The crystallization of the homologous recombination mediators Swi5 and Sfr1 from fission yeast is reported.
Keywords: homologous recombination mediators, Swi5, Sfr1, fission yeast
Abstract
The assembly of the presynaptic filament of recombinases represents the most important step in homologous recombination. The formation of the filament requires assistance from mediator proteins. Swi5 and Sfr1 have been identified as mediators in fission yeast and these proteins form a complex that stimulates strand exchange. Here, the expression, purification and crystallization of Swi5 and its complex with an N-terminally truncated form of Sfr1 (ΔN180Sfr1) are presented. Analytical ultracentrifugation of the purified samples showed that Swi5 and the protein complex exist as tetramers and heterodimers in solution, respectively. Swi5 was crystallized in two forms belonging to space groups C2 and R3 and the crystals diffracted to 2.7 Å resolution. Swi5–ΔN180Sfr1 was crystallized in space group P21212 and the crystals diffracted to 2.3 Å resolution. The crystals of Swi5 and Swi5–ΔN180Sfr1 are likely to contain one tetramer and two heterodimers in the asymmetric unit, respectively.
1. Introduction
Homologous recombination is crucial for achieving genetic diversity during meiosis and for the repair of damaged DNA owing to double-strand breaks during mitosis (Camerini-Otero & Hsieh, 1995 ▶; Bianco et al., 1998 ▶; Symington, 2002 ▶). The central reaction in recombination is the exchange of strands between two homologous DNA molecules, which is catalyzed by the RecA family of ATPases. In eukaryotes, the RecA-like recombinases Rad51 and Dmc1 initiate strand exchange by forming a helical nucleoprotein filament on single-stranded DNA (ssDNA) which promotes subsequent pairing and strand-exchange reactions (Cox, 2007 ▶; Sung et al., 2000 ▶). Although the presynaptic filaments are the essential functional units in homologous recombination (Masson & West, 2001 ▶; Sehorn et al., 2004 ▶), presynaptic filament assembly is particularly prone to interference by replication factor A (RPA), which binds ssDNA. Proteins that are capable of overcoming the inhibitory effect of RPA on filament assembly are referred to as mediators (Sung & Klein, 2006 ▶). Swi5 and Sfr1 have been identified as mediators in Schizosaccharomyces pombe (Akamatsu et al., 2003 ▶).
Swi5 is a small protein that is evolutionarily conserved from yeast to humans and Sfr1 (Swi5-dependent recombination repair protein 1) has been identified as a protein that interacts with Swi5 (Akamatsu et al., 2003 ▶). Recent studies have revealed that the Swi5–Sfr1 complex stimulates both Rad51-mediated and Dmc1-mediated strand-exchange reactions by activating the recombinases directly. Thus, Swi5–Sfr1 acts as an activator of the recombinases (Haruta et al., 2007 ▶; Kurokawa et al., 2008 ▶). Determining the crystal structures of Swi5 and Sfr1 is of importance because such molecular information will provide a better understanding of the function of Swi5 and Sfr1 in homologous recombination. However, no structural studies of Swi5 and Sfr1 have been conducted. Here, the expression, purification and crystallization of Swi5 and of its complex with ΔN180Sfr1 are reported. The results show that Swi5 forms a tetramer, whereas the complex exists as a heterodimer with a 1:1 stoichiometry in solution.
2. Materials and methods
2.1. Limited proteolysis
After purification of full-length Swi5–Sfr1 using the previously published protocol (Haruta et al., 2006 ▶), the sample (5 mg ml−1) was digested with trypsin or α-chymotrypsin for various times on ice at a 1:250 protease:protein ratio. Digestion was terminated by boiling the samples with SDS sample buffer (50 mM Tris–HCl pH 6.8, 1% SDS, 10% glycerol, 0.01% BPB, 1.6% β-mercaptoethanol). The proteolytic products were resolved by SDS–PAGE and visualized by Coomassie Brilliant Blue staining. The proteolytic fragments were in-gel digested by trypsin and identified by electrospray ionization mass spectrometry.
2.2. Protein expression and purification
The cDNA of swi5 was subcloned into a pET11a vector (Novagen). Swi5 was overexpressed by Escherichia coli BL21 (DE3) Codon Plus RIL cells (Stratagene) transformed with the plasmid. The E. coli cells were grown at 310 K to an OD600 of ∼0.5 in LB medium containing 50 µg ml−1 ampicillin, 34 µg ml−1 chloramphenicol and 0.2% glucose. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM and the culture was incubated overnight at 291 K. The harvested cell pellet was suspended in buffer 1 (50 mM Tris–HCl pH 7.8, 500 mM NaCl, 2 mM EDTA, 1 mM DTT) with 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and the cells were disrupted by sonication. After the addition of polyethyleneimine to a final concentration of 0.05%, the lysate was centrifuged to remove the insoluble fraction. Ammonium sulfate was added to the supernatant to a 50% saturation concentration and the sample was stirred for 30 min on ice and subsequently centrifuged. The pellet was suspended in buffer 2 (50 mM Tris–HCl pH 7.8, 100 mM NaCl, 1 mM EDTA, 1 mM DTT) and loaded onto Q Sepharose (GE Healthcare). The sample was eluted by buffer 3 (50 mM Tris–HCl pH 7.8, 250 mM NaCl, 1 mM EDTA, 2 mM DTT), diluted and loaded onto HiTrap Heparin (GE Healthcare) after being passed through HiTrap SP. The eluted fraction containing Swi5 was passed through HiTrap Q and loaded onto a Superdex 200 26/60 (GE Healthcare) column equilibrated with buffer 4 (5 mM Tris–HCl pH 7.8, 200 mM NaCl, 1 mM DTT, 2 mM EDTA). The fractions containing the target protein were collected and concentrated to ∼20 mg ml−1.
For the expression and purification of Swi5–Sfr1 containing residues 178–299 of Sfr1 (ΔN177Sfr1), a previously published protocol (Haruta et al., 2006 ▶) was used with minor modifications.
For crystallization, a His6 tag and a TEV protease recognition site were attached to the N-terminal end of Sfr1 containing residues 181–299 (ΔN180Sfr1). After Swi5 and ΔN180Sfr1 has been coexpressed using the same protocol (Haruta et al., 2006 ▶), the cell pellet was suspended in buffer A (50 mM HEPES–NaOH pH 7.0, 500 mM NaCl, 10 mM imidazole, 1 mM DTT) and sonicated. After centrifugation (40 000g, 277 K, 30 min), the supernatant was loaded onto Ni-Sepharose (GE Healthcare). After washing the Ni-Sepharose material with buffer A containing 20 mM imidazole, the target protein was eluted using buffer A containing 200 mM imidazole. Following the addition of TEV protease at a ratio of 1:20 (protease:protein), the sample was incubated on ice overnight. The sample was diluted, passed through a HiTrap Q column and loaded onto a HiTrap SP column. The eluted fractions containing Swi5–ΔN180Sfr1 were collected and passed through a HisTrap HP column. The sample was subsequently loaded onto a Superdex 75 16/60 column equilibrated with buffer consisting of 10 mM HEPES–NaOH pH 7.0, 200 mM NaCl and 1 mM DTT. The peak fractions were collected and concentrated to 15 mg ml−1.
Purified samples were frozen in liquid nitrogen and stored at 193 K until use.
2.3. Analytical ultracentrifugation
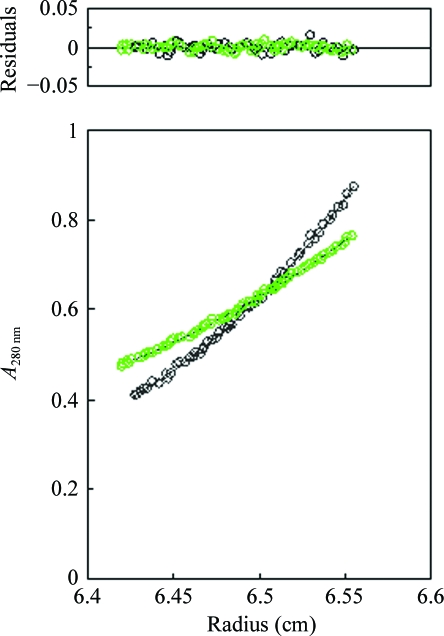
Sedimentation-equilibrium experiments were carried out in cells with a six-channel centrepiece and quartz windows. The protein concentrations used were 1.3, 2.5 and 5.0 mg ml−1 for Swi5 and 0.5, 1.3 and 2.2 mg ml−1 for Swi5–ΔN177Sfr1. Data were obtained at 10 000, 15 000 and 20 000 rev min−1. Data analysis was performed by global analysis of data sets obtained at different loading concentrations and rotor speeds using ULTRASPIN (MRC Centre for Protein Engineering, Cambridge, England; Fig. 1 ▶). The experiments were performed at 293 K. The reference buffers contained 5 mM Tris–HCl pH 7.8, 200 mM NaCl, 1 mM DTT and 2 mM EDTA for Swi5 and 20 mM Tris–HCl pH 7.5, 200 mM NaCl, 10% glycerol, 1 mM DTT and 1 mM EDTA for Swi5–ΔN177Sfr1.
Figure 1.
Sedimentation-equilibrium analysis of Swi5 and Swi5–ΔN177Sfr1. Sedimentation-equilibrium data are shown with the residuals from the best fit to a single ideal species. The plots show data for Swi5 at 2.5 mg ml−1 and 15 000 rev min−1 (black) and for Swi5–ΔN177Sfr1 at 1.3 mg ml−1 and 15 000 rev min−1 (green).
2.4. Crystallization
Initial crystallization trials were performed by the sitting-drop vapour-diffusion method in 96-well or 24-well plates at 293 and 277 K using a series of crystallization kits (Hampton Research and Qiagen).
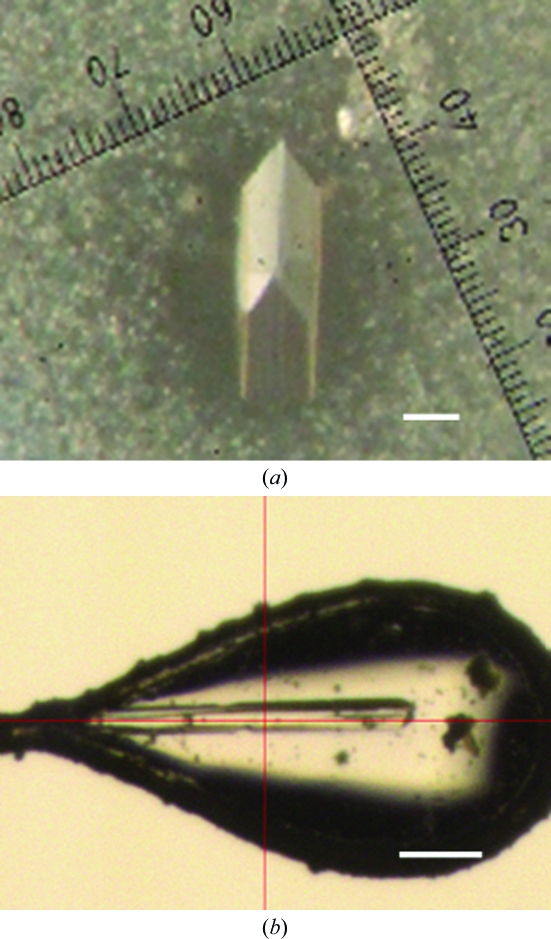
The best crystals of Swi5 were obtained by mixing 10 mg ml−1 Swi5 with a reservoir solution consisting of 20% MPD, 0.1 M Tris–HCl pH 8.0 and 24.5 mM octyl β-d-glucoside. Crystals grew to dimensions of approximately 1.0 × 0.2 × 0.1 mm in a few weeks and could be directly cryocooled (Fig. 2 ▶).
Figure 2.
Crystals of (a) Swi5 (space group C2) and (b) Swi5–ΔN180Sfr1. The scale bars correspond to 100 µm.
In contrast, the best crystals of Swi5–ΔN180Sfr1 were obtained by mixing 1 µl 7.5 mg ml−1 Swi5–ΔN180Sfr1, 0.9 µl reservoir solution consisting of 15% PEG MME 2000, 0.2 M sodium acetate pH 5.5 and 0.20 M ammonium sulfate, and 0.1 µl 6 M ammonium nitrate and 0.1 M sodium acetate pH 4.6 at 277 K. The needle-shaped crystals grew to dimensions of approximately 0.8 × 0.02 × 0.02 mm (Fig. 2 ▶). The crystals were transferred to a reservoir solution containing 20% glycerol for cryocooling.
2.5. Data collection and processing
Diffraction data were collected at 100 K on beamline BL-17A at the Photon Factory (PF), Tsukuba, Japan using an ADSC Quantum 270 detector and on beamline NW12A at the Photon Factory Advanced Ring (PF-AR) using an ADSC Quantum 210r detector. Diffraction images were indexed, integrated and scaled using the HKL-2000 package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
The results of the limited proteolysis revealed that the C-terminal region (residues ∼178−299) of Sfr1 is stable but the N-terminus is cleaved (data not shown). To investigate the properties of purified Swi5 and Swi5–ΔN177Sfr1 in solution, analytical ultracentrifugation experiments were performed. Sedimentation-equilibrium analysis showed that the molecular masses of Swi5 (9.7 kDa monomer) and of its complex with ΔN177Sfr1 (14.1 kDa monomer) were 37 and 27 kDa, respectively. These results suggest that Swi5 forms a tetramer and Swi5–ΔN177Sfr1 forms a 1:1 complex in solution (Fig. 1 ▶).
Swi5 was crystallized in two crystal forms that belonged to space groups R3 and C2 using the same crystallization condition. The R3 crystals, with unit-cell parameters a = b = 159.2, c = 69.4 Å, diffracted to 2.8 Å resolution and the C2 crystals, with unit-cell parameters a = 177.1, b = 57.8, c = 64.4 Å, β = 96.7°, diffracted to 2.7 Å resolution (Table 1 ▶). Assuming that one tetramer was present in the asymmetric unit, the Matthews coefficients for the R3 and C2 crystals were 4.34 and 4.21 Å3 Da−1 and the solvent contents were 71.8 and 70.8%, respectively (Matthews, 1968 ▶).
Table 1. X-ray diffraction data for Swi5 and the Swi5–ΔN180Sfr1 complex.
Values in parentheses are for the outer resolution shell.
| Swi5 form 1 | Swi5 form 2 | Swi5–ΔN180Sfr1 complex | |
|---|---|---|---|
| X-ray source | PF-AR NW12A | PF-AR NW12A | PF BL-17A |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 |
| Space group | R3 | C2 | P21212 |
| Unit-cell parameters | |||
| a (Å) | 159.2 | 177.2 | 88.2 |
| b (Å) | 159.2 | 57.9 | 128.7 |
| c (Å) | 69.4 | 64.4 | 60.0 |
| β (°) | 96.7 | ||
| Resolution (Å) | 50–2.8 (2.90–2.80) | 50–2.7 (2.80–2.70) | 50–2.3 (2.38–2.3) |
| Reflections | 181760 | 61664 | 194271 |
| Unique reflections | 16071 | 15572 | 28641 |
| Completeness (%) | 99.5 (98.5) | 86.4 (62.1) | 99.9 (99.9) |
| Rmerge† (%) | 5.9 (47.4) | 5.8 (26.3) | 7.6 (33.5) |
| Mean I/σ(I) | 11.3 (5.4) | 10.4 (4.5) | 15.5 (5.6) |
R
merge = 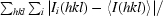
 , where Ii(hkl) is the intensity of reflection hkl,
, where Ii(hkl) is the intensity of reflection hkl,  is the sum over all measured reflections and
is the sum over all measured reflections and 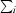 is the sum over i measurements of a reflection.
is the sum over i measurements of a reflection.
We failed to crystallize the full-length Swi5–Sfr1 complex. According to the results of limited proteolysis, a series of N-terminal truncated mutants of Sfr1 were prepared and we succeeded in crystallizing Swi5–ΔN180Sfr1. The Swi5–ΔN180Sfr1 crystals belonged to space group P21212, with unit-cell parameters a = 88.2, b = 128.7, c = 60.0 Å. The crystals diffracted to 2.3 Å resolution (Table 1 ▶). The calculated Matthews coefficient was 3.57 Å3 Da−1, which corresponds to the presence of two heterodimers in the asymmetric unit and a solvent content of 65.5% (Matthews, 1968 ▶).
Acknowledgments
We thank the beamline staff for data collection at the Photon Factory and Dr Kawasaki for the electrospray ionization mass spectrometry at the Graduate School of Nanobiosciences, Yokohama City University. We also thank MEXT for support through a Target Proteins Research Program grant and Grants-in-Aid for Scientific Research.
References
- Akamatsu, Y., Dziadkowiec, D., Ikeguchi, M., Shinagawa, H. & Iwasaki, H. (2003). Proc. Natl Acad. Sci. USA, 100, 15770–15775. [DOI] [PMC free article] [PubMed]
- Bianco, P. R., Tracy, R. B. & Kowalczykowski, S. C. (1998). Front. Biosci.3, D570–D603. [DOI] [PubMed]
- Camerini-Otero, R. D. & Hsieh, P. (1995). Annu. Rev. Genet.29, 509–552. [DOI] [PubMed]
- Cox, M. M. (2007). Nature Rev. Mol. Cell Biol.8, 127–138. [DOI] [PubMed]
- Haruta, N., Akamatsu, Y., Tsutsui, Y., Kurokawa, Y., Murayama, Y., Arcangioli, B. & Iwasaki, H. (2007). DNA Repair, 7, 1–8. [DOI] [PubMed]
- Haruta, N., Kurokawa, Y., Murayama, Y., Akamatsu, Y., Unzai, S., Tsutsui, Y. & Iwasaki, H. (2006). Nature Struct. Mol. Biol.13, 823–830. [DOI] [PubMed]
- Kurokawa, Y., Murayama, Y., Haruta-Takahashi, N., Urabe, I. & Iwasaki, H. (2008). PLoS Biol.6, e88. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Masson, J. Y. & West, S. C. (2001). Trends Biochem. Sci.26, 131–136. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Sehorn, M. G., Sigurdsson, S., Bussen, W., Unger, V. M. & Sung, P. (2004). Nature (London), 429, 433–437. [DOI] [PubMed]
- Sung, P. & Klein, H. (2006). Nature Rev. Mol. Cell Biol.7, 739–750. [DOI] [PubMed]
- Sung, P., Trujillo, K. M. & Van Komen, S. (2000). Mutat. Res.451, 257–275. [DOI] [PubMed]
- Symington, L. S. (2002). Microbiol. Mol. Biol. Rev.66, 630–670. [DOI] [PMC free article] [PubMed]