pET-15b-mEAG PAS were grown in Luria broth liquid medium supplemented with ampicillin (100 mg l−1) in 4 l baffled flasks at 310 K with agitation. The optical density of the culture measured at 600 nm (OD600nm) was monitored during cell growth; protein expression was induced by the addition of IPTG to the medium (to a final concentration of 0.5 mM) when the OD600nm reached 0.8. After a 3 h incubation, cells were harvested by centrifugation (20 min, 4785g, 277 K), resuspended in buffer A (50 mM Tris–HCl pH 8.0, 10 mM imidazole, 150 mM NaCl, 1 mM DTT and protease inhibitors: 1 mM PMSF, 1 µg ml−1 leupeptin and 1 µg ml−1 pepstatin), disrupted by sonication and centrifuged (45 min, 23 500g, 277 K) to remove unbroken cells and cell debris. The supernatant was loaded onto a column packed with 5 ml Ni–NTA beads (Qiagen) equilibrated with buffer A. Washes were performed with 20 mM Tris–HCl pH 8.0, 150 mM NaCl (buffer B) supplemented with 20 mM imidazole and the protein was eluted with buffer B supplemented with 150 mM imidazole. The eluted fractions were pooled, thrombin was added at a 1:500 ratio (thrombin:fusion protein) in order to cleave the affinity tag and the mixture was dialysed overnight at 277 K against 20 mM Tris–HCl pH 8.0, 150 mM NaCl and 5 mM DTT (buffer C) using a 3500 Da molecular-weight cutoff membrane. Thrombin digestion generated a protein domain that included residues 27–136 of mEAG with the N-terminal amino-acid extension GSHM just before the first residue of the PAS domain. The protein was further purified by size-exclusion chromatography using a Sephadex 200 column equilibrated in buffer C at 277 K and a flow rate of 0.5 ml min−1. Fractions containing mEAG PAS were pooled and concentrated to a final concentration of 10 mg ml−1 using a 5000 Da molecular-weight cutoff device.
2.2. Purification and methylation of recombinant fruit-fly ELK PAS domain
The open reading frame encoding residues 11–136 of the fruit-fly ELK potassium channel (dmELK; UniGene Dm.4335) was amplified by PCR with Pfu Taq polymerase (Fermentas) according to the manufacturer’s specifications. The PCR product was cloned into pET-15b as described in §2.1; the resulting construct (pET-15b-dmELK PAS) encodes a 6×His N-terminal tag and a thrombin cleavage site (corresponding to the amino-acid sequence MGSSHHHHHHSSGLVPRGSHM) followed by residues 11–136 of the dmELK channel, as confirmed by DNA sequencing. E. coli BL21 (DE3) cells harbouring the pET-15b-dmELK PAS were grown in the auto-inducing media ZYM-5052 (Studier, 2005 ▶) supplemented with ampicillin (100 mg l−1) in 4 l baffled flasks at 310 K with agitation until the OD600nm reached 1.0. The temperature in the shaker-incubator was then shifted to 291 K and the cultures were allowed to grow for approximately 18 h. The cells were harvested by centrifugation (20 min, 4785g, 277 K), resuspended in 100 mM HEPES pH 8.0 and 300 mM NaCl (buffer D) with protease inhibitors (1 mM PMSF, 1 µg ml−1 leupeptin and 1 µg ml−1 pepstatin), lysed using a cell cracker (Emulsiflex C5, Avestin) and centrifuged (45 min, 32 800g, 277 K) to remove unbroken cells and cell debris. Supernatant was loaded onto a column packed with 5 ml Talon beads (Clontech) equilibrated with buffer D. Washes were performed with buffer D supplemented with 20 mM imidazole and elution was carried out by increasing the imidazole concentration to 200 mM. The eluted fractions were pooled, thrombin was added at a 1:500 ratio (thrombin:fusion protein) in order to cleave the purification tag and the mixture was dialysed overnight at 277 K against 20 mM HEPES pH 8.0, 150 mM NaCl and 5 mM DTT using a 3500 Da molecular-weight cutoff membrane. Proteolysis generated a polypeptide that included residues 11–136 of dmELK and an N-terminal extension with amino-acid sequence GSHM. Methylation of the lysine residues in dmELK PAS was performed overnight at 277 K according to Shaw et al. (2007 ▶); the reaction was stopped by size-exclusion chromatography in a Sephadex 200 column equilibrated with buffer C running at 0.5 ml min−1 at 277 K. Fractions containing dmELK PAS were pooled and concentrated to a final concentration of 10 mg ml−1 using a 5000 Da molecular-weight cutoff device.
2.3. Crystallization, data collection and processing
2.3.1. mEAG PAS
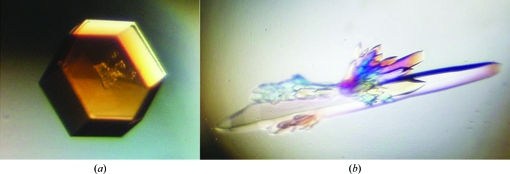
Initial crystallization conditions were screened at 277 K using commercial Emerald BioSystems, Hampton Research and Qiagen formulations. Sitting-drop Cryschem plates were manually filled with 300 µl precipitant solution in the reservoir and 2 µl drops; the drops were formed of 1 µl precipitant solution plus 1 µl mEAG PAS solution (at 10 mg ml−1 in buffer C). Two crystallization conditions were identified: one that resulted in the formation of hexagonal shaped crystals (1.28 M ammonium sulfate, 0.2 M lithium sulfate, 0.1 M Tris–HCl pH 8.5) and a second condition that resulted in long needles (2 M sodium/potassium phosphate, 0.2 M lithium sulfate, 0.1 M CAPS pH 10.5). Both conditions were optimized by fine-grid screening and larger crystals were obtained at 277 K by mixing 2 µl mEAG PAS solution at 10 mg ml−1 in buffer C with 2 µl precipitant solution (crystal form A, 1.3 M ammonium sulfate, 0.25 M lithium sulfate, 0.1 M Tris–HCl pH 8.5; crystal form B, 1.3 M sodium phosphate, 0.9 M potassium phosphate, 0.1 M lithium sulfate, 0.1 M CAPS pH 10.0) and equilibrating against a reservoir volume of 500 µl in sitting-drop plates (Fig. 1 ▶). Typically, both crystal forms took 1–2 weeks to grow.
Figure 1.
(a) Single crystal of EAG PAS from Mus musculus belonging to space group P3221. Approximate crystal dimensions were 300 µm in width and 60 µm in height. (b) Multiple crystals of EAG PAS from M. musculus belonging to space group P3121. The approximate crystal dimensions of the largest crystal were 500 µm in length and 70 µm in thickness.
Cryoprotection of both crystal forms was achieved by quick transfers between solutions consisting of the crystallization reservoir solution plus increasing concentrations of glycerol [in 5%(v/v) steps] up to a final concentration of 20%(v/v). The crystals were then cooled and stored in liquid nitrogen before data collection. X-ray diffraction data for mEAG PAS were collected on beamline ID14-EH2 (ADSC Quantum 4 detector) at the European Synchrotron Radiation Facility (ESRF, Grenoble, France). For crystal form A two data sets, each composed of 130 images representing 1° oscillation, were collected at 100 K from a single crystal. To record the high-angle diffraction spots (at 1.85 Å) the first data set was collected with a crystal-to-detector distance of 160 mm and 8 s exposure time per image. To record the very strong low-angle diffraction spots a second data set was collected from the same crystal with a crystal-to-detector distance of 234 mm and an exposure time of 2 s. For crystal form B, 90 images, each representing 1° oscillation with 2 s exposure time at a crystal-to-detector distance of 235 mm, were recorded from a single crystal. Diffraction from crystal form B extended to 2.35 Å resolution, with strong reflections observed in the outer diffraction shell; despite an 〈I/σ(I)〉 of 1 in the highest resolution bin, we chose to process the data to the limit of resolution with the purpose of evaluating the contribution of these reflections to the map and structure quality during the refinement stage. Diffraction data sets were processed with MOSFLM (Leslie, 2006 ▶) and SCALA (Collaborative Computational Project, Number 4, 1994 ▶). The structure of mEAG PAS crystal form A was solved by molecular replacement with Phaser (McCoy et al., 2007 ▶) using the PAS domain of Phot-LOV1 from Chlamydomonas reinhardtii (PDB code 1n9n; Fedorov et al., 2003 ▶) as a search model. A partially refined structure was obtained with REFMAC5 (Murshudov et al., 1997 ▶), which was then used to determine the molecular-replacement solution of mEAG PAS crystal form B.
2.3.2. Methylated dmELK PAS
Initial crystallization conditions were defined using the same protocol and materials as described above for the mEAG PAS domain. One single crystal was observed in a condition consisting of 0.1 M MES pH 6.5, 30%(w/v) PEG 5000 MME and 0.2 M ammonium sulfate. Optimization of the condition was performed by fine-grid screening using sitting-drop Cryschem plates manually filled with 500 µl precipitant solution in the reservoir and drops made up of 1.5 µl methylated dmELK PAS (at 10 mg ml−1 in buffer C) plus 1.5 µl precipitant solution. Better looking crystals were obtained within 3 d in 0.1 M MES pH 6.5, 30%(w/v) PEG 5000 MME, 0.35 M ammonium sulfate.
Crystals were cryoprotected, cooled and stored as described in §2.3.1. Two data sets were collected at 100 K from a single crystal on the ESRF ID14-EH1 beamline equipped with an ADSC Quantum Q210 detector (crystal-to-detector distance of 203 mm). In the high-resolution data set diffraction extended to 2.0 Å and each image corresponded to a 1° crystal oscillation with 25 s exposure; in the low-resolution data set each image corresponded to 5 s exposure. Data sets were processed as described in §2.3.1. The structure was solved by molecular replacement with Phaser using the partially refined structure of mEAG PAS as a search model.
3. Results and discussion
3.1. Crystallization of mEAG and dmELK PAS
Purified mEAG PAS 27–136 was easily crystallized in two different precipitant solutions, resulting in hexagonal shaped crystals (form A) and rod-like crystals (form B) which diffracted to 1.85 and 2.35 Å, respectively. Data-collection statistics are given in Table 1 ▶.
Table 1. Crystal and data-collection statistics.
Values in parentheses are for the last shell.
| Crystal form | mEAG PAS form A | mEAG PAS form B | dmELK PAS |
|---|---|---|---|
| Crystal data | |||
| Matthews coefficient VM (Å3 Da−1) | 3.7 | 3.8 | 4.8 |
| Solvent content (%) | 67 | 68 | 73 |
| Crystal system | Hexagonal | Hexagonal | Cubic |
| Space group | P3221 | P3121 | P4132 |
| Unit-cell parameters | |||
| a = b (Å) | 91.28 | 66.89 | 150.30 |
| c (Å) | 172.25 | 166.63 | 150.30 |
| α = β (°) | 90.0 | 90.0 | 90.0 |
| γ (°) | 120.0 | 120.0 | 90.0 |
| No. of molecules in unit cell (Z) | 24 | 12 | 48 |
| Data-collection details | |||
| Diffraction source | ESRF | ESRF | ESRF |
| Wavelength (Å) | 0.9330 | 0.9330 | 0.9334 |
| Detector | ADSC Quantum 4 | ADSC Quantum 4 | ADSC Quantum Q210 |
| Temperature (K) | 100 | 100 | 100 |
| Resolution range (Å) | 43–1.85 (1.95–1.85) | 50–2.35 (2.48–2.35) | 61–2.00 (2.11–2.00) |
| No. of unique reflections | 71849 (10400) | 18792 (12709) | 39431 (5615) |
| No. of observed reflections | 563953 (57622) | 137516 (16045) | 577984 (39457) |
| Multiplicity | 7.8 (5.5) | 7.3 (5.9) | 14.7 (7.0) |
| Overall B factor from Wilson plot (Å2) | 26 | 49 | 27 |
| Completeness (%) | 100 (100) | 100 (100) | 99.5 (99.4) |
| Redundancy | 7.8 (5.5) | 7.3 (5.9) | 14.7 (7.0) |
| 〈I/σ(I)〉 | 7.5 (1.3) | 7.5 (1.0) | 6.4 (1.4) |
| Rmerge (%) | 7.0 (56.3) | 9.1 (74.0) | 8.9 (55.0) |
| Data-processing software | CCP4 package, MOSFLM, SCALA | ||
In contrast, attempts to crystallize dmELK PAS were not successful. Literature reports have described changes in the crystallization properties of proteins after lysine methylation. We followed the protocol described above and assessed the completeness of the methylation reaction by mass spectrometry. Sparse-matrix crystallization trials with the methylated protein resulted in the formation of crystals; a possible reason for this effect is discussed below. In these crystallization conditions we noticed the establishment of different liquid phases owing to the presence of salt and PEG at high concentrations. The presence of multiple phases during pipetting to crystallization plates made it harder to reproduce these crystals; therefore, during the improvement of crystal quality we optimized the preparation of the precipitant solution by mixing the precipitant components, vortexing and heating. The optimized crystals diffracted to 2.0 Å resolution on a synchrotron beamline.
3.2. Structure determination
All three PAS-domain structures were solved by molecular replacement using Phaser. It was during this procedure that final determination of the space group was achieved. Crystal form A of mEAG PAS has P3221 symmetry with four molecules in the asymmetric unit, while crystal form B of this same construct presents two molecules per asymmetric unit in space group P3121. The packing arrangement of protein molecules in the dmELK crystals was determined to belong to space group P4132, with two molecules in the asymmetric unit. Analysis of the packing of these two molecules revealed that six methylated lysines of a total of 24 appear to be involved in crystal contacts and may be the reason for the improved crystallizability of the modified protein. All crystal structures are under refinement.
Acknowledgments
We acknowledge the ESRF for providing access to X-ray beamlines and thank the ESRF staff for support during data collection. This work was supported by grants from FCT (PTDC/QUI/66171/2006) and EMBO (installation grant) awarded to JHMC.
References
- Bauer, C. K. & Schwarz, J. R. (2001). J. Membr. Biol.182, 1–15. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Fedorov, R., Schlichting, I., Hartmann, E., Domratcheva, T., Fuhrmann, M. & Hegemann, P. (2003). Biophys. J.84, 2474–2482. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- McIntosh, B. E., Hogenesch, J. B. & Bradfield, C. A. (2010). Annu. Rev. Physiol.72, 625–645. [DOI] [PubMed]
- Morais Cabral, J. H., Lee, A., Cohen, S. L., Chait, B. T., Li, M. & Mackinnon, R. (1998). Cell, 95, 649–655. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Shaw, N., Zhao, M., Cheng, C., Xu, H., Saarikettu, J., Li, Y., Da, Y., Yao, Z., Silvennoinen, O., Yang, J., Liu, Z.-J., Wang, B.-C. & Rao, Z. (2007). Nature Struct. Mol. Biol.14, 779–784. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif.41, 207–234. [DOI] [PubMed]
- Taylor, B. L. & Zhulin, I. B. (1999). Microbiol. Mol. Biol. Rev.63, 479–506. [DOI] [PMC free article] [PubMed]