The crystallization and primary crystallographic analysis of B. megaterium tyrosinase is reported. The purified protein was crystallized in the absence or presence of zinc ions to a resolution of 2.0 Å.
Keywords: tyrosinases, Bacillus megaterium, type 3 copper enzymes
Abstract
Tyrosinases are type 3 copper enzymes that are involved in the production of melanin and have two copper ions in the active site. Here, the crystallization and primary analysis of a tyrosinase from Bacillus megaterium is reported. The purified protein was crystallized in the absence or presence of zinc ions and the crystals diffracted to a resolution of 2.0 Å. Crystals obtained in the presence of zinc belonged to space group P212121, while crystals grown in the absence of zinc belonged to space group P21. In both space groups the asymmetric unit contained a dimer, with minor differences in the crystal density and in packing interactions.
1. Introduction
Tyrosinases (EC 1.14.18.1) are copper-containing enzymes that are ubiquitously distributed in all domains of life. Molecular oxygen is used by tyrosinases to catalyze two different enzymatic reactions: (i) the ortho-hydroxylation of monophenols to o-diphenols (monophenolase activity) and (ii) the oxidation of o-diphenols to o-quinones (diphenolase activity) (Burton, 2003 ▶; Claus & Decker, 2006 ▶). Tyrosinases show a significantly higher specific activity for the oxidation of o-diphenols than for the hydroxylation of monophenols (Hernandez-Romero et al., 2006 ▶). Tyrosinases belong to a larger family of proteins named the type 3 copper proteins, which include catechol oxidases (exhibiting only diphenolase activity) and oxygen-binding haemocyanins from molluscs and arthropods (Itoh & Fukuzumi, 2007 ▶). Tyrosinase is perhaps the most intensely studied enzyme of this family because of its role in skin pigmentation, on account of the diseases that result from its activity at incorrect levels, and because of its role in the browning of fruits and vegetables (Rosenzweig & Sazinsky, 2006 ▶). The tyrosinases from Streptomyces nigrifaciens, S. glaucescens, Pseudomonas putida F6 and Bacillus thuringiensis are all monomers, while the Vibrio tyrosinaticus and Thermomicrobium roseum tyrosinases are dimers (Kong et al., 2000 ▶; Liu et al., 2004 ▶; McMahon et al., 2007 ▶). There are numerous biotechnological applications for this enzyme, including the detoxification of phenol-containing waste waters and soils and as a biosensor for monitoring the presence and concentration of phenols (Burton, 2003 ▶; Claus & Decker, 2006 ▶).
The active site of tyrosinases consists of two Cu atoms (typically called CuA and CuB), each coordinated by three conserved histidine residues located within a four-helical bundle (Claus & Decker, 2006 ▶; Decker et al., 2007 ▶). Although the mechanism of tyrosinases and other type 3 copper proteins has been addressed by a wide variety of (bio)chemical, spectroscopic and structural methods, the observed functional differences between tyrosinase and catechol oxidase are far from being understood at the molecular level (Decker et al., 2006 ▶; Gerdemann et al., 2002 ▶). To date, two crystal structures of catechol oxidase (Klabunde et al., 1998 ▶; Virador et al., 2010 ▶), three of haemocyanin (Cuff et al., 1998 ▶; Gaykema et al., 1984 ▶; Hazes et al., 1993 ▶), one of arthropod prophenoloxidase (Li et al., 2009 ▶) and the structure of a tyrosinase from S. castaneoglobisporus crystallized in complex with a copper-binding caddie protein (Matoba et al., 2006 ▶) have been determined.
We recently isolated and characterized a tyrosinase from the soil bacterium B. megaterium (TyrBm) with an exceptionally high activity in water-miscible organic solvents (Shuster & Fishman, 2009 ▶). TyrBm has 40% sequence identity to tyrosinase from S. castaneoglobisporus; however, B. megaterium does not contain a gene encoding a caddie protein, possibly indicating differences in assembly of the active site. Preliminary results indicated that both monomeric and dimeric species of TyrBm are present and active in solution. In vivo, the protein is excreted to the environment and its conformation under these conditions is not yet known. Here, we describe the crystallization and preliminary crystallographic analysis of TyrBm.
2. Materials and methods
2.1. Purification for crystallization
Tyrosinase-producing B. megaterium was isolated in our laboratory from soil and the gene (accession No. ACC86108) was cloned in Escherichia coli BL21 as described previously (Shuster & Fishman, 2009 ▶). Briefly, the gene cloned with a C-terminal His6 tag into the pET9d vector (Novagen, Darmstadt, Germany) was overexpressed in E. coli cells, resulting in full-length protein ending with the sequence SKRSSHHHHHH. The cells were grown overnight at 310 K in 0.5 l TB medium [1.2% tryptone, 2.4% yeast extract, 0.4%(w/v) glycerol and 90 mM potassium phosphate buffer] and harvested by centrifugation (8000g for 10 min at room temperature). Cells were treated twice with a pressure cell (French Press, Spectronic Instruments Inc., Rochester, New York, USA) and cell debris was removed by centrifugation (16 000g for 20 min at 288 K). The supernatant containing TyrBm was applied onto an Ni2+-bound affinity column and eluted with an appropriate buffer (20 mM Tris–HCl pH 7.5, 500 mM NaCl and 500 mM imidazole). The molecular mass of the purified TyrBm was approximately 35 kDa as determined by SDS–PAGE, confirming the calculated value (Shuster & Fishman, 2009 ▶). Further purification was performed using anion-exchange (AEC) HPLC (PL-SAX 1000 Å, Polymer Laboratories, Varian Inc., Palo Alto, California, USA). The protein was eluted using a 0–1 M linear gradient of NaCl in 50 mM Tris–HCl pH 8. Fractions were collected and analyzed and were confirmed to be enriched in TyrBm by SDS–PAGE and by an activity assay measuring the oxidation of l-DOPA to dopachrome at 475 nm (Shuster & Fishman, 2009 ▶). The fractions containing TyrBm were combined, dialyzed against 50 mM Tris pH 8 buffer and then concentrated to 2 mg ml−1 by ultrafiltration using an Amicon Ultra 10K centrifugal filter (Millipore, Carrigtwohill, Ireland).
2.2. Crystallization and data collection
Crystallization trials were performed using the hanging-drop vapour-diffusion method with a Crystal Screen I kit (Hampton Research, Aliso Viejo, California, USA) at 293 K. Drops consisting of 2 µl protein solution (2 mg ml−1) and 2 µl of each trial mixture were equilibrated against a 600 µl reservoir of the trial mixture.
3. Results and discussion
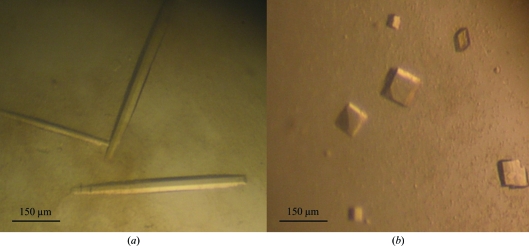
Initial crystallization trials with protein purified according to Shuster & Fishman (2009 ▶) yielded very small crystals within 1 d using 18%(w/v) PEG 8000, 0.2 M zinc acetate, 0.1 M cacodylic acid pH 6.5 at 293 K. In order to improve the crystal quality, AEC-HPLC was performed and the pH and temperature were changed to pH 5.6–6.1 and 289 K, respectively. The protein crystallized well under the modified conditions and large rod-shaped crystals (type 1) were obtained after 7–10 d and reached maximum dimensions of ∼500 × 50 × 50 µm (Fig. 1 ▶ a). Recognizing that the presence of a large excess of Zn ions might have an effect on the TyrBm structure (perhaps competing with the copper ions in the active site), crystallization in the absence of Zn ions was attempted. Using a modified precipitant solution (without the zinc acetate; pH 6.5, 293 K), cube-shaped crystals (type 2) were obtained after 14–21 d. The dimensions of the type 2 crystals were typically ∼80 × 80 × 80 µm (Fig. 1 ▶ b).
Figure 1.
Crystals of TyrBm. (a) Type 1 rod-shaped crystals that belonged to space group P212121. (b) Type 2 cubic crystals that belonged to space group P21.
X-ray diffraction data were collected on beamlines ID14-1 and ID23-1 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. All data were indexed, integrated, scaled and merged using MOSFLM and SCALA (Leslie, 1992 ▶). Crystal parameters and data-collection statistics are summarized in Table 1 ▶. The type 1 and type 2 crystals diffracted to a maximal resolution of 2.0–2.3 Å and analysis indicated that the type 1 crystals belonged to space group P212121, with unit-cell parameters a = 51.2, b = 84.5, c = 146.3 Å, while the type 2 crystals belonged to space group P21, with unit-cell parameters a = 47.8, b = 78.7, c = 85.6 Å, β = 105.3°. Assuming two molecules in the asymmetric unit, the Matthews coefficients for the two space groups are 2.2 and 2.0 Å3 Da−1, with solvent contents of 44 and 40.8%, respectively (Matthews, 1968 ▶).
Table 1. Summary of crystal parameters and data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Type 1 | Type 2 | |
|---|---|---|
| No. of crystals | 1 | 1 |
| Beamline | ESRF ID23-1 | ESRF ID14-1 |
| Wavelength (Å) | 1.000 | 0.933 |
| Detector | CCD | CCD |
| Crystal-to-detector distance (mm) | 241.95 | 239.28 |
| Rotation range per image (°) | 1 | 1 |
| Total rotation range (°) | 120 | 70 |
| Exposure time per image (s) | 2 | 2 |
| Resolution range (Å) | 50–2.0 (2.1–2.0) | 40–2.2 (2.3–2.2) |
| Space group | P212121 | P21 |
| Unit-cell parameters (Å, °) | a = 51.2, b = 84.5, c = 146.3 | a = 47.8, b = 78.5, c = 85.6, β = 101.6 |
| Total No. of measured intensities | 155304 | 60695 |
| Unique reflections | 42991 | 29606 |
| Multiplicity | 3.6 (3.8) | 2.1 (1.9) |
| Mean I/σ(I) | 7.7 (1.6) | 8.3 (2.4) |
| Completeness (%) | 98.1 (98.4) | 95.1 (87.3) |
| Rmerge† | 0.09 | 0.10 |
| Overall B factor from Wilson plot (Å2) | 25.3 | 12.4 |
R
merge = 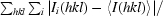
 , where Ii(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of the reflections.
, where Ii(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of the reflections.
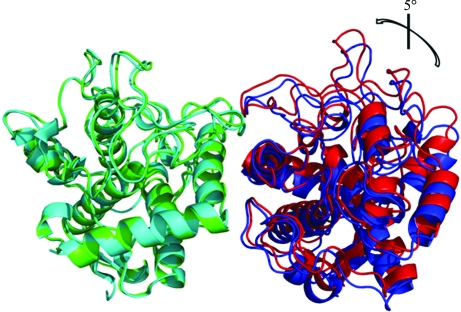
Molecular replacement was performed on a complete data set from a single type 1 crystal with Phaser (McCoy, 2007 ▶) using the structure of the full-length S. castaneoglobisporus tyrosinase monomer (PDB code 1wx2; 40% sequence identity; Matoba et al., 2006 ▶). Molecular replacement resulted in a single solution with a log-likelihood gain (LLG) of 664.33 and a Z score of 28.67. Following replacement of the amino-acid sequence of the model with that of TyrBm, the unrefined model was used to solve the structure of the type 2 crystal. The initial R work and R free values obtained after rigid-body refinement using REFMAC (Murshudov et al., 1997 ▶; Skubák et al., 2004 ▶) were 0.47 and 0.48, respectively. 10% of the reflections were used to calculate R free for both data sets. In the preliminary structures of the two crystal forms a movement of the dimer is observed, explaining the different space groups obtained (Fig. 2 ▶). The presence of Zn ions apparently affects the interactions of the two monomers in the type 1 crystals, inducing a rotation of ∼5°. A full description of the TyrBm structure and the resulting functional characterization will be described elsewhere.
Figure 2.
Preliminary structures of TyrBm: type 1 (cyan and blue) and type 2 (green and red) asymmetric units. All monomers superimpose equally (r.m.s.d. of ∼0.3–0.4 Å). In the figure the Cα atoms of subunits A (cyan and green, respectively) of the two crystal forms were superimposed using the algorithm embedded in PyMOL (http://www.pymol.org/), showing that the position of the type 2 monomer B (red) is rotated by about 5° in comparison to the type 1 monomer B (blue). This rotation induces the change in packing that modifies the crystal lattice.
Acknowledgments
This work was supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities, grant number 535/07.
References
- Burton, S. G. (2003). Trends Biotechnol.21, 543–549. [DOI] [PubMed]
- Claus, H. & Decker, H. (2006). Syst. Appl. Microbiol.29, 3–14. [DOI] [PubMed]
- Cuff, M. E., Miller, K. I., van Holde, K. E. & Hendrickson, W. A. (1998). J. Mol. Biol.278, 855–870. [DOI] [PubMed]
- Decker, H., Schweikardt, T., Nillius, D., Salzbrunn, U., Jaenicke, E. & Tuczek, F. (2007). Gene, 398, 183–191. [DOI] [PubMed]
- Decker, H., Schweikardt, T. & Tuczek, F. (2006). Angew. Chem. Int. Ed. Engl.45, 4546–4550. [DOI] [PubMed]
- Gaykema, W. P. J., Hol, W. G. J., Vereijken, J. M., Soeter, N. M., Bak, H. J. & Beintema, J. J. (1984). Nature (London), 309, 23–29.
- Gerdemann, C., Eicken, C. & Krebs, B. (2002). Acc. Chem. Res.35, 183–191. [DOI] [PubMed]
- Hazes, B., Magnus, K. A., Bonaventura, C., Bonaventura, J., Dauter, Z., Kalk, K. H. & Hol, W. G. (1993). Protein Sci.2, 597–619. [DOI] [PMC free article] [PubMed]
- Hernandez-Romero, D., Sanchez-Amat, A. & Solano, F. (2006). FEBS J.273, 257–270. [DOI] [PubMed]
- Itoh, S. & Fukuzumi, S. (2007). Acc. Chem. Res.40, 592–600. [DOI] [PubMed]
- Klabunde, T., Eicken, C., Sacchettini, J. C. & Krebs, B. (1998). Nature Struct. Biol.5, 1084–1090. [DOI] [PubMed]
- Kong, K.-H., Hong, M.-P., Choi, S.-S., Kim, Y.-T. & Cho, S.-H. (2000). Biotechnol. Appl. Biochem.31, 113–118. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Li, Y., Wang, Y., Jiang, H. & Deng, J. (2009). Proc. Natl Acad. Sci. USA, 106, 17002–17006. [DOI] [PMC free article] [PubMed]
- Liu, N., Zhang, T., Wang, Y. J., Huang, Y. P., Ou, J. H. & Shen, P. (2004). Lett. Appl. Microbiol.39, 407–412. [DOI] [PubMed]
- Matoba, Y., Kumagai, T., Yamamoto, A., Yoshitsu, H. & Sugiyama, M. (2006). J. Biol. Chem.281, 8981–8990. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491. [DOI] [PubMed]
- McCoy, A. J. (2007). Acta Cryst. D63, 32–41. [DOI] [PMC free article] [PubMed]
- McMahon, A. M., Doyle, E. M., Brooks, S. & O’Connor, K. E. (2007). Enzyme Microb. Technol.40, 1435–1441.
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Rosenzweig, A. C. & Sazinsky, M. H. (2006). Curr. Opin. Struct. Biol.16, 729–735. [DOI] [PubMed]
- Shuster, V. & Fishman, A. (2009). J. Mol. Microbiol. Biotechnol.17, 188–200. [DOI] [PubMed]
- Skubák, P., Murshudov, G. N. & Pannu, N. S. (2004). Acta Cryst. D60, 2196–2201. [DOI] [PubMed]
- Virador, V. M., Reyes Grajeda, J. P., Blanco-Labra, A., Mendiola-Olaya, E., Smith, G. M., Moreno, A. & Whitaker, J. R. (2010). J. Agric. Food Chem.58, 1189–1201. [DOI] [PubMed]