The carbonic anhydrase αCA1 from C. reinhardtii is a dimeric class αCA enzyme with post-translational glycosylation at three asparagine residues and proteolytic removal of a short peptide. The mature enzyme has been crystallized, MAD data have been collected to 1.88 Å resolution and a preliminary solution of the crystal structure has been obtained.
Keywords: carbonic anhydrases, Chlamydomonas reinhardtii
Abstract
Carbonic anhydrases (CAs) are ubiquitously distributed and are grouped into three structurally independent classes (αCA, βCA and γCA). Most αCA enzymes are monomeric, but αCA1 from Chlamydomonas reinhardtii is a dimer that is uniquely stabilized by disulfide bonds. In addition, during maturation an internal peptide of 35 residues is removed and three asparagine residues are glycosylated. In order to obtain insight into the effects of these structural features on CA function, wild-type C. reinhardtii αCA1 has been crystallized in space group P65, with unit-cell parameters a = b = 134.3, c = 120.2 Å. The crystal diffracted to 1.88 Å resolution and a preliminary solution of its crystal structure has been obtained by the MAD method.
1. Introduction
Carbonic anhydrase (CA) was first identified as an enzyme that assists in the rapid interconversion of carbon dioxide and carbonic acid (Meldrum & Roughton, 1933 ▶). Since then, carbonic anhydrases have been found to be widely distributed as essential proteins in many types of organisms. The enzymes belong to three classes (αCA, βCA and γCA) that apparently originate from different structural genes (Hewett-Emmett & Tashian, 1996 ▶; Roberts et al., 1997 ▶; Tripp et al., 2001 ▶; So et al., 2004 ▶).
Almost all αCAs are monomeric, with a central core composed of a β-sheet of ten peptide strands (Kannan et al., 1975 ▶). However, there are some exceptions. αCA from the alga Chlamydomonas reinhardtii is known to be a dimer (Ishida et al., 1993 ▶; Kamo et al., 1990 ▶) and αCA from the bacterium Rhodopseudomonas palustris (Puskás et al., 2000 ▶) and the human isozyme αCA-XII (Whittington et al., 2001 ▶) are also dimeric, while the human isozyme αCA-IX is tetrameric (Alterio et al., 2009 ▶). The latter three CAs are membrane-associated proteins.
βCAs have two subtypes with different active-site sequence conservations: ‘plant’-type and ‘cab’-type (Smith & Ferry, 1999 ▶; Kimber & Pai, 2000 ▶). βCA from the archaeon Methanobacterium thermoautotrophicum (‘cab’-type) is dimeric (Strop et al., 2001 ▶), while βCA from the red alga Porphyridium purpureum (‘plant’-type) is pseudo-tetrameric (Mitsuhashi et al., 2000 ▶) and βCA from the dicotyledonous plant Pisum sativum is octameric (Kimber & Pai, 2000 ▶).
γCA is trimeric (Alber & Ferry, 1994 ▶) and the active site is formed in the interface between two neighbouring subunits, with each subunit consisting of a seven-turn left-handed β-helix (Kisker et al., 1996 ▶).
Recently, it has been found that CAs have several isozymes that are distributed in different tissues (Chegwidden, 2000 ▶). For instance, in C. reinhardtii (Cr) three Cr-αCA isozymes and five Cr-βCA isozymes have been reported. Cr-αCA1 and possibly Cr-αCA21 are found in the periplasm (Coleman & Grossman, 1984 ▶; Kimpel et al., 1983 ▶; Yagawa et al., 1986 ▶), Cr-αCA3 and Cr-βCA8 are found in the chloroplast (Karlsson et al., 1998 ▶; van Hunnik et al., 2001 ▶; Villarejo et al., 2002 ▶) and Cr-βCA6 is localized in the thylakoid lumen (Parker et al., 2008 ▶).
Cr-αCA1 is predominantly expressed under practical conditions (Coleman & Grossman, 1984 ▶; Yang et al., 1985 ▶), while the expression of Cr-αCA2 has not yet been confirmed in vivo. However, the two Cr-αCAs have been assigned as having the following properties: Cr-αCA1 is the major CA in the presence of light in low-CO2 conditions, while Cr-αCA2 may be expressed in high-CO2 conditions (Fujiwara et al., 1990 ▶).
In addition to its dimeric quaternary structure, Cr-αCA1 displays unique post-translational modifications and disulfide bonds that may be essential for biological processes. Glycosylation occurs at three asparagine residues: Asn101, Asn135 and Asn297 (Ishida et al., 1993 ▶). Most other αCAs are not known to be glycosylated and a few adopt glycosylation sites that differ from those of Cr-αCA1 (Whittington et al., 2004 ▶; Alterio et al., 2009 ▶). Moreover, Cr-αCA1 is cleaved into two peptides (short and long) by the elimination of an internal 35-residue peptide (Ishida et al., 1993 ▶). It is noteworthy that these 35 residues (residues 306–340) do not exist in corresponding regions of other αCA genes. Finally, Cr-αCA1 forms four disulfide bonds: Cys61–Cys264, Cys194–Cys198 and Cys296–Cys351 (which link the large and small peptides) and Cys21–Cys21 (which cross-links the dimer interface). The latter three are specific to C. reinhardtii.
In order to obtain an insight into the structural and functional contributions of N-glycosylation and peptide cleavage, wild-type Cr-αCA1 has been isolated, purified and crystallized for X-ray analysis.
2. Materials and methods
2.1. Protein purification
Isolation of Cr-αCA1 from C. reinhardtii was performed according to the previously reported procedure (Fujiwara et al., 1990 ▶; Fukuzawa et al., 1990 ▶). To accelerate the cell cycle of Cr, Cr cells were cultivated in a 40 l tank under photoautotrophic conditions in air and homogenized using a French press. The centrifuged supernatant was fractionated with 30 and 65% ammonium sulfate saturated solutions. The crude sample solution was initially loaded onto a Fast Flow column (GE Healthcare) with DEAE-cellulose (Seikagaku Corp.), followed by Phenyl-650M Toyopearl chromatography (Tosoh Corp.) and finally Superdex G-200 gel filtration (GE Healthcare). Low and High Gel Filtration Calibration kits (GE Healthcare) were used to calibrate the gel-filtration column. The eluted fractions were monitored by UV absorption at 280 nm and analyzed by SDS–PAGE, which was carried out using a Phast System (GE Healthcare) with a gradient of 8–12%(w/v) acrylamide gel. Protein bands were stained with Coomassie Brilliant Blue R-350. A broad-range marker (APRO Science) was used to calibrate the molecular weights. The enzymatic activity was confirmed by the method of Yang et al. (1985 ▶). The protein concentration was assayed using a dye-binding protein-assay kit (Bio-Rad). For crystallization, the proteins were concentrated to 20 mg ml−1 in 25 mM Tris–HCl pH 7.5 using centrifugal filter devices (Amicon and Microcon Ultracel 15 10k or YM-10 membrane from Millipore Co.).
2.2. Crystallization
Crystallizations were carried out using the hanging-drop vapour-diffusion method by mixing equal volumes (2 µl) of protein solution and reservoir solution and equilibrating the mixed solutions against 700 µl reservoir solution in a 24-well plate (Stem Corp.) at 293 K. Initial conditions were surveyed using more than 290 conditions from commercially available matrices from Hampton Research and Emerald BioSystems. The conditions under which crystalline precipitates appeared were further optimized by changing the concentrations of the protein, precipitant and salt and by changing the pH of the buffer. For crystallization, siliconized (L-25; Fuji Systems Corp.) circular plain glass cover slides (diameter 18 mm; Matsunami Glass Ind. Ltd) and high-vacuum grease (Dow Corning Toray Co. Ltd) were used.
2.3. X-ray diffraction experiment
X-ray diffraction experiments were performed on beamline AR-NW12 of the Photon Factory (PF; Tsukuba, Japan). A crystal was soaked for 30 s in reservoir solution containing 20% glycerol and mounted on a CryoLoop (Hampton Research) prior to flash-cooling. To apply multiple anomalous dispersion (MAD/SAD) methods for phase estimation, three wavelengths (referred to as peak, edge and remote) were selected based on the X-ray absorption fine structure (XAFS) of Zn atoms bound to the protein in the crystal. The diffraction patterns were recorded at 100 K on a Quantum 210 CCD detector positioned at 129.8, 86.3 and 86.3 mm from the crystal for the peak, edge and remote wavelengths, respectively. Each frame was taken with an exposure time of 10 s and with a 1° oscillation step in the range 0–180°. Bragg spots were indexed and intensities were estimated by integrating around the spots. Intensity data were then scaled and merged using the HKL-2000 package (Otwinowski & Minor, 1997 ▶). These data were further converted to structure-factor amplitudes using TRUNCATE from the CCP4 suite (Collaboration Computational Project, Number 4, 1994 ▶). For preliminary phasing combined with structure modelling, the programs SHARP/autoSHARP (Vonrhein et al., 2007 ▶) and ARP/wARP (Perrakis et al., 1999 ▶) were applied to the MAD data sets.
3. Results and discussion
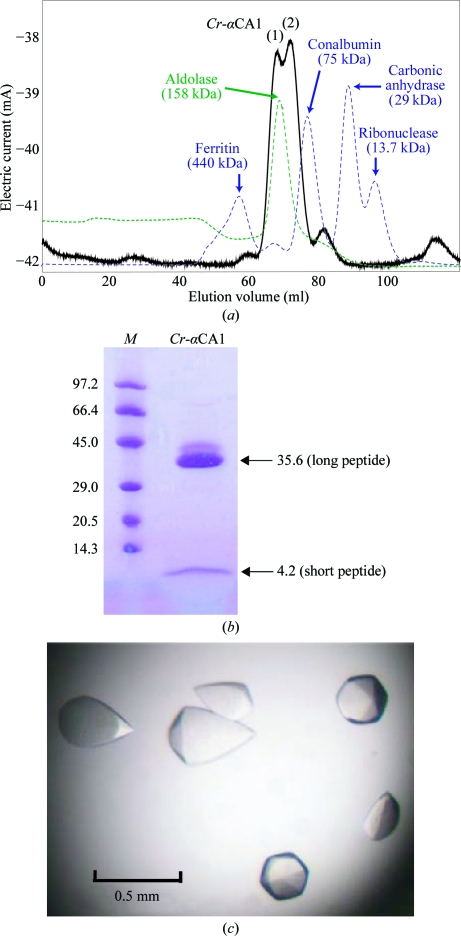
Wild type Cr-αCA1 is a disulfide-linked homodimer with a molecular weight of 79.6 kDa (Ishida et al., 1993 ▶). During size-exclusion chromatography, the elution profile was slightly split (Fig. 1 ▶ a); this was always experienced and suggests that Cr-αCA1 dimers may adhere to also form a tetrameric state (Kamo et al., 1990 ▶; Ishida et al., 1993 ▶). However, reducing SDS–PAGE (Fig. 1 ▶ b) showed only the expected bands of 35.6 and 4.2 kDa for the long and short peptides, respectively, as expected. The purified proteins from the two peaks also both showed the enzymatic activity expected for Cr-αCA1. Therefore, the fractions containing the two peaks were merged and used for crystallization.
Figure 1.
(a) Chromatogram of the gel-filtration step, (b) SDS–PAGE of purified Cr-αCA1 and (c) crystals obtained under the optimized conditions (see text). The left lane in (b) labelled M contains molecular-weight markers (labelled in kDa).
Crystals suitable for X-ray experiments were obtained under the following optimized conditions: equal volumes of 10 mg ml−1 Cr-αCA1 protein solution in 25 mM Tris–HCl pH 7.5 and reservoir solution containing 1.0 M ammonium sulfate and 100 mM Tris–HCl pH 8.5 were mixed and equilibrated against 700 µl reservoir solution at 293 K. The crystals exhibited a hexagonal bipyramidal shape, with maximum dimensions of 0.5 × 0.3 × 0.3 mm, as shown in Fig. 1 ▶(c).
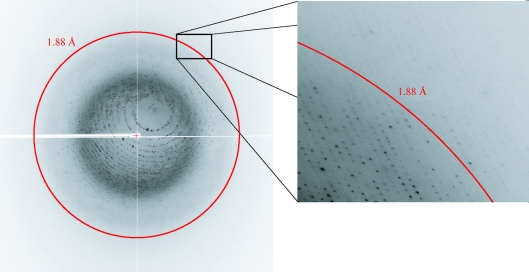
The anomalous scattering factors (f′ and f″) of the Zn atoms in the crystal were derived (Evans & Pettifer, 2001 ▶) from XAFS data obtained prior to data collection and three wavelengths were chosen and used for data collection: 1.28254 Å (peak), 1.28297 Å (edge) and 1.00 Å (remote). The diffraction intensity data collected at the three wavelengths were processed to 2.10, 2.00 and 1.88 Å resolution, respectively. Higher resolution diffraction was observed in some images (Fig. 2 ▶), but weaker diffraction in other crystal orientations warranted a more conservative resolution cutoff. The statistics of the intensity and crystallographic data are given in Table 1 ▶. The wild-type Cr-αCA1 in the apo form crystallized in a hexagonal lattice, with unit-cell parameters a = b = 134.3, c = 120.1 Å and space group P61 or P65.
Figure 2.
X-ray diffraction pattern of a Cr-αCA1 crystal. The Quantum 210 CCD detector was positioned 129.8 mm from the crystal.
Table 1. Statistics of the observed intensity and crystallographic data at three wavelengths.
| Peak | Edge | Remote | |
|---|---|---|---|
| Wavelength (Å) | 1.28254 | 1.28297 | 1.00 |
| Resolution (Å) | 50–2.00 (2.07–2.00) | 50–2.10 (2.18–2.10) | 50–1.88 (1.95–1.88) |
| Space group | P65 | P65 | P65 |
| Unit-cell parameters (Å) | |||
| a = b | 134.3 | 134.4 | 134.2 |
| c | 120.2 | 120.2 | 120.1 |
| Observed reflections | 885376 | 762127 | 1055288 |
| Unique reflections | 82859 | 71757 | 99548 |
| Completeness (%) | 99.9 (100) | 99.9 (100) | 99.9 (100) |
| Redundancy | 10.7 (10.2) | 10.6 (10.1) | 10.6 (10.0) |
| I/σ(I) | 57.5 (10.0) | 57.6 (11.5) | 60.0 (8.7) |
| Rmerge† (%) | 8.1 (30.2) | 8.1 (27.1) | 8.2 (31.4) |
R
merge = 100 × 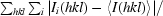
 .
.
MAD calculations gave a best solution containing two Zn atoms in the asymmetric unit. The Matthews coefficient V M (Matthews, 1968 ▶), estimated by assuming that the asymmetric unit contains a dimer, had a high value of 3.9 Å3 Da−1, which is equivalent to 68.7% solvent content. This assumption was confirmed by preliminary structure determination, which assigned more than 88% of the dimer residues in the asymmetric unit and also resolved the space group as P65. The larger dimer of dimers suggested by gel filtration is not found in the crystals, since crystallographic symmetry in P65 cannot generate such a complex. Full refinement of the atomic parameters and structure analysis are in progress and will be reported elsewhere, together with structural insights into the functional properties of the dimeric enzyme.
Acknowledgments
This work was supported in part by Grants-in-Aid for the Protein3000 Research Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We thank S. Kuramitsu for organizing the research group in the program and N. Igarashi and S. Wakatsuki for facilities and help during data collection.
Footnotes
The two corresponding genes are aligned in tandem in the genome (Fujiwara et al., 1990 ▶).
References
- Alber, B. E. & Ferry, J. G. (1994). Proc. Natl Acad. Sci. USA, 91, 6906–6913.
- Alterio, V., Hilvo, M., Di Fiore, A., Supuran, C. T., Pan, P., Parkkila, S., Scaloni, A., Pastorek, J., Pastorekova, S., Pedone, C., Scozzafava, A., Monti, S. M. & De Simone, G. (2009). Proc. Natl Acad. Sci. USA, 106, 16233–16238. [DOI] [PMC free article] [PubMed]
- Chegwidden, W. R. (2000). The Carbonic Anhydrases: New Horizons. Basel: Birkhäuser Verlag.
- Coleman, J. R. & Grossman, A. R. (1984). Proc. Natl Acad. Sci. USA, 81, 6049–6053. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Evans, G. & Pettifer, R. F. (2001). J. Appl. Cryst.34, 82–86.
- Fujiwara, S., Fukuzawa, H., Tachiki, A. & Miyachi, D. (1990). Proc. Natl Acad. Sci. USA, 87, 9779–9783. [DOI] [PMC free article] [PubMed]
- Fukuzawa, H., Fujiwara, S., Yamamoto, Y., Dionisio-Sese, M. L. & Miyachi, D. (1990). Proc. Natl Acad. Sci. USA, 87, 4383–4387. [DOI] [PMC free article] [PubMed]
- Hewett-Emmett, D. & Tashian, R. E. (1996). Mol. Phylogenet. Evol.5, 50–77. [DOI] [PubMed]
- Hunnik, E. van, Livne, A., Pogenberg, V., Spijkerman, E., van den Ende, H., Garcia, E. M., Sültemeyer, D. & de Leeuw, J. W. (2001). Plant Physiol.94, 284–290. [DOI] [PubMed]
- Ishida, A., Muto, A. & Miyachi, S. (1993). Eur. J. Biochem.214, 9–16. [DOI] [PubMed]
- Kamo, T., Shimogawara, K., Fukuzawa, H., Muto, S. & Miyachi, S. (1990). Eur. J. Biochem.192, 557–562. [DOI] [PubMed]
- Kannan, K. K., Notstrand, B., Fridborg, K., Loevgren, S., Ohlsson, A. & Petef, M. (1975). Proc. Natl Acad. Sci. USA, 72, 51–55. [DOI] [PMC free article] [PubMed]
- Karlsson, J., Clarke, A. K., Chen, Z.-Y., Hugghins, S. Y., Park, Y.-I., Husic, H. D., Moroney, J. V. & Samuelsson, G. (1998). EMBO J.17, 1208–1216. [DOI] [PMC free article] [PubMed]
- Kimber, M. S. & Pai, E. F. (2000). EMBO J.19, 1407–1418. [DOI] [PMC free article] [PubMed]
- Kimpel, D. L., Togasaki, R. K. & Miyachi, S. (1983). Plant Cell Physiol.24, 255–259.
- Kisker, C., Schindelin, H., Alber, B. E., Ferry, J. G. & Rees, D. C. (1996). EMBO J.15, 2323–2330. [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Meldrum, H. & Roughton, F. (1933). J. Physiol.80, 113–142. [DOI] [PMC free article] [PubMed]
- Mitsuhashi, S., Mizushima, T., Yamashita, E., Yamamoto, M., Kumasaka, T., Moriyama, H., Ueki, T., Miyachi, S. & Tsukihara, T. (2000). J. Biol. Chem.275, 5521–5526. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Parker, M. S., Mock, T. & Armbrust, E. V. (2008). Annu. Rev. Genet.42, 619–645. [DOI] [PubMed]
- Perrakis, A., Morris, R. M. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed]
- Puskás, L. G., Inui, M., Zahn, K. & Yukawa, H. (2000). Microbiology, 146, 2957–2966. [DOI] [PubMed]
- Roberts, S. B., Lane, T. W. & Morel, F. M. M. (1997). J. Phycol.33, 845–850.
- Smith, K. S. & Ferry, J. G. (1999). J. Bacteriol.181, 6247–6253. [DOI] [PMC free article] [PubMed]
- So, A. K.-C., Epsie, G. S., Williams, E. B., Shively, J. M., Heinhorst, S. & Cannon, G. C. (2004). J. Bacteriol.186, 623–630. [DOI] [PMC free article] [PubMed]
- Strop, P., Smith, K. S., Iverson, T. M., Ferry, J. G. & Rees, D. C. (2001). J. Biol. Chem.276, 10299–10305. [DOI] [PubMed]
- Tripp, B. C., Smith, K. & Ferry, J. G. (2001). J. Biol. Chem.276, 48615–48618. [DOI] [PubMed]
- Villarejo, A., Shutora, T., Moshkin, O., Forssén, M., Klimov, V. V. & Samuelsson, G. (2002). EMBO J.21, 1930–1938. [DOI] [PMC free article] [PubMed]
- Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. (2007). Methods Mol. Biol.364, 215–230. [DOI] [PubMed]
- Whittington, D. A., Grubb, J. H., Waheed, A., Shah, G. N., Sly, W. S. & Christianson, D. W. (2004). J. Biol. Chem.279, 7223–7228. [DOI] [PubMed]
- Whittington, D. A., Waheed, A., Ulmasov, B., Shah, G. N., Grubb, J. H., Sly, W. S. & Christianson, D. W. (2001). Proc. Natl Acad. Sci. USA, 98, 9545–9550. [DOI] [PMC free article] [PubMed]
- Yagawa, Y., Aizawa, K., Yang, S.-Y. & Miyachi, S. (1986). Plant Cell Physiol.27, 215–221.
- Yang, S.-Y., Tsuzuki, M. & Miyachi, S. (1985). Plant Cell Physiol.26, 25–34.