GenX, a lysyl-tRNA synthetase paralogue from Escherichia coli, has been overexpressed in E. coli, purified by three chromatographic steps and cocrystallized with a lysyl adenylate analogue (LysAMS) by the hanging-drop vapour-diffusion method using PEG 4000 as a precipitant.
Keywords: GenX, lysyl-tRNA synthetase paralogue, translation elongation factor P
Abstract
GenX, a lysyl-tRNA synthetase paralogue from Escherichia coli, was overexpressed in E. coli, purified by three chromatographic steps and cocrystallized with a lysyl adenylate analogue (LysAMS) by the hanging-drop vapour-diffusion method using PEG 4000 as a precipitant. The GenX–LysAMS crystals belonged to the triclinic space group P1, with unit-cell parameters a = 54.80, b = 69.15, c = 94.08 Å, α = 95.47, β = 106.51, γ = 90.46°, and diffracted to 1.9 Å resolution. Furthermore, GenX was cocrystallized with translation elongation factor P (EF-P), which is believed to be a putative substrate of GenX, and LysAMS using PEG 4000 and ammonium sulfate as precipitants. The GenX–EF-P–LysAMS crystals belonged to the monoclinic space group P21, with unit-cell parameters a = 105.93, b = 102.96, c = 119.94 Å, β = 99.4°, and diffracted to 2.5 Å resolution. Structure determination of the E. coli GenX–LysAMS and GenX–EF-P–LysAMS complexes by molecular replacement was successful and structure refinements are now in progress.
1. Introduction
Aminoacyl-tRNA synthetases (aaRSs) catalyze the esterification of a specific amino acid to the 3′-terminal adenosine of tRNA (Schimmel, 1987 ▶). The proper connection between the amino acid and the tRNA is based on strict recognition by the 20 kinds of aaRS. The aaRSs have been partitioned into two classes (class I and class II) on the basis of conserved sequences and characteristic structure motifs (Cusack et al., 1990 ▶; Eriani et al., 1990 ▶). Class I aaRSs have two conserved motifs and generally form a monomer or dimer, while class II aaRSs have three conserved motifs and form a dimer or tetramer. Whole-genome analyses have revealed that aaRS paralogues, many with unknown functions, exist in various species (Schimmel & Ribas de Pouplana, 2000 ▶; Novoa et al., 2010 ▶). GenX is a paralogue of class II lysyl-tRNA synthetase (LysRS), which exists as a dimer, and is similar to the C-terminal catalytic domain of LysRS (Kong et al., 1991 ▶; Kaniga et al., 1998 ▶; Peng et al., 2001 ▶; Bailly & de Crécy-Lagard, 2010 ▶). GenX lacks the tRNA anticodon-binding domain of LysRS, which indicates that GenX by itself does not play the role of a classical aaRS. Studies using a database of known and predicted protein interactions, as well as a comparative genomic analysis, led to the hypothesis that GenX may interact with translation elongation factor P (EF-P; von Mering et al., 2005 ▶; Bailly & de Crécy-Lagard, 2010 ▶), which has been reported to be an essential protein for the stimulation of peptidyl transferase activity in bacteria (Glick & Ganoza, 1975 ▶, 1979 ▶; Ganoza et al., 2002 ▶). Crystallographic analyses revealed that EF-P adopts a tRNA-like L-shaped structure (Hanawa-Suetsugu et al., 2004 ▶; Blaha et al., 2009 ▶). However, no biochemical or structural studies supporting the functional relevance of the interaction between GenX and EF-P have been reported.
Therefore, to investigate whether EF-P is a substrate of GenX and whether EF-P is a binding partner of GenX, we attempted to crystallize the complex of GenX and EF-P. Here, we report the crystallization and preliminary X-ray crystallographic analysis of GenX from Escherichia coli complexed with a lysyl-AMP analogue, 5′-O-[N-(l-lysyl)sulfamoyl]adenosine (LysAMS), and of GenX complexed with E. coli EF-P and LysAMS. This is the first report of the crystallization of an aaRS paralogue–translation factor complex.
2. Materials and methods
2.1. Cloning, expression and purification
The genX gene (978 bp) encoding E. coli GenX was PCR-amplified using Pyrobest DNA polymerase (Takara) from E. coli MC4100 genomic DNA as a template and was cloned into the pET28c vector (Novagen). E. coli strain BL21 (DE3) cells transformed with recombinant GenX were incubated at 293 K in 1 l Luria–Bertani (LB) broth with shaking. When the culture reached an OD600 of 0.6, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce recombinant protein expression. After an additional 16 h culture at 293 K, the cells were harvested by centrifugation and suspended in 50 ml buffer A (50 mM potassium phosphate buffer pH 7.4 containing 150 mM NaCl, 5 mM β-mercaptoethanol, 10% glycerol and 50 mM imidazole) with protease inhibitors (Complete EDTA-free; Roche Diagnostics). After sonication, the cellular debris was removed by centrifugation (20 000g for 30 min at 277 K) and the supernatant thus obtained was loaded onto a HisTrap HP column (5 ml; GE Healthcare) pre-equilibrated with buffer A. The column was washed with 50 ml buffer A and GenX was eluted with buffer B (50 mM potassium phosphate buffer pH 7.4 containing 150 mM NaCl, 5 mM β-mercaptoethanol, 10% glycerol and 300 mM imidazole). The GenX-containing fractions were pooled, dialyzed against buffer C (50 mM potassium phosphate buffer pH 7.5 containing 10% glycerol and 1 mM DTT) and applied onto a HiTrap Heparin HP column (5 ml; GE Healthcare) to remove cationic contaminants as well as the basic proteins from E. coli. The flowthrough fractions were collected and applied onto a HiTrap Q (5 ml; GE Healthcare) column. The protein was eluted with a linear gradient of 0–0.25 M NaCl. The GenX-containing fractions were pooled and dialyzed against buffer D (10 mM Na HEPES buffer pH 7.5 containing 5 mM MgCl2, 150 mM NaCl and 10 mM β-mercaptoethanol). The purified GenX was concentrated to 18 mg ml−1 with a Centricon YM-30 filter (Millipore). The GenX protein used for crystallization contained a vector-derived histidine tag and a linker sequence (GSSHHHHHHSSGLVPRGSH) at the N-terminus. The procedures for the cloning of the efp gene (567 bp) encoding E. coli EF-P and the expression and purification using a HisTrap HP column of the EF-P protein were the same as those used for GenX. The histidine tag was cleaved with thrombin at 277 K for 16 h and the EF-P protein was applied onto a HiTrap Q column equilibrated with buffer C. The protein was eluted with a linear gradient of 0–0.3 M NaCl. The EF-P-containing fractions were pooled and dialyzed against buffer D. The purified EF-P was concentrated to 10 mg ml−1 with a Centricon YM-10 filter. The EF-P protein used for crystallization contained three extra amino-acid residues (GSH), which were part of the linker sequence, at the N-terminus. The homogeneity of the proteins was confirmed by SDS–PAGE and the protein concentration was determined by the Bradford method using the Coomassie Plus Protein Assay reagent (Thermo Scientific). All of the purification procedures described above were conducted at 277 K.
2.2. Crystallization
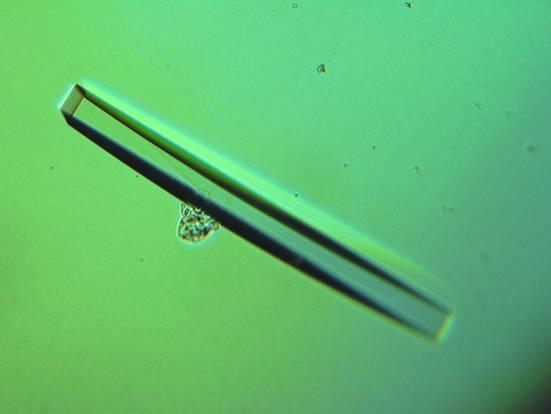
For the crystallization of the GenX–LysAMS complex, the aminoacyl adenylate substrate analogue LysAMS was added to the GenX protein solution (10 mg ml−1) to a final concentration of 5 mM. The initial crystallization conditions were determined by means of the hanging-drop vapour-diffusion method at 293 K using Crystal Screen, Crystal Screen 2 and Natrix (Hampton Research). The volume of the reservoir solution was 500 µl and the drops contained 1 µl each of the GenX–LysAMS mixture solution and the reservoir solution. Crystals were obtained using polyethylene glycol as a precipitant. The initial crystallization conditions were further refined by changing the pH and the concentration of the precipitants and by screening additives. The most promising crystals were obtained by mixing 1 µl protein solution with 1 µl reservoir solution containing 0.1 M Na HEPES pH 6.5 and 20%(v/v) PEG 4000 (Fig. 1 ▶). The triangular plate-like crystals grew to maximum dimensions of 0.6 × 0.2 × 0.05 mm in 2–5 d.
Figure 1.
A crystal of GenX complexed with LysAMS.
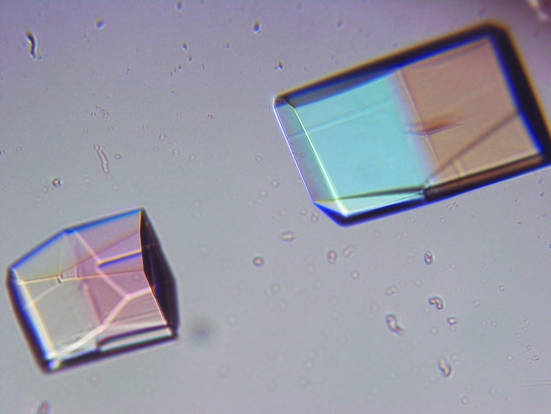
For cocrystallization of the GenX–EF-P–LysAMS complex, GenX and EF-P were mixed in a 1:1 molar ratio, with each at a final concentration of 0.25 mM (9 and 5 mg ml−1, respectively), and LysAMS was added to the protein solution to a final concentration of 5 mM. Crystals were obtained using polyethylene glycol and ammonium sulfate as precipitants. The most promising crystals were obtained by mixing 1 µl protein solution with 1 µl reservoir solution containing 0.1 M sodium cacodylate pH 6.5, 30%(v/v) PEG 4000 and 0.2 M ammonium sulfate (Fig. 2 ▶). The plate-like crystals grew to maximum dimensions of 0.3 × 0.2 × 0.05 mm in 4–5 d. To confirm that the crystals contained both proteins, the crystals were washed with reservoir solution several times, dissolved in SDS–PAGE sample buffer and analyzed by SDS–PAGE. The crystal solution obtained by dissolving the crystals yielded two protein bands corresponding to GenX and EF-P upon SDS–PAGE (Fig. 3 ▶).
Figure 2.
Crystals of the GenX–EF-P–LysAMS complex.
Figure 3.
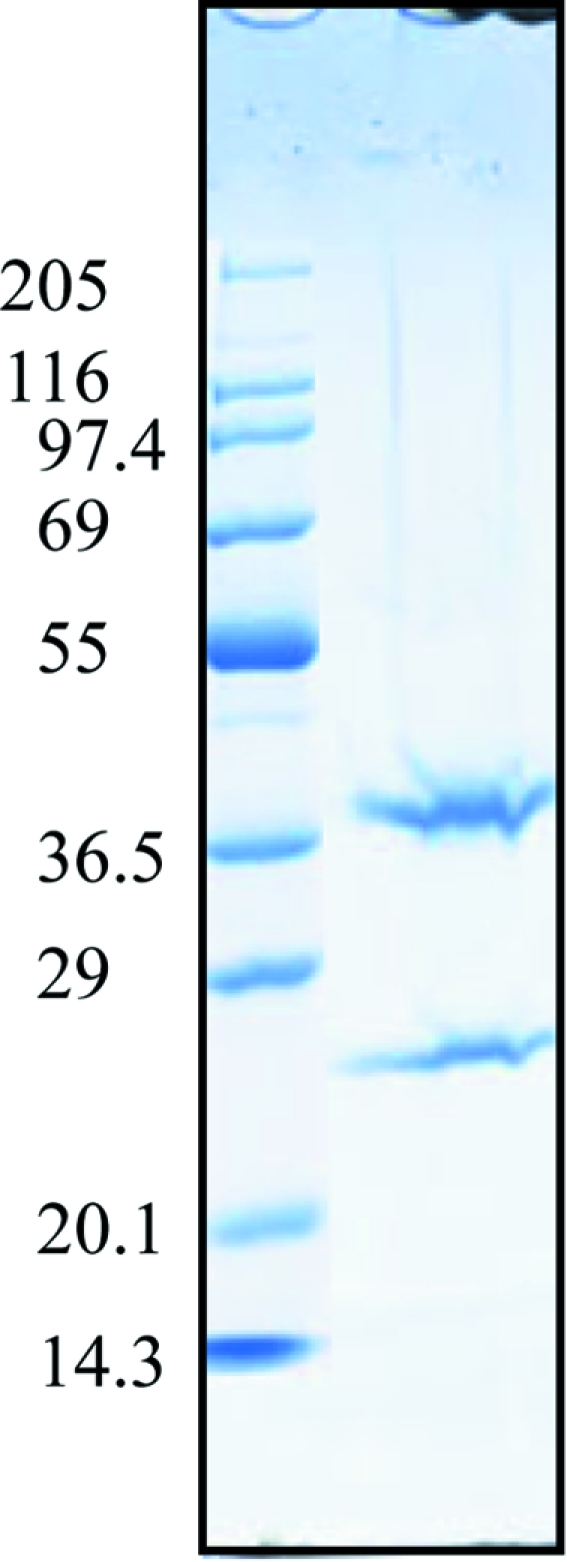
SDS–PAGE analysis of the GenX–EF-P–LysAMS complex crystals. Lane 1, marker proteins; the size of each protein (kDa) is shown on the left. Lane 2, dissolved crystals.
2.3. X-ray data collection and processing
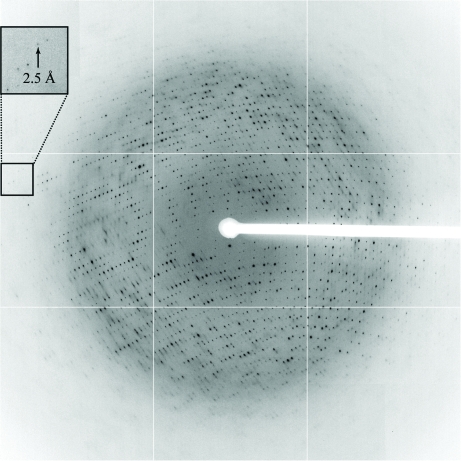
Before measurement, the crystals were soaked in a cryoprotection solution by stepwise transfer and flash-cooled in liquid nitrogen. The cryoprotection solution for the GenX–LysAMS complex crystal consisted of 0.12 M Na HEPES buffer pH 6.5, 24%(v/v) PEG 4000 and 20%(v/v) PEG 400; that for the GenX–EF-P–LysAMS complex crystal consisted of 0.1 M sodium cacodylate buffer pH 6.5, 30%(v/v) PEG 4000, 0.2 M ammonium sulfate and 10% trehalose. The data set for the GenX–LysAMS complex crystal, which diffracted to 1.9 Å resolution, was collected on Photon Factory beamline BL-5A using an ADSC Quantum 315 CCD detector. A total of 360 frames were collected with a crystal-to-detector distance of 249 mm, an oscillation angle of 1° and an exposure time of 1 s per frame at a wavelength of 1.0000 Å at 100 K in a cold nitrogen stream. The data set for the GenX–EF-P–LysAMS complex crystal, which diffracted to 2.5 Å resolution, was collected on SPring-8 beamline BL41XU (Fig. 4 ▶) using an ADSC Q315 detector. A total of 180 frames were collected with a crystal-to-detector distance of 350 mm, a 1° oscillation range and an exposure time of 0.5 s per frame at a wavelength of 1.0000 Å at 100 K in a cold nitrogen stream. The data were processed with HKL-2000 (Otwinowski & Minor, 1997 ▶). Other crystallographic calculations were performed with the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶). The statistics of the data collections are summarized in Table 1 ▶.
Figure 4.
X-ray diffraction image from a GenX–EF-P–LysAMS complex crystal. The small arrow indicates the diffraction limit, which corresponds to 2.5 Å.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| GenX–LysAMS complex | GenX–EF-P–LysAMS complex | |
|---|---|---|
| X-ray source | Photon Factory BL-5A | SPring-8 BL41XU |
| Wavelength (Å) | 1.0000 | 1.0000 |
| Space group | P1 | P21 |
| Unit-cell parameters (Å, °) | a = 95.47, b = 69.15, c = 64.08, α = 95.47, β = 106.51, γ = 90.46 | a = 105.93, b = 102.96, c = 119.94, α = 90, β = 99.4, γ = 90 |
| Resolution (Å) | 50–1.9 (1.97–1.9) | 50–2.5 (2.59–2.5) |
| Observed reflections | 364770 | 300059 |
| Unique reflections | 99019 | 82592 |
| Redundancy | 3.7 | 3.6 |
| Completeness (%) | 95.4 (77.9) | 93.8 (67.4) |
| I/σ(I) | 17.15 (2.85) | 21.28 (2.38) |
| Rmerge (%) | 8.6 (27.8) | 5.8 (35.7) |
3. Results and discussion
The GenX and EF-P proteins from E. coli were successfully overexpressed in E. coli BL21 (DE3) and purified to homogeneity; 160 and 60 mg protein was obtained per litre of LB medium, respectively. Crystallization of GenX and of the GenX–EF-P complex was only successful in the presence of LysAMS. Gel-filtration assays demonstrated that GenX exists as a dimer in solution, while the GenX dimer did not form a stable complex with EF-P (data not shown). The GenX–LysAMS complex was crystallized in reservoir solution containing PEG 4000 as a precipitant (Fig. 1 ▶). The GenX–LysAMS crystals belonged to the triclinic space group P1, with unit-cell parameters a = 54.80, b = 69.15, c = 94.08 Å, α = 95.47, β = 106.51, γ = 90.46°. The asymmetric unit contained four GenX–LysAMS molecules, with a corresponding crystal volume per protein weight (V M) of 2.18 Å3 Da−1 and a solvent content of 43.56%. Molecular replacement was performed using the MOLREP program (Collaborative Computational Project, Number 4, 1994 ▶) with reflections in the 50.0–3.0 Å resolution range. Since E. coli GenX shares 30% sequence identity with the catalytic domain of E. coli LysRS, we created a homology model of the GenX structure from the catalytic domain of LysRS (PDB code 1lyl; Onesti et al., 1995 ▶) using the 3D-PSSM automatic fold-recognition server (http://www.sbg.bio.ic.ac.uk/~3dpssm). Using this structure as a search model, the program placed four GenX monomers in the asymmetric unit, with correlation and R factors of 22.3% and 55.3%, respectively. The R free factor decreased to 54.5, 50.0 and 49.1% after initial rigid-body refinement, subsequent minimization and individual B-factor refinement using CNS (Brünger et al., 1998 ▶), respectively. Four GenX monomers form two dimers, in a similar manner to the typical quaternary structure of the catalytic domain of LysRS (Onesti et al., 1995 ▶; Cusack et al., 1996 ▶; Desogus et al., 2000 ▶). Further structure refinement is now in progress.
The GenX–EF-P–LysAMS complex was crystallized in reservoir solution containing PEG 4000 and ammonium sulfate as precipitants (Fig. 2 ▶). The GenX–EF-P–LysAMS complex crystals belonged to the monoclinic space group P21, with unit-cell parameters a = 105.93, b = 102.96, c = 119.94 Å, α = γ = 90.0°, β = 99.4°. The asymmetric unit contained four GenX–EF-P–LysAMS molecules, with a corresponding crystal volume per protein weight (V M) of 2.69 Å3 Da−1 and a solvent content of 54.34%. The initial molecular replacement was performed with MOLREP, using the partially refined structure of the E. coli GenX dimer as a search model. The program successfully placed two dimers in the asymmetric unit. After initial refinement with CNS, electron density corresponding to EF-P was observed in the F o − F c map. The E. coli EF-P structure, which was predicted from that of Thermus thermophilus EF-P (PDB code 1ueb; Hanawa-Suetsugu et al., 2004 ▶) using the 3D-PSSM server, was manually docked to the GenX dimer according to the F o − F c electron density. E. coli EF-P shares 46% sequence identity with T. thermophilus EF-P. We then performed a second molecular replacement with the structure of the GenX–EF-P heterodimer as the search model. Molecular replacement using MOLREP with reflections in the 50.0–3.0 Å resolution range yielded a solution and the program placed four GenX–EF-P heterodimers in the asymmetric unit, with correlation and R factors of 49.2% and 55.6%, respectively. The R free factor for the GenX–EF-P–LysAMS complex was 53.3, 46.8 and 44.4% after initial rigid-body refinement, subsequent minimization and individual B-factor refinement using CNS, respectively. Structure refinement of the model is currently under way and details of the structural analyses will be published elsewhere. The structure of the GenX–EF-P complex will provide new structural insights into the functional relevance of the interaction between GenX and EF-P.
Acknowledgments
We thank the staff of beamline BL41XU at SPring-8 and the staff of the BL-5A beamline at the Photon Factory for technical assistance. We also thank Drs Shun-ichi Sekine, Takuhiro Ito, Ryuya Fukunaga (The University of Tokyo) and Toru Sengoku (RIKEN) for assisting with data collection and for helpful discussions. We are grateful to Azusa Ishii and Tomoko Nakayama for clerical assistance. This work was supported in part by Grants-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, the Targeted Proteins Research Program (TPRP) and the RIKEN Structural Genomics/Proteomics Initiative (RSGI) of the National Project on Protein Structural and Functional Analyses, MEXT.
References
- Bailly, M. & de Crécy-Lagard, V. (2010). Biol. Direct, 5, 3. [DOI] [PMC free article] [PubMed]
- Blaha, G., Stanley, R. E. & Steitz, T. A. (2009). Science, 325, 966–970. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cusack, S., Berthet-Colominas, C., Hartlein, M., Nassar, N. & Leberman, R. (1990). Nature (London), 347, 249–255. [DOI] [PubMed]
- Cusack, S., Yaremchuk, A. & Tukalo, M. (1996). EMBO J.15, 6321–6334. [PMC free article] [PubMed]
- Desogus, G., Todone, F., Brick, P. & Onesti, S. (2000). Biochemistry, 39, 8418–8425. [DOI] [PubMed]
- Eriani, G., Delarue, M., Poch, O., Gangloff, J. & Moras, D. (1990). Nature (London), 347, 203–206. [DOI] [PubMed]
- Ganoza, M. C., Kiel, M. C. & Aoki, H. (2002). Microbiol. Mol. Biol. Rev.66, 460–485. [DOI] [PMC free article] [PubMed]
- Glick, B. R. & Ganoza, M. C. (1975). Proc. Natl Acad. Sci. USA, 72, 4257–4260. [DOI] [PMC free article] [PubMed]
- Glick, B. R. & Ganoza, M. C. (1979). Eur. J. Biochem.97, 23–28. [DOI] [PubMed]
- Hanawa-Suetsugu, K., Sekine, S., Sakai, H., Hori-Takemoto, C., Terada, T., Unzai, S., Tame, J. R., Kuramitsu, S., Shirouzu, M. & Yokoyama, S. (2004). Proc. Natl Acad. Sci. USA, 101, 9595–9600. [DOI] [PMC free article] [PubMed]
- Kaniga, K., Compton, M. S., Curtiss, R. III & Sundaram, P. (1998). Infect. Immunol.66, 5599–5606. [DOI] [PMC free article] [PubMed]
- Kong, L., Fromant, M., Blanquet, S. & Plateau, P. (1991). Gene, 108, 163–164. [DOI] [PubMed]
- Mering, C. von, Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., Jouffre, N., Huynen, M. A. & Bork, P. (2005). Nucleic Acids Res.33, D433–D437. [DOI] [PMC free article] [PubMed]
- Novoa, E. M., Castro de Moura, M., Orozco, M. & Ribas de Pouplana, R. (2010). FEBS Lett.584, 460–466. [DOI] [PubMed]
- Onesti, S., Miller, A. D. & Brick, P. (1995). Structure, 3, 163–176. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Peng, W.-T., Banta, L. H., Charles, T. C. & Nester, E. W. (2001). J. Bacteriol.183, 36–45. [DOI] [PMC free article] [PubMed]
- Schimmel, P. (1987). Annu. Rev. Biochem.56, 125–158. [DOI] [PubMed]
- Schimmel, P. & Ribas de Pouplana, R. (2000). Trends Biochem. Sci.25, 207–209. [DOI] [PubMed]