Summary
The bacterium Caulobacter crescentus has morphologically and functionally distinct cell poles that undergo sequential changes during the cell cycle. We show that the PopZ oligomeric network forms polar ribosome exclusion zones that change function during cell cycle progression. The parS/ParB chromosomal centromere is tethered to PopZ at one pole prior to the initiation of DNA replication. During polar maturation, the PopZ-centromere tether is broken, and the PopZ zone at that pole then switches function to act as a recruitment factor for the ordered addition of multiple proteins that promote the transformation of the flagellated pole into a stalked pole. Stalked pole assembly, in turn, triggers the initiation of chromosome replication, which signals the formation of a new PopZ zone at the opposite cell pole, where it functions to anchor the newly duplicated centromere that has traversed the long axis of the cell. We propose that pole-specific control of PopZ function co-ordinates polar development and cell cycle progression by enabling independent assembly and tethering activities at the two cell poles.
Introduction
The cell poles of rod-shaped microorganisms act as landmarks for site-specific assembly of protein complexes that play a central role in the regulation of the cell cycle and cell behaviour (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009, Shapiro et al., 2009). Therefore, an understanding of how cell pole structure and function changes during the cell cycle is necessary for defining the regulatory networks that underlie cell cycle progression. Here, we report a polar sub-cellular domain that changes function during the C. crescentus cell cycle and show that this functional switch is an integrated component of the spatial and temporal control of chromosome replication initiation and centromere tethering.
Caulobacter crescentus divides asymmetrically, producing a swarmer cell with a polar flagellum and a stalked cell with a membranous polar stalk (Fig. 1A). In the predivisional cell, these morphologically distinct poles are comprised of differing sets of polar signalling proteins, and they direct the swarmer and stalked cell progeny onto different developmental pathways after cell division (Goley et al., 2007). Proteins at the stalked pole signal immediate entry into S-phase, whereas those at the flagellar pole promote cell cycle delay. This delay is subsequently relieved by the transformation of the flagellar pole into a new stalked pole during cell cycle progression (Fig. 1A, step 1). Thus, C. crescentus cells establish and maintain two unique programs of polar composition and activity within a single contiguous cytoplasm, and moreover, they undergo cell cycle controlled polar reprogramming such that polar identity is transformed in association with DNA replication. One aspect of polar reprogramming involves the synthesis of proteins for building a new stalked pole, and this process is initiated by signals that emanate from the flagellar pole shortly after cell division (Radhakrishnan et al., 2008). However, the mechanisms that are responsible for recruiting these proteins to the correct pole are poorly understood, as is the sequence of events that connects chromosome replication with polar development.
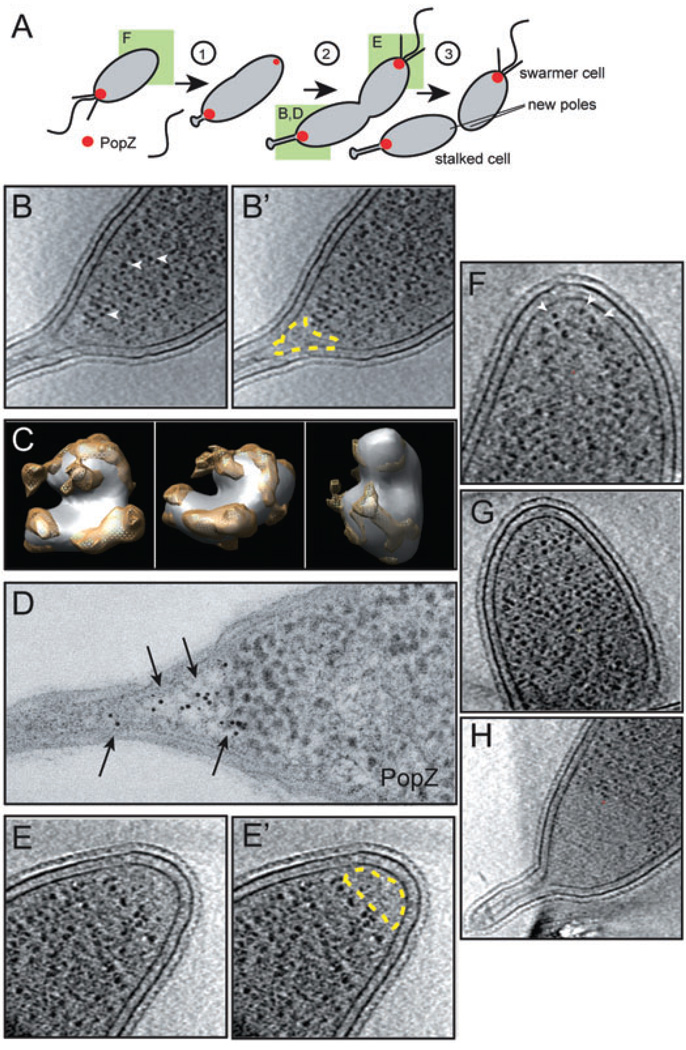
Fig. 1. PopZ establishes a ribosome exclusion zone at the cell poles.
A. A schematic of cell division in C. crescentus. PopZ localization is represented in red, and the poles shown in other panels are denoted by a green box.
B. An individual section from a three-dimensional reconstruction of a Cryo-EM tomogram of a wild-type cell preserved in vitreous ice, showing a stalked pole, with the ribosome exclusion zone outlined in yellow in B′. Examples of individual ribosomes are marked by arrowheads.
C. The high contrast globular particles observed in Cryo-EM tomograms are ribosomes. The averaged structure of the dense globular particles found in C. crescentus cytoplasm (white) is overlayed on the structure the E. coli ribosome (yellow) revealing similarity in size and shape. Three different angles of rotation are shown.
D. Immuno-gold labelling of PopZ (arrows) at the stalked pole of a wild-type cell.
E. A Cryo-EM section of a flagellated pole, with the ribosome exclusion zone outlined in yellow in E′. The flagellar motor was clearly visualized in a different section of this data set (not shown).
F. A new pole in a swarmer cell (Cryo-EM). Dense, ribosome studded cytoplasm extends to the membrane (arrowheads), an observation repeated in over 30 cells.
G. A cell pole in a ΔpopZ cell (Cryo-EM), strain GB255, with ribosome studded cytoplasm extending to the membrane.
H. A stalked pole of a PopZ overproducing cell (Cryo-EM), strain GB123 (top panel), exhibiting an enlarged ribosome exclusion zone. PopZ overproduction was stimulated by growth in 0.3% xylose for 4 h prior to freezing.
The chromosome is physically connected to the cell poles by tethering to the PopZ polar organizing protein (Bowman et al., 2008; Ebersbach et al., 2008). PopZ interacts directly with the chromosome partitioning protein ParB, which specifically associates with parS DNA sequences (Easter and Gober, 2002) in a centromere-like complex (Toro et al., 2008). This tethering mechanism ensures that the chromosome is arranged with its centromere at the cell pole; when PopZ is depleted, unanchored centromeres drift in the cytoplasm. Chromosome replication and segregation are temporally correlated with the swarmer to stalked cell transition (Fig. 2A), and these events are also associated with a change in the localization pattern of PopZ (Fig. 1A). Swarmer cells have one chromosome, and a single polar focus of PopZ anchors the centromere to the flagellar pole. During the swarmer to stalked cell transition, a second focus of PopZ appears at the pole opposite the developing stalk, and when the newly replicated centromere is translocated to this pole, PopZ is there to anchor it in place. The close correlation between chromosome dynamics and PopZ localization implies that these events are co-ordinated. Here, we show that DNA replication initiation is a trigger for the accumulation of the PopZ tether at the new cell pole.
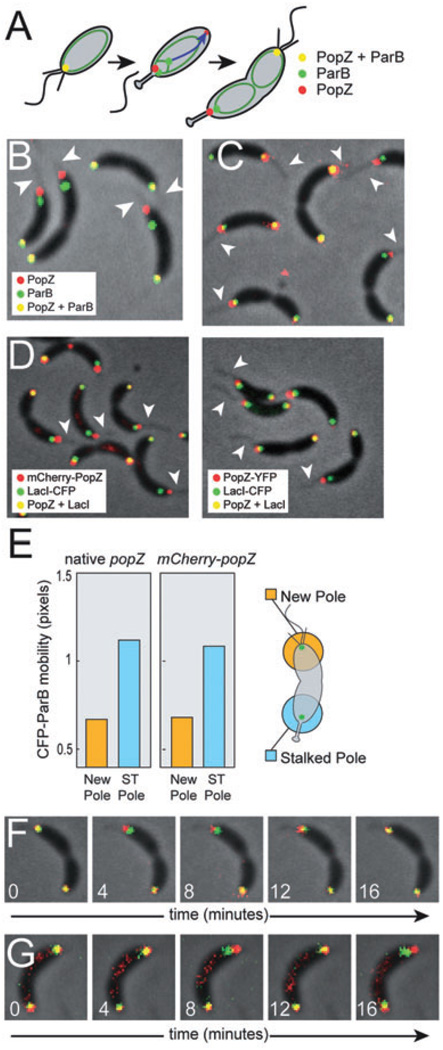
Fig. 2. The centromere tethering function of PopZ is lost at the stalked pole.
A. Schematic of ParB and PopZ localization during chromosome replication and centromere translocation. The chromosome is represented as a green oval, and the direction of movement during translocation is indicated by a blue arrow.
B. Cells of strain GB301, producing PopZ-YFP (red) and CFP-ParB (green). Fluorescence channels are overlayed on the phase-contrast image. Visible stalks are indicated by arrowheads. Fluorescence signals were considered ‘partially overlapping’ when 0–50% of the area of the smaller diameter signal was contained within the area of the larger diameter signal.
C. Cells of strain GB529, producing mCherry-PopZ (in red) and CFP-ParB (in green), processed as in (B).
D. In FROS (Straight et al., 1996; Viollier et al., 2004), exogenously expressed LacI-CFP binds to tandem arrays of lac operator sites inserted at a specific location on the chromosome, here a centromere proximal sequence. This labelling was performed in the context of mCherry-PopZ expression (left panel, strain GB593) and PopZ-YFP expression (right panel, strain GB594). Images were processed as in B, and visible stalks are marked by arrowheads.
E. Quantification of the motion of CFP-ParB foci at flagellated and stalked poles in time lapse movies of strain MT190 and GB529. Position was determined by calculating the weighted centroid of a fluorescent focus, and distance was measured in pixels, with a measured pixel width of approximately 80 nm. The plot shows average displacement after a four-minute interval. In three separate experiments, ~60 cells of each strain were analysed over four consecutive time intervals, yielding a total of over 700 data points. The standard error on the determination of the mean value was negligibly small.
F and G. Individual frames from time lapse movies of strain GB529 (F) and GB301 (G). Time (in minutes) is indicated. The CFP-ParB focus (green) moves back-and-forth in the vicinity of the stalked pole (top), but remains stationary at the flagellar pole (bottom). All experiments were performed in M2G medium.
In addition to being defective in centromere tethering, cells depleted of PopZ do not form stalks and fail to localize the polar transmembrane histidine kinases CckA and DivJ, suggesting that PopZ has multiple roles in polar assembly and function (Bowman et al., 2008; Ebersbach et al., 2008). PopZ is thought to play a functional role in the recruitment of these proteins, as there is biochemical evidence for physical association and, when PopZ is highly overproduced, it has the dramatic effect of accumulating as a large polar mass that recruits DivJ, CckA and ParB over an extended area (Ebersbach et al., 2008). DivJ and CckA are part of a complex circuit that controls the timing of DNA replication through modulation of the master regulatory protein CtrA (Bowers et al., 2008), and their mis-localization in ΔpopZ cells is likely to be a contributing factor in the growth defect of this strain. These results show that a polar recruitment function of PopZ lies upstream of the control mechanism for chromosome replication, and raise the possibility that PopZ is a multifunctional protein that is subject to temporal and spatial control. Stalked pole proteins must be recruited to only one pole when PopZ is localized to both poles, and the assembly of polar multi-protein complexes must be integrated with the mechanism for chromosome tethering. We show here that PopZ accomplishes multiple activities by switching function during the swarmer to stalked cell transition, and that this spatially controlled switch is a driving force in the transitional phases of polar development.
The exaggerated accumulation of PopZ following overproduction can be explained by its ability to undergo self-assembly. As a monomer, PopZ is an 18 kDa protein, but it exists as a homotypic polymer of at least 300 kDa in cells, and is capable of undergoing further assembly in vitro into a network like structure connected by trimeric junctions (Bowman et al., 2008). Following overproduction in vivo, a large polar plug of accumulated PopZ is readily visible in cryo-electron micrographs (Ebersbach et al., 2008). However, this massive accumulation is not relevant to polar development in normal cells. Here, we demonstrate that a polar ribosome-free zone exists in wild-type cells, and that the size of the PopZ polar zones can be modulated by adjusting PopZ expression. Furthermore, we show at nanometer resolution that DivJ is entirely confined within the PopZ zone at the stalked pole, suggesting that PopZ generates a sub-cellular domain that includes scaffolding function.
Results
PopZ forms a ribosome exclusion zone at the cell poles
To define the polar deposition of PopZ at normal expression levels, we preserved wild-type cells by freezing in vitreous ice and collected three-dimensional Cryo-EM tomographic data sets. Clearly, distinguishable regions of low cytoplasmic density were visible at stalked poles (Fig. 1B and B′), far smaller in size from those observed in mutants overproducing PopZ (Ebersbach et al., 2008). This distinct cytoplasmic region in wild-type cells was characterized by a sharp reduction in the concentration of high contrast globular particles that are abundant in normal cytoplasm. Averaging techniques were used to determine the shape of the globular particles (Fig. 1C), and provided sufficient resolution to conclude that they are ribosomes. We then used anti-PopZ immuno-gold labelling to compare the area of the ribosome exclusion zone to the distribution of PopZ protein at the stalked pole (Fig. 1D). Gold particle labelling was highly concentrated across the low density region, and only present at background levels in the adjacent cytoplasm. This experiment compliments the lack of targeted, high-resolution protein labelling in cryo-EM tomograms by providing direct evidence that the ribosome exclusion zone is comprised of PopZ. Ribosome exclusion zones were also observed at the flagellated pole in predivisional cells (Fig. 1E and E′), but not at the new pole opposite the flagellum in swarmer cells (Fig. 1F, Fig. S1 and Movies S1–4). Over the course of repeated Cryo-EM observation of 30 swarmer cells and a greater number of stalked and predivisional cells, we found that the locations of these zones paralleled the distribution of PopZ-YFP previously reported (Bowman et al., 2008; Fig. 1A). Furthermore, these ribosome exclusion zones were absent in popZ deletion strains (Fig. 1G), and the extent of the ribosome exclusion zone was significantly increased by the overproduction of PopZ (Fig. 1H and Ebersbach et al., 2008), indicating that the cellular level of PopZ determines the size of these sub-cellular regions. From these data, we conclude that PopZ is the primary structural component of a large-scale physical feature that distinguishes the cytoplasm at cell poles from the rest of the cytoplasm.
PopZ function switches from centromere tethering to stalked pole development
The ParB partitioning protein binds to both the parS chromosomal centromere sequence and to PopZ (Bowman et al., 2008; Toro et al., 2008). We noted previously that the CFP-ParB labelled centromere nearest to the newly formed stalked pole was often separated from the focus of PopZ-YFP (Bowman et al., 2008), indicating that centromere tethering may be lost during the flagellar to stalked pole transition (Fig. 2A). Here, we quantified the colocalization of PopZ-YFP and CFP-ParB in cells that had completed centromere segregation and polar transformation, and found that one of the centromeres was fully separated from polar PopZ-YFP in 51% of those cells, and the two markers showed partial overlap in an additional 19% of cells. In nearly every case where a stalk was visible, the separation of the centromere from PopZ-YFP occurred at the stalked end of the cell (Fig. 2B). To control for the potential influence of the fluorescent protein tag on PopZ activity, we created a second strain in which PopZ was modified by the addition of mCherry at the N-terminus. In this strain (Fig. 2C), we observed that the CFP-ParB marked centromere was fully separated from the focus of mCherry-PopZ at 27% of stalked poles and the two markers showed partial overlap in an additional 24% of cells. Thus, more frequent separation was observed when PopZ was tagged with YFP rather than mCherry, possibly due to variable effects of the fluorescent protein tags on PopZ activity. To control for the potential influence of the CFP tag on ParB localization and activity, we used an additional method for centromere labelling, in which a centromere proximal sequence was labelled by the FROS Fluorescent Repressor-Operator System (Fig. 2D). Again, we observed that the CFP labelled centromere region was fully separated from the focus of mCherry-PopZ at many stalked poles, while the two markers were colocalized at the pole opposite the stalk.
To observe centromere tethering without the influence of fluorescently tagged PopZ, we tracked the movement of CFP-ParB foci in hundreds of cells in a native popZ background (Fig. 2E and Movie S5). We found that the CFP-ParB labelled centromeres at the pole opposite the stalk were nearly stationary, moving an average distance of less than one pixel width (80 nm) over an interval of 4 min. However, the centromeres at the stalked pole exhibited more than twofold greater motion, consistent with the loss of polar attachment observed in Fig. 2B and C. We performed the same analysis in cells that expressed mCherry-PopZ, and found that the motion of the CFP-ParB centromeres was nearly identical in this strain (Fig. 2D, Movie S6). These observations indicate that CFP-ParB polar anchoring and dynamic movement is unperturbed by the expression of mCherry-PopZ, and that the observed behaviours of these labelled proteins are reliable indicators of the native state. Time lapse videos of CFP-ParB movement relative to either mCherry-PopZ or PopZ-YFP (Fig. 2E and F; Movies S6 and S7) showed that the CFP-ParB focus exhibited dynamic movement at the stalked pole, but not at the pole opposite the stalk. Based on these detailed analyses of centromere dynamics, we conclude that cell cycle progression includes a mechanism for breaking the interaction between PopZ and ParB/parS at the developing stalked pole.
To compare the timing of centromere release with the changes in protein composition at the stalked pole, we created triple-labelled strains that express CFP-ParB in addition to fluorescently tagged versions of PopZ and the DivJ transmembrane histidine kinase, the latter used as a marker of stalked pole development (Wheeler and Shapiro, 1999). Synchronized swarmer cells were allowed to proceed through the cell cycle, and images were collected every 20 min. In the PopZ-YFP, CFP-ParB, DivJ-mCherry background (Fig. 3A and B), there was a significant increase in the fraction of cells exhibiting CFP-ParB separation from polar PopZ-YFP in the first 40 min of the experiment, concomitant with the exit of cells from G1 phase. Beyond 40 min, the separation of CFP-ParB from PopZ-YFP reached a plateau. In an mCherry-PopZ, CFP-ParB, DivJ-YFP background (Fig. S2), we observed a similarly timed increase in centromere detachment followed by a plateau, although the PopZ-YFP background strain exhibited a higher frequency of visible CFP-ParB separation than the mCherry-PopZ strain. In both strain backgrounds, the fraction of polar DivJ (tagged with either YFP or mCherry) also increased during the first 40 min, but this was just the initial phase of a steady upward trend (Fig. 3B). These results indicate that the centromere becomes detached from PopZ during development of the stalked pole, usually before its maturation is complete.
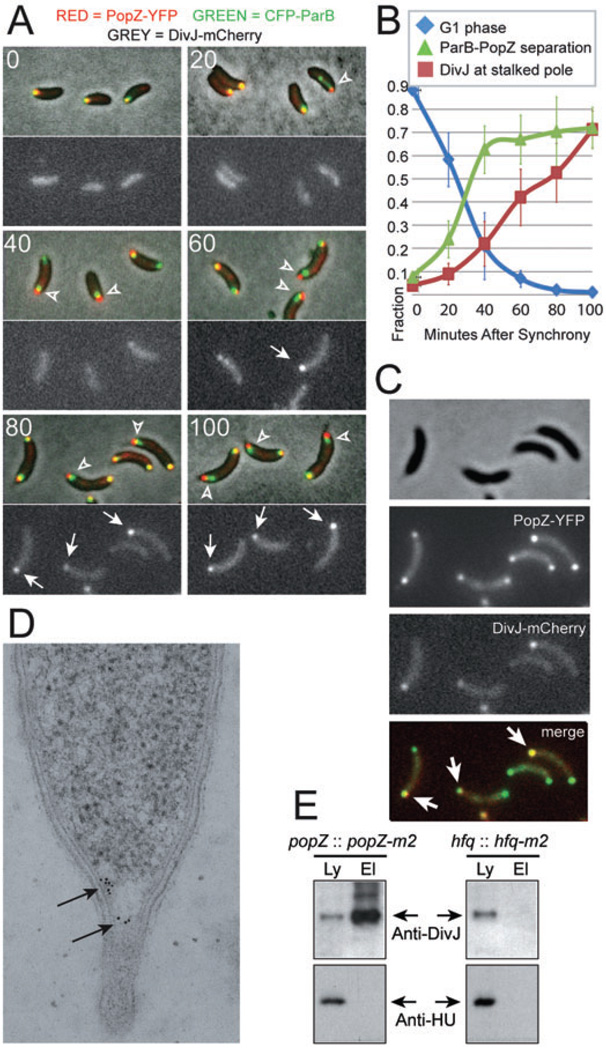
Fig. 3. Centromere detachment occurs before stalked pole development is complete.
A. Swarmer cells were isolated from strain GB433, expressing CFP-ParB (green), PopZ-YFP (red) and DivJ-mCherry (grey image, lower panels), and grown in M2G medium. At the indicated time (in minutes), an aliquot of cells was removed any analysed by fluorescence microscopy. Representative examples are shown. CFP-ParB and PopZ-YFP were considered separated when less than 50% of the area of the smaller diameter signal was contained within the area of the larger diameter signal (arrowheads). Polar DivJ-mCherry foci are marked by arrows.
B. Quantification of the localization patterns observed in (A). Cells with no DivJ-mCherry focus and colocalized PopZ-YFP/CFP-ParB were scored as swarmer cells. Over 200 cells per time point from each of three separate trials were counted. Error bars represent the standard deviation between trials.
C. Colocalization of PopZ-YFP (green) and DivJ-mCherry (red) at the stalked pole (arrows) in strain GB433. For all experiments, cells were grown in M2G medium.
D. Immuno-gold labelling of DivJ at a stalked pole in a wild-type cell. The gold particles (arrows) are limited to area of low ribosome density at the base of the pole.
E. A co-immunoprecipitation assay shows that DivJ and PopZ are in close molecular proximity. Strains GB135 (popZ-m2) and LS4379 (hfq-m2) were treated with DSP cross-linker prior to lysis in detergent buffer. Samples of the lysate (Ly) and affinity purified PopZ-M2 protein complexes (El) were probed with anti-DivJ and anti-HU antibodies by immunoblotting.
After being recruited to the stalked pole, DivJ-mCherry colocalized with PopZ (Fig. 3C). We used immuno-gold labelling to obtain more precise localization information on DivJ with respect to the ribosome exclusion zone established by PopZ (Fig. 1), and found that the distribution of DivJ was always contained within the region of low ribosome density at the stalked pole (Fig. 3D). To determine if DivJ molecules are in close proximity to PopZ, we performed a co-immunoprecipitation experiment in which the endogenous popZ coding sequence was replaced by epitope tagged popZ-m2. When the cell lysates were treated with a 12 angstrom cross-linker, affinity purification of PopZ-M2 and associated factors readily pulled down DivJ protein, suggesting that these proteins are part of a mutliprotein complex (Fig. 3E). To demonstrate the specificity of this interaction, we show that PopZ-M2 does not co-precipitate with an unrelated cytoplasmic protein (HU), and that DivJ is not pulled down when the M2 tag is fused to an irrelevant protein (Hfq).
PopZ mediates ordered assembly of SpmX and DivJ at the stalked pole
Robust polar localization of DivJ requires both PopZ (Ebersbach et al., 2008) and the membrane spanning protein SpmX (Radhakrishnan et al., 2008). The polar localization of SpmX precedes DivJ, and is the earliest known protein to be recruited to the developing stalked pole. Co-immunoprecipitation experiments have shown that DivJ interacts with both SpmX (Radhakrishnan et al., 2008) and PopZ (Fig. 3E). To determine if SpmX and PopZ act in parallel to recruit DivJ or if one lies upstream of the other, we observed the localization pattern of PopZ-YFP in a ΔspmX strain. As shown in Fig. 4A, the absence of SpmX had no effect on the localization pattern of PopZ, indicating that PopZ polar accumulation is independent of SpmX. To determine if PopZ is required for the polar accumulation of SpmX, we observed SpmX-mCherry localization in a ΔpopZ background (Fig. 4B). In the absence of PopZ, the cells became filamentous and SpmX polar localization was severely impaired. The ratio of polar SpmX-mCherry signal intensity to delocalized background was reduced by nearly ninefold compared with wild-type control (Fig. 4C). The level of SpmX-mCherry protein in the ΔpopZ background was nearly the same as in wild-type (Fig. 4D), indicating that the reduction in polar fluorescence was caused by mis-localization rather than a reduction in protein levels. To control for the potential effect of cell length on SpmX-mCherry localization, we utilized a strain background in which filamentation can be induced by depleting the division protein FtsZ (Wang et al., 2001). In the absence of FtsZ expression, SpmX-mCherry retained strong polar localization in filamentous cells (Fig. 4E), indicating that cell length does not influence SpmX localization. We repeated these experiments in minimal medium and observed that the ΔpopZ strains had similar defects in SpmX-mCherry localization (Fig. S3).
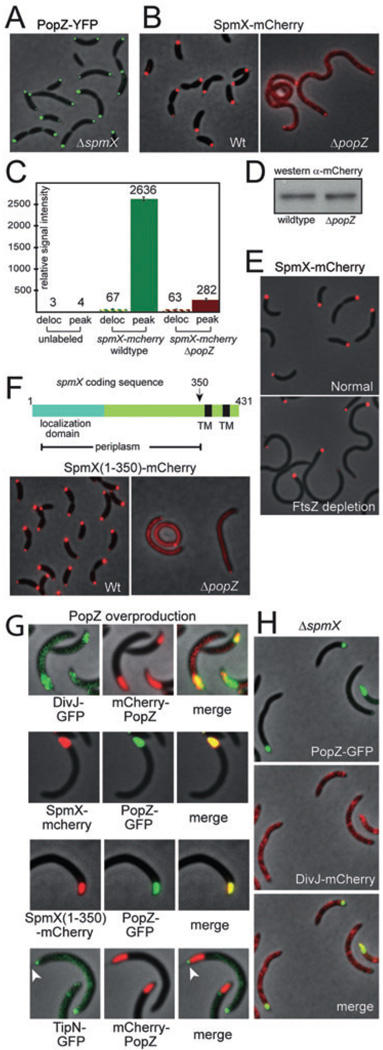
Fig. 4. PopZ recruits SpmX to the stalked pole through a periplasmic intermediary.
A. Localization of PopZ-YFP in ΔspmX cells (strain GB507).
B. Localization of SpmX-mCherry in wild-type (strain GB378) and ΔpopZ cells (strain GB387).
C. Quantification of the data in (B), as described in Experimental procedures. Peak refers to the intensity of the localized signal; Deloc refers to the median intensity of the delocalized signal across the cell body.
D. A Western blot showing the cellular levels of SpmX-mCherry protein in native popZ and ΔpopZ backgrounds.
E. Localization of SpmX-mCherry in FtsZ depletion strain GB574. Cells were grown in PYE medium supplemented with 0.03% xylose, then washed and grown for 3.5 hours in the same medium (upper panel) or PYE without xylose (lower panel).
F. A schematic of the spmX coding sequence (upper panel), with transmembrane domains (TM) indicated in black. The N-terminal localization domain (blue) is predicted to be exported to the periplasm. Localization (lower panel) of SpmX(1–350)-mCherry in wild-type (strain GB440) and ΔpopZ cells (strain GB447). SpmX(1–350)-mCherry expression was induced by growth in 50 µM vanillate for two hours prior to analysis.
G. Effects of PopZ overproduction on the localization of polar proteins. Top panel: DivJ-GFP localization during mCherry-PopZ overproduction (strain GB449). Middle panels: SpmX-mCherry and SpmX(1–350)-mCherry localization during PopZ-GFP overproduction (Strains GB430 and GB453). Lower panel: TipN-GFP localization during mCherry-PopZ overproduction (strain GB450). To achieve PopZ overproduction, cells were stimulated by growth in 0.3% xylose for 5 h prior to analysis.
H. DivJ-mCherry remains delocalized in a ΔspmX mutant in the context of PopZ-GFP overproduction (strain GB514). For PopZ-GFP overproduction, cells were stimulated as in (F). For all experiments, strains were grown in PYE medium and coloured fluorescence images are placed on a phase-contrast background.
The strong effect of the PopZ network on SpmX localization is surprising, considering that PopZ is cytoplasmic and the localization determinant in SpmX is a periplasmic muramidase domain that is necessary and sufficient for polar accumulation (Radhakrishnan et al., 2008). To determine if the recruitment activity of PopZ is directed towards the periplasmic portion of SpmX, we utilized a truncated mutant that contains only the periplasmic portion of the protein (Fig. 4F). This truncated protein [SpmX(1–350)-mCherry] exhibited strong polar localization when expressed in wild-type cells, but was mostly de-localized in the absence of PopZ, indicating that the influence of PopZ on polar assembly extends into the periplasmic compartment.
We asked how the creation of enlarged ribosome exclusion zones through PopZ overproduction (see Fig. 1H) affects polar assembly of SpmX and DivJ. The distribution of DivJ-GFP in our mCherry-PopZ overproducing strain was expanded to cover the entire region filled by mCherry-PopZ (Fig. 4G), consistent with the results of Ebersbach et al. (2008). The polar distributions of SpmX-mCherry and SpmX(1–350)-mCherry were similarly expanded to fill the enlarged polar zone resulting from the overproduction of PopZ-GFP (Fig. 4G, middle panels), indicating that the recruitment of a periplasmic localization determinant also occurred under these conditions. To test the specificity of PopZ recruitment, we expressed TipN-GFP, a protein that normally localizes to the pole opposite the stalk (Huitema et al., 2006; Lam et al., 2006), in the context of mcherry-PopZ overpdoduction. We found that TipN-GFP was excluded from the area of mCherry-PopZ overproduction, even when both proteins were present at the same pole (Fig. 4G, arrowheads), suggesting that recruitment by PopZ is limited to a subset of polar proteins. To determine if polar development follows an order of assembly at these expanded cell poles, DivJ-mCherry was expressed in a ΔspmX/PopZ overproducing strain (Fig. 4H). DivJ-mCherry failed to accumulate at the poles, indicating that the over-accumulation of polar PopZ cannot compensate for the loss of SpmX. We therefore conclude that PopZ lies upstream of SpmX, the earliest known marker of stalked pole development, and that PopZ recruits DivJ through the ordered pathway PopZ→SpmX→DivJ.
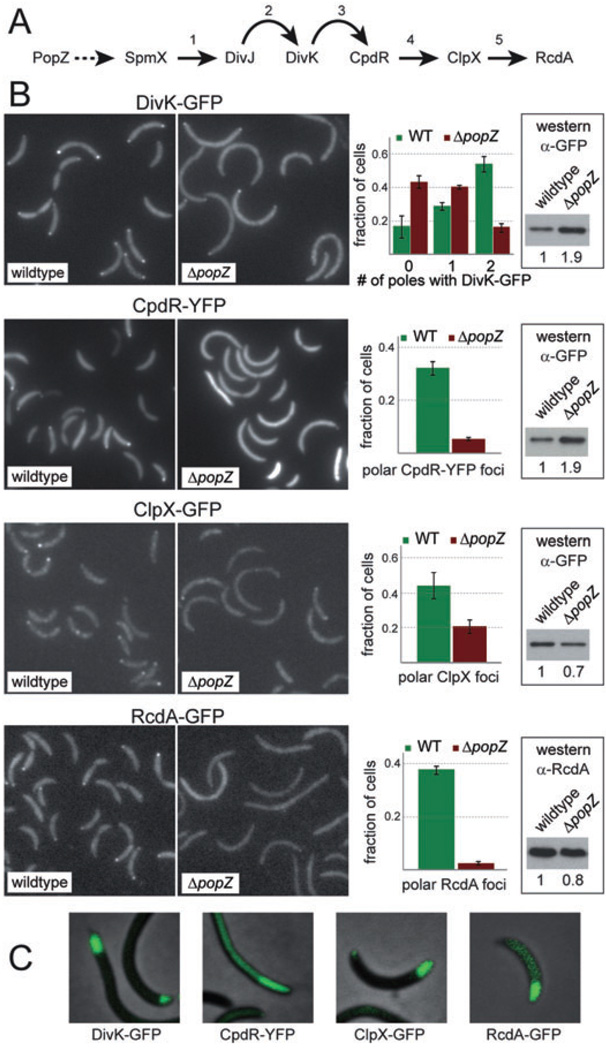
PopZ is required for the assembly of a multi-protein protease complex at the stalked pole
After being recruited to the cell pole by SpmX, the DivJ histidine kinase is part of a complex phospho-signalling pathway governing further stalked pole assembly and function (Fig. 5A). The substrate for DivJ is the polar single domain response regulator DivK, and a result of DivJ/DivK signalling is assembly of the multi-protein ClpXP protease complex at the stalked pole, mediated by the polar localization factor CpdR (Iniesta and Shapiro, 2008). The polar protease complex clears the CtrA master regulator from the cell, allowing the initiation of DNA replication. It is known that DivK is delocalized in the absence of DivJ (Jacobs et al., 2001), and that the protease complex is delocalized in the absence of DivK and CpdR (Iniesta et al., 2006; Iniesta and Shapiro, 2008). Since this regulatory pathway is strongly associated with polar localization, and PopZ lies at the top of the cascade, we predicted that the polar recruitment of all proteins in the pathway would be disrupted in the absence of PopZ. To test this, we observed the localization of DivK-GFP, CpdR-YFP, RcdA-GFP and ClpX-GFP in ΔpopZ and wild-type backgrounds, and found that the polar accumulation of all four markers was sharply reduced (Fig. 5B). Polar localization of TipN and PleC, two flagellar pole proteins that are not part of the stalked pole assembly pathway, was retained in the ΔpopZ background (our unpublished observations and Ebersbach et al., 2008), indicating that the loss of PopZ has a specific effect on the recruitment of staked pole proteins. Although the loss of PopZ partially affected the cellular levels of some marker proteins, this cannot explain the reduction in polar localization, since the levels of DivK-GFP and CpdR-YFP were increased, and the levels of ClpX-GFP and RcdA-GFP were decreased by no more than 30%. Similar defects in protein localization were obtained when the experiment was performed in rich medium (Fig. S4), although in this medium the loss of PopZ also resulted in a significant reduction of ClpX-GFP and RcdA-GFP protein levels.
Fig. 5. PopZ mediates multi-protein complex assembly at the developing stalked pole.
A. An outline of the ClpXP protease localization and assembly pathway during stalked pole development. Curved arrows indicate control through phosphosignalling and straight arrows indicate physical protein interactions that are required for polar localization. The dotted line from PopZ to SpmX is inferred. References: 1: Radhakrishnan et al. (2008); 2: Jacobs et al. (2001); 3: Iniesta and Shapiro (2008); 4: Iniesta et al. (2006); 5: Mcgrath et al. (2006).
B. Localization of DivK-GFP, CpdR-YFP, ClpX-GFP and RcdA-GFP in wild-type and ΔpopZ cells grown in M2G medium. For each fluorescent protein, representative fluorescence image panels are placed beside quantified data from at least two independent experiments, each involving more than 200 cells, with error bars representing the standard deviation between trials. At the far right of each panel is a Western blot showing the cellular levels of the fluorescently tagged protein under these conditions. Below each blot, the numbers refer to the quantification and normalization of band intensities, with wild-type level set to 1. Reading from top left, the strains used were GB229, GB459, LS4250, GB460, LS4183, GB457, LS4191 and GB458.
C. Effects of PopZ overproduction on DivK-GFP, CpdR-YFP, ClpX-GFP and RcdA-GFP localization. Strains GB281, GB235, GB233 and GB265 were stimulated by growth in PYE medium supplemented with 0.3% xylose for eight hours prior to analysis.
If PopZ contributes to the polar localization of these proteins, then the expansion of the polar zone through PopZ overproduction (as in Fig. 1H and Fig. 4G) should have corresponding effects on the their polar recruitment. Indeed, the localization patterns of DivK-GFP, CpdR-YFP, RcdA-GFP and ClpX-GFP were expanded after PopZ was overproduced (Fig. 5C), indicating that all components of this stalked pole assembly pathway are recruited to the enlarged polar zone. Together, these results demonstrate that PopZ plays a central role in stalked pole development, acting as a keystone in an assembly pathway that recruits many proteins to the incipient stalked pole and influencing the organization of proteins in both the periplasmic and cytoplasmic compartments. Importantly, all of the polar markers showed some residual polar localization in the absence of PopZ, indicating that polar localization is influenced by other factors. It is therefore likely that polar assembly involves a network of protein interactions, facilitated in large part by the presence of PopZ. The polymeric nature of PopZ may allow it to act like a scaffold in this process.
The ‘off switch’ for centromere tethering is independent of SpmX expression, protease recruitment and stalk production
Stalked pole proteins are recruited by PopZ during the same period of time in which centromere detachment occurs. It is therefore possible that the ‘off switch’ for centromere tethering is flipped as PopZ becomes saturated by interactions with newly arrived stalked pole proteins, displacing a lower affinity interaction between PopZ and ParB. To test this model, we asked if the detachment of centromeric foci from PopZ could be blocked in strains that are defective in the stalked pole assembly pathway. We tested two different polar assembly mutants: a ΔcpdR mutant that fails to recruit the ClpX protease complex (Iniesta et al., 2006), and a pleC∷Tn5 insertion mutant in which SpmX expression is abolished, DivJ does not localize to the poles and stalk assembly is inhibited (Sommer and Newton, 1989; Radhakrishnan et al., 2008). Each strain expressed CFP-ParB for locating the centromere and mCherry-PopZ for marking the poles. In both of the mutant strains, many of the cells that had completed centromere translocation exhibited a normal pattern of centromere attachment and detachment, wherein CFP-ParB and mCherry-PopZ were colocalized at one pole but clearly separated at the opposite pole (Fig. 6A and B). Thus, the centromere tethering function of PopZ can be turned off even when the recruitment and assembly of stalked pole protein complexes does not occur.
Fig. 6. DNA replication is not required for centromere detachment, but is required for the accumulation of PopZ at the new cell pole.
A and B. Defects in stalked pole development do not prevent the detachment of CFP-ParB foci from polar mCherry-PopZ. CFP-ParB (green) and mCherry-PopZ (red) localization is shown in a ΔcpdR background, strain GB559 (A), and in a pleC∷Tn5 background, strain GB566 (B). Arrowheads mark poles where CFP-ParB foci are not colocalized with mCherry-PopZ.
C. DnaA depletion does not prevent the detachment of CFP-ParB foci from polar mCherry-PopZ. Swarmer cells (left panel) isolated from strain GB569 were grown in M2X or M2G media supplemented with 50 µM vanillate for normal growth (top panel) or growth under DnaA depletion (lower panel) respectively. Images were collected after 90 min of growth in liquid media. The diagrams and percentages below each image indicate the percentage of cells containing single foci of mCherry-PopZ and CFP-ParB that overlapped by > 50% (left) or by < 50% (right).
D. Constitutively active CtrA does not prevent the detachment of CFP-ParB from polar mCherry-PopZ. Swarmer cells were isolated from strain GB532 were grown in M2G or M2X media supplemented with 50 µM vanillate for normal growth (top panel) or growth under CtrA D51EΔ3Ω production (lower panel) respectively. Image collection and quantification was as in (C).
E. The detachment of CFP-ParB from PopZ-YFP is maintained after DNA replication is blocked with novobiocin treatment. Swarmer cells isolated from strain GB529 were grown in M2G supplemented with 50 µM vanillate in the absence or presence of 100 µg ml−1 novobiocin, and images were collected and quantified as in (C).
F. Western blots showing the level of mCherry-PopZ in whole cell lysates taken from the experiments in (C) to (E). mCherry-PopZ is partially degraded and runs as a doublet. For each experiment, combined band intensity was normalized with respect to the control.
DNA replication initiation is required for the accumulation of PopZ at the pole opposite the stalk, but not for the release of the centromere from the stalked pole
To determine if DNA replication initiation mediates the switch in PopZ function at the stalked pole, we measured centromere positioning under conditions in which DNA replication was blocked. We utilized conditional production of DnaA, a protein that forms an essential part of the replication initiation complex (Mott and Berger, 2007). DnaA production was controlled by deleting the native dnaA coding sequence and inserting a second copy of the dnaA coding sequence under control of the inducible xylX promoter (Hottes et al., 2005). In the presence of xylose, swarmer cells produced DnaA and exhibited normal chromosome replication and segregation, including the separation of CFP-ParB centromeric foci from mCherry-PopZ at the stalked pole (Fig. 6C, upper arrow). In the absence of xylose, dnaA was depleted and DNA replication failed to initiate. We observed that the single CFP-ParB/centromeric focus in many of these cells had become visibly detached from polar mCherry-PopZ (Fig. 6C, lower arrow), suggesting that centromere detachment is independent of chromosome replication initiation through DnaA. In addition, we found that the depletion of DnaA completely blocked the accumulation of a new focus of mCherry-PopZ at the pole opposite the stalk. In this experiment, mcherry-popZ expression was maintained by placing the gene under control of the vanillate inducible vanA promoter, and the level of mCherry-PopZ at the stalked pole increased over time (not shown), indicating that the defect in mCherry-PopZ localization cannot be attributed to a lack of available protein. Thus, DnaA activity is required for initiating PopZ accumulation at the new cell pole.
Because DnaA is a global transcriptional regulator in addition to its role in initiating DNA replication (Hottes et al., 2005), we sought to avoid the pleiotropic effects of DnaA inhibition by creating a second strain in which chromosome replication could be controlled without direct perturbation of DnaA. In this strain, DNA replication could be inhibited by blocking the degradation and deactivation of CtrA, a negative regulator of DNA replication, through the overproduction of a constitutively active and non-degradable form of the protein (Domian et al., 1997). When this mutant form of CtrA was expressed in swarmer cells, all retained a single focus of CFP-ParB after 90 min of growth, consistent with a complete block in DNA replication. We observed that the centromeric foci were still able to separate from mCherry-PopZ at the cell pole (Fig. 6D, lower arrow) at a frequency that is comparable to stalked poles under normal conditions (Fig. 2). Again, the accumulation of mCherry-PopZ at the pole opposite the stalk was completely inhibited. A third independent method of blocking DNA replication initiation was treatment with the antibiotic novobiocin, which inhibits the activity of DNA gyrase (Fig. 6E). Consistent with our other observations, the blockage of DNA replication with novobiocin permitted the release of the centromere from the flagellated pole, but blocked the accumulation of a new focus of mCherry-PopZ at the new pole.
Together, the experiments in Fig. 6C–E show that the detachment of CFP-ParB centromeric foci from polar mCherry-PopZ occurs independent of DNA replication initiation, and is controlled by a signal that lies upstream of DnaA and CtrA function. Thus, the ‘off switch’ for the centromere tethering function of PopZ responds to a signal that occurs at the earliest stages of the swarmer to stalked cell transition. Additionally, all three methods for blocking DNA replication initiation caused a nearly complete block in the accumulation of mCherry-PopZ at the new cell pole. To determine if this is caused by a replication-dependent effect on mCherry-PopZ production, we measured the cellular levels of mCherry-PopZ in our replication inhibition experiments, and found that it was reduced by a minimum of 0% and a maximum of 24% compared with controls (Fig. 6F). The levels of untagged PopZ were similarly affected (not shown). Since all three methods for inhibiting DNA replication yielded similar results, we conclude that the failure of mCherry-PopZ to accumulate at the new pole cannot be explained by a reduction in mCherry-PopZ production. We propose that the initiation of chromosome replication is a regulatory step that lies in between the release of the centromere tether at the developing stalked pole and the accumulation of PopZ at the other end of the cell. This suggests a stepwise order of events in chromosome dynamics: the centromere is released from the developing stalked pole in time for but independent of the initiation of replication, and chromosomal replication is then required for assembly of the PopZ tether that receives a copy of the newly replicated centromere at the opposite pole.
Discussion
PopZ activity is under temporal and spatial control
The asymmetry exhibited by C. crescentus is in large part attributable to differences in composition and activity of the two cell poles. In addition to being morphologically distinguishable (a stalk versus a flagellum), opposing sets of signalling molecules at either pole control differential timing of S-phase in the stalked cell and swarmer cell progeny following cell division (Goley et al., 2007). It is therefore not surprising that an unbiased screen for localized proteins in C. crescentus (Werner et al., 2009) found that over 10% of the predicted coding sequences yielded proteins with distinct sub-cellular locations, and that approximately 90% of the pole localized proteins were limited to only one pole. The PopZ localization factor is among the minority that can simultaneously localize to both cell poles. Previous reports have shown that PopZ has multiple functions (Bowman et al., 2008; Ebersbach et al., 2008), but have not addressed whether the timing or the location of those functions is changed to accommodate programmed asymmetry during cell cycle progression. We show here that cells maintain polar asymmetry with respect to PopZ by achieving temporal and spatial control of its function. The modulation of PopZ activity underlies sequential steps in cell cycle progression, as shown in the integrated model in Fig. 7. As the swarmer cell matures, the interaction between ParB and PopZ is broken, with subsequent release of the centromere from its tether. At this time, the recruitment of stalked pole proteins by PopZ is activated, initiating an assembly pathway for a polar multi-protein complex that degrades the CtrA replication inhibitor, allowing the initiation of DNA replication. Chromosome replication creates two copies of parS, and also cues the formation of a PopZ ribosome-free zone at the opposite cell pole. One of the parS/centromere complexes is translocated to the opposite cell pole, and although the timing of its arrival varies with respect to the appearance of a visible focus of PopZ (Bowman et al., 2008), the centromere is firmly tethered in place with the accumulation of the PopZ matrix.
Fig. 7.
A description of PopZ activity during the cell cycle. Details are described in the text.
Building the stalked pole
Upon the differentiation of a swarmer cell into a stalked cell and entry into S phase, ParB/parS is released from polar PopZ, but the PopZ polymeric network remains as a permanent fixture at that pole (Fig. 7). The persistence of PopZ is surprising, because it has lost its function as the centromere anchor, and because almost all of the known flagellated pole proteins are lost during polar remodelling at the swarmer to stalked cell transition (Goley et al., 2007). We report here that a switch in function underlies the persistence of the polar PopZ zone, whereby the centromere tethering function is replaced by a new role as stalked pole assembly factor. Seven different stalked pole proteins (SpmX, DivJ, DivK, CpdR, RcdA, ClpX and CckA) are known to require PopZ for robust polar localization (Fig. 5; and for DivJ and CckA, Ebersbach et al., 2008). In serving as the centromere tether at the flagellar pole, PopZ is in place well before these stalked pole proteins are expressed at the swarmer to stalked cell transition, and is ideally positioned to act as a landmark for the initiation of stalked pole assembly. As assembly proceeds, the arrival of the ClpXP protease promotes the degradation of flagellar pole components (Tsai and Alley, 2001) as well as the CtrA master regulator, thereby relieving the inhibition of DNA replication initiation (Jenal and Fuchs, 1998; Mcgrath et al., 2006).
Previous studies indicate that the stalked pole proteins shown in this study are recruited in a linear pathway, beginning with SpmX (Fig. 5A). Our data are also consistent with an alternative assembly model, in which the PopZ scaffold has specific affinity for many stalked pole proteins, and future experiments will determine the direct targets for PopZ interaction.
We show that PopZ is required for robust polar accumulation of SpmX, the earliest known marker of stalked pole development. SpmX localization occurs through the activity of a periplasmic domain, indicating that cytoplasmic PopZ communicates with SpmX indirectly. This could occur through a transmembrane protein that binds to PopZ and SpmX, or, as suggested by the homology of the SpmX localization domain with peptidoglycan-binding lysozyme/muramidases (Radhakrishnan et al., 2008), PopZ may affect SpmX recruitment by influencing cell wall synthesis or modification. Peptidoglycan incorporation is concentrated at the developing stalked pole (Aaron et al., 2007; Divakaruni et al., 2007); however, the role of PopZ in this activity remains to be determined.
Localization of PopZ to the new pole
Following stalked pole assembly and the initiation of chromosome replication, a copy of the ParB/parS centromere travels to the opposite end of the cell. Here, we show that DNA replication initiation is also a cue for the assembly of a new zone of PopZ that tethers the translocated centromere to the pole (Fig. 7). We found that novobiocin treatment was as effective as DnaA depletion or CtrA overproduction in inhibiting PopZ accumulation at the new pole. As novobiocin is an inhibitor of DNA gyrase, and affects DNA replication directly by preventing the expansion of the replication bubble (Baker et al., 1987), these results suggest that the trigger for PopZ polar accumulation is dependent on replication itself, and not the transcriptional activities of DnaA or CtrA. Second, we can exclude the possibility that the trigger is driven only by the production of new PopZ protein, since mCherry-popZ expression was controlled by vanillate induction and not by the native popZ promoter.
Previous studies argue that either PopZ localizes to the new pole by a passive mechanism in which it collects in relatively empty space that is unoccupied by the nucleoid (Ebersbach et al., 2008), or that it is localized by a more directed process that relies, at least in part, on the actin-like cytoskeletal protein MreB (Bowman et al., 2008). In this study, the polar accumulation of PopZ is completely blocked by DnaA depletion (Fig. 6C), even though cell elongation continues under these conditions and presumably creates more open space that could accommodate passive PopZ accumulation. We therefore favour the hypothesis that the site-directed accumulation of PopZ is mediated through a mechanism that includes MreB, and occurs either in conjunction with DNA replication initiation or downstream of this event. Further analysis of PopZ localization determinants may shed light on the complex relationship between MreB, the establishment of cell polarity (Gitai et al., 2004), and other early events in the cell cycle (Shebelut et al., 2009).
Temporal and spatial control of PopZ activity
Following chromosome replication and segregation, both of the centromeres remain in close proximity to a cell pole (Fig. 7) and the presence of the PopZ ribosome-free zone at these locations suggests that physical attachment occurs at both cell poles. However, in closely comparing the localizations of ParB and PopZ, we show that the centromere is often detached from the stalked pole, revealing a novel and unexpected aspect of polar asymmetry. This implies that the general orientation of the chromosomes can be maintained by providing a single anchoring point for the translocating centromere during chromosome segregation. We presume that the bulkiness of the chromosome is sufficient to prevent the disorganization of the unanchored chromosome at the stalked pole, although we cannot rule out the possibility that there remains some limited and short-lasting contact between PopZ and ParB at this location.
A better understanding of how polar development is related to PopZ function will require additional knowledge of its structure. Our observation that PopZ assembles into a large-scale macromolecular complex (Fig. 1) suggests that it serves as a physical scaffold for centromere tethering followed by multi-protein complex assembly. Due to its small size, PopZ is markedly different than typical scaffolding proteins of eukaryotic organisms, which usually have several protein interaction domains (Dhanasekaran et al., 2007). PopZ appears to have only two domains, and is therefore unlikely to carry out its recruitment functions through multiple protein interactions. The simplest model for the recruitment of at least seven different stalked pole proteins by PopZ is hierarchical, in which PopZ acts near the top of a cascade of protein interactions that leads to stalked pole assembly.
However, our experiments uncovered evidence that this model is too simple, and that PopZ likely cooperates with other factors to achieve robust polar localization at the stalked pole. As shown in Figs 4 and 5, SpmX and DivK were still present at the poles in the absence of PopZ, though at greatly reduced levels compared with wild-type. Similarly, Ebersbach et al. (2008) reported that the polar localization of DivJ-YFP and CckA-CFP was sharply reduced, but still present as weak and transient polar foci in cells lacking PopZ. Additionally, we observed that the mislocalization phenotypes in the absence of PopZ were stronger in rich peptone-yeast extract (PYE) medium than in M2G minimal medium, suggesting that the influence of PopZ activity is somewhat context dependent. It is possible that PopZ works together with additional factors to interact with target proteins during the initial stages of stalked pole recruitment. Alternatively, the mesh-like PopZ network could act as a barrier that restricts the diffusion of polar markers away from the pole after they have been attracted there by some other mechanism. For example, monomeric proteins might diffuse into the PopZ network and then become trapped after assembling into larger complexes and becoming too large to escape through the pores. A diffusion barrier mechanism has been proposed for eukaryotic cytoskeletal scaffolding proteins in generating neuronal cell polarity (Nishimura et al., 2007).
Genome-scale analysis suggests that there are over 150 distinct polypeptides that are localized to the pole during part of the cell cycle (Werner et al., 2009), and although this group includes many proteins that are critical for cell cycle regulation (Collier and Shapiro, 2007), knowledge of how these proteins are localized is incomplete. Our analysis shows that some of these proteins are localized by associating with a large polymeric network, assembled from subunits of PopZ. Compared with receptor-based localization in which the primary interaction is a two dimensional membrane surface, the PopZ network may offer the advantages of increased surface area for accommodating many binding partners and increased freedom for protein interactions within three-dimensional space. Furthermore, the relative accessibility of a porous network may facilitate rapid modification, making it suitable as a multifunctional platform.
Experimental procedures
C. crescentus strains, cell growth and synchronization
All C. crescentus strains used in this study are derived from the synchronizeable wild-type strain CB15N, and were grown at 28°C in PYE medium (Poindexter, 1964), or M2 minimal medium (Johnson and Ely, 1977) supplemented with d-glucose (0.2% in M2G) or d-xylose (0.3% in M2X). When appropriate, cells were stimulated by the addition of Na-vanillate at 50 µM 45 min before the start of the experiment. Strains were analysed at mid-exponential phase of growth. Small-scale cell synchronization for microscopic analysis was described by Tsai and Alley (2001). Descriptions of strains and plasmids used in this study are given in Tables 1 and 2, respectively, and the plasmid backbones are described (Thanbichler et al., 2007) where no other references are given. The details of strain and plasmid construction with relevant primer sequences are available upon request.
Table 1.
C. crescentus strains.
|
C. crescentus strains |
Relevant genetic markers, plasmids and/or description |
Construction, source or reference |
|---|---|---|
| CB15N | Synchronizeable derivative of wild-type strain CB15 | Evinger and Agabian (1977) |
| GB123 | pGB122 | pGB122 into CB15N |
| GB135 | popZ∷popZ-m2 | Bowman et al. (2008) |
| GB212 | divJ-PdivJdivJ-mcherry | pGB211 integrated into CB15N |
| GB229 | pMR20divk-gfp | pMR20divk-gfp into CB15N |
| GB233 | xylX∷PxylX-clpX-gfp; pGB172 | pGB172 into LS4183 |
| GB235 | pCpdR-YFP; pGB172 | pGB172 into LS4250 |
| GB255 | popZ∷Δ | Bowman et al. (2008) |
| GB265 | rcdA∷rcdA-gfp; pGB172 | pGB172 into LS4191 |
| GB281 | pMR20divk-gfp; pGB122 | pGB122 into GB229 |
| GB283 | parB∷cfp-parB; pGB274 | pGB274 into MT190 (Thanbichler and Shapiro, 2006) |
| GB301 | parB∷cfp-parB; vanA∷PvanA-popZ-yfp | Bowman et al. (2008) |
| GB378 | spmX∷spmX-mcherry | Radhakrishnan et al. (2008) |
| GB387 | spmX∷spmX-mcherry; popZ∷Δ | Transduction of SpecR from GB255 into GB378 |
| GB430 | spmX∷spmX-mcherry; pGB126 | pGB126 into GB378 |
| GB433 | parB∷cfp-parB; vanA∷PvanA-popZ-yfp; divJ-PdivJ-mcherry | Transduction of TetR from GB212 into GB301 |
| GB440 | vanA∷PvanA-spmX(1–350)-mcherry | pGB446 integrated into CB15N |
| GB447 | vanA∷PvanA-spmX(1–350)-mcherry; popZ∷Δ | pGB446 integrated into GB255 |
| GB449 | divJ∷PdivJ-divJ-gfp; pGB435 | pGB435 into GB212 |
| GB450 | tipN∷tipN-gfp; pGB435 | pGB435 into tipN-gfp (Huitema et al., 2006) |
| GB453 | vanA∷PvanA-spmX(1–350)-mcherry; pGB126 | pGB126 into GB440 |
| GB457 | xylX∷PxylX-clpX-gfp; popZ∷Δ | Transduction of KanR from LS4183 into GB255 |
| GB458 | rcdA∷rcdA-gfp; popZ∷Δ | Transduction of KanR from LS4191 into GB255 |
| GB459 | pMR20divk-gfp; popZ∷Δ | pMR20divk-gfp into GB255 |
| GB460 | pCpdR-YFP; popZ∷Δ | pCpdR-YFP into GB255 |
| GB507 | vanA∷PvanA-popZ-yfp; spmX∷Δ | Transduction of GentR from GB175 (Bowman et al., 2008) in DspmX (Radhakrishnan et al., 2008) |
| GB514 | divJ-PdivJ-mcherry; spmX∷Δ; pGB126 | pGB126 and transduction of TetR from GB212 into ΔspmX (Radhakrishnan et al., 2008) |
| GB529 | parB∷cfp-parB; vanA∷PvanA-mCherry-popZ | pJP44 into MT139 |
| GB532 | parB∷cfp-parB; vanA∷PvanA-mCherry-popZ; pLS3781 | pLS3781 into GB529 |
| GB559 | parB∷cfp-parB; vanA∷PvanA-mCherry-popZ; cpdR∷Δ | SpecR from LS4177 (Iniesta et al., 2006) transduced into GB529 |
| GB566 | parB∷cfp-parB; vanA∷PvanA-mCherry-popZ; pleC∷Tn5 | Transduction of pleC∷Tn5 into GB529 |
| GB569 | parB∷cfp-parB; vanA∷PvanA-mCherry-popZ; dnaA∷Δ; xylX∷PxylX-dnaA | Transduction of KanR and SpecR from GM2471 (Gorbatyuk and Marczynski, 2001) into GB529 |
| GB574 | ftsZ∷PxylX-ftsZ; spmX∷spmX-mcherry | Transdunction of KanR from YB1585 (Wang et al., 2001) into GB378 |
| GB593 | cc0006∷ (lacO)n; xylX∷ PxylX-lacI-cfp; vanA∷PvanA-mCherry-popZ | pGB539 introduced into MT15 (Viollier et al., 2004) |
| GB594 | cc0006∷ (lacO)n; xylX∷ PxylX-lacI-cfp; vanA∷PvanA- popZ-yfp | Transduction of GentR from GB301 into MT15 (Viollier et al., 2004) |
| LS4183 | xylX∷PxylX-clpX-gfp | Mcgrath et al. (2006) |
| LS4191 | rcdA∷rcdA-gfp | Mcgrath et al. (2006) |
| LS4250 | pCpdR-YFP | Iniesta et al. (2006) |
| LS4379 | hfq∷hfq-m2 | Landt et al. (2008) |
| MT190 | parB∷cfp-parB | Thanbichler and Shapiro (2006) |
Table 2.
Plasmids.
| Name | Description | Backbone | Source |
|---|---|---|---|
| pCpdR-YFP | low copy plasmid PcpdR-cpdR-yfp | pMR11 | Iniesta et al. (2006) |
| pGB122 | high copy plasmid PxylX-popZ | pBXMCS-2 | This study |
| pGB126 | high copy plasmid PxylX-popZ-gfp | pBXMCS-2 | This study |
| pGB172 | high copy plasmid PxylX-popZ | pBXMCS-4 | This study |
| pGB211 | PdivJ-divJ-mcherry integrates at divJ locus | pMCS-5 | This study |
| pGB274 | high copy plasmid PxylX-popZ-yfp | pBXMCS-2 | This study |
| pGB435 | high copy plasmid PxylX-mcherry-popZ | pBXMCS-4 | This study |
| pGB446 | PvanA-spmX(1–350)-mcherry integrates at vanA locus | pVCHYC-5 | This study |
| pGB539 | PvanA-mCherry-popZ integrates at vanA locus | pVCHYN-4 | This study |
| pJP44 | PvanA-mCherry-popZ integrates at vanA locus | pVCHYN-2 | J. Ptacin, unpublished |
| pLS3781 | high copy plasmid PxylX-ctrAD51EΔ3Ω | pJS14 | Domian et al. (1997) |
| pMR20divk-gfp | low copy plasmid PxylX-divk-gfp | pMR20 | Jacobs et al. (2001) |
Cryo-EM tomography and ribosome analysis
Specimen preparation, tomographic data acquisition and image analysis were performed as described (Comolli et al., 2009). We collected and analysed 25 tomographic datasets of wild-type cells, 5 of ΔpopZ and 9 of PopZ overproducing cells. Comolli et al. (2009) also describes the techniques for three-dimensional reconstruction of ribosomal particles. In this analysis, the average of 39 subvolumes of selected dense globular particles was compared with the structure of the Escherichia coli translating ribosome (EMBD-ID 1070) resampled and filtered to approximately 5 nm resolution.
Immuno-electron microscopy
Bacterial cultures were grown to mid/late log phase (OD600 = 0.5–0.7), induced with xylose when required, and fixed in 0.01% acrolein, 2% glutaraldehyde (diluted in 0.01 M cacodylate buffer, pH 7.4) for 15 min (5 min at room temperature, then 10 min in ice). Fixed cells were equilibrated in cacodylate buffer and plated on sapphire discs (Guild Opt., Amherst, NH, USA) coated with poly(ethyleneimine) (Sigma-Aldrich, Saint Louis, MO, USA). For each high pressure freezing session, two sapphire discs were sandwiched together and processed according to the manufacturer’s instructions (HPF-010, Bal-Tec, Balzers, Liechtenstein), freeze-substituted in Araldite/Epon, infiltrated and embedded as described by Matsko and Mueller (2005). Sections (80 nm thick) were cut from the blocks and, to facilitate antibody penetration, etched in a freshly prepared solution of sodium hydroxide (1% of a saturated solution of sodium hydroxide made in 100% ethanol). Etched sections were labelled O/N with primary antibodies then incubated with gold-conjugated secondary antibodies 6 nm or 10 nm gold particles for detection at the electron microscope. Prior to viewing, the labelled sections were post-fixed with 2% glutaraldehyde and stained in 2% uranyl-acetate for 5 min and Sato lead for 1 min. The electron micrographs were shot on a JEOL 1200 EX microscope (JEOL Ltd, Tokyo, Japan).
Gel electrophoresis, Western blotting and co-immunoprecipitation
Analytical sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and native gel electrophoresis were performed using a 12% or 4–20% Tris-glycine gel (Bio-Rad), respectively, followed by electrophoretic transfer to a PVDF membrane (Millipore). Immunoblotting was with polyclonal anti-PopZ, anti-DivJ, anti-mCherry or anti-RcdA antibodies at a concentration of 1:10 000; polyclonal anti-HU and monoclonal anti-GFP (Roche) at 1:3000; and detection with chemiluminescent substrate (Pierce). Band intensity was quantified using ImageJ. The co-immunoprecipitation assay was performed as described by Bowman et al. (2008).
Light and fluorescence microscopy
Cells were immobilized on a 1.25% agarose pad containing growth media. For time lapse experiments, the edges of the coverslip and pad were sealed as described by Thanbichler and Shapiro (2006) to prevent evaporation. Images were collected using a Leica DM6000B microscope with a Hamamatsu C9100 EM-CCD camera and a 100× PH3 PlanApo 1.40 NA objective, driven by independently designed software (Michael Fero, unpublished). The filter sets used for YFP, CFP and mCherry imaging were Semrock model 2427A, 2432A and 4040B respectively.
Computational image analyses: To quantify SpmX-mCherry fluorescence, an automated program separated individual cells and, for each cell, measured the intensity of the highest peak (assuming a Gaussian profile) and the delocalized signal intensity by calculating the median value of pixel intensity across the whole cell, excluding the area of peak intensity. These values were then subtracted by the local background signal. To quantify the motion of CFP-ParB foci, we first corrected for systematic shifts between frames using ImagePro software (Media Cybernetics). In the analysis of wild-type cells, foci at the flagellated pole were differentiated from foci at the stalked pole by observing stalks in the corresponding phase-contrast image, and a mask was used to exclude all foci of one type. We developed a MATLAB application based on the program ‘SPtrack1.0’ to determine the centres of fluorescent peaks by calculating the weighted centroid, and movement (in pixels) was calculated by measuring the Euclidean distance travelled by the centre of a peak over a given time interval.
Supplementary Material
Acknowledgements
We thank Patrick Viollier for kindly providing strains, and also Esteban Toro, Antonio Iniesta and other members of the Shapiro lab for advice and critical reading of the manuscript. This work was supported by National Institute of Health Grants GM073001 to H.H.M and L.S., GM51426 and GM32506 to L.S., 5P41RR004050-16 to M.H.E. and F32GM080008 to G.B.; and by Department of Energy Grants DE-FG02-05ER64136 to L.S. and H.H.M, and DE-FG02-05ER64136 to M.H.E.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Baker TA, Funnell BE, Kornberg A. Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J Biol Chem. 1987;262:6877–6885. [PubMed] [Google Scholar]
- Bowers LM, Shapland EB, Ryan KR. Who’s in charge here? Regulating cell cycle regulators. Curr Opin Microbiol. 2008;11:547–552. doi: 10.1016/j.mib.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, Shapiro L. Spatial complexity and control of a bacterial cell cycle. Curr Opin Biotechnol. 2007;18:333–340. doi: 10.1016/j.copbio.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J. 2009;3:159–167. doi: 10.1038/ismej.2008.99. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran DN, Kashef K, Lee CM, Xu H, Reddy EP. Scaffold proteins of MAP-kinase modules. Oncogene. 2007;26:3185–3202. doi: 10.1038/sj.onc.1210411. [DOI] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Easter J, Jr, Gober JW. ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol Cell. 2002;10:427–434. doi: 10.1016/s1097-2765(02)00594-4. [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Iniesta AA, Shapiro L. Cell cycle regulation in Caulobacter: location, location, location. J Cell Sci. 2007;120:3501–3507. doi: 10.1242/jcs.005967. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol Microbiol. 2001;40:485–497. doi: 10.1046/j.1365-2958.2001.02404.x. [DOI] [PubMed] [Google Scholar]
- Hottes AK, Shapiro L, McAdams HH. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol. 2005;58:1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Iniesta AA, Shapiro L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc Natl Acad Sci USA. 2008;105:16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci USA. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Hung D, Shapiro L. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc Natl Acad Sci USA. 2001;98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC, Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genet. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L. Small non-coding RNAs in Caulobacter crescentus. Mol Microbiol. 2008;68:600–614. doi: 10.1111/j.1365-2958.2008.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- Matsko N, Mueller M. Epoxy resin as fixative during freeze-substitution. J Struct Biol. 2005;152:92–103. doi: 10.1016/j.jsb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Akiyama H, Komada M, Kamiguchi H. betaIV-spectrin forms a diffusion barrier against L1CAM at the axon initial segment. Mol Cell Neurosci. 2007;34:422–430. doi: 10.1016/j.mcn.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Poindexter JS. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shebelut CW, Jensen RB, Gitai Z. Growth conditions regulate the requirements for Caulobacter chromosome segregation. J Bacteriol. 2009;191:1097–1100. doi: 10.1128/JB.00862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer JM, Newton A. Turning off flagellum rotation requires the pleiotropic gene pleD: pleA, pleC, and pleD define two morphogenic pathways in Caulobacter crescentus. J Bacteriol. 1989;171:392–401. doi: 10.1128/jb.171.1.392-401.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro E, Hong SH, McAdams HH, Shapiro L. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci USA. 2008;105:15435–15440. doi: 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Alley MR. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jones BD, Brun YV. A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol Microbiol. 2001;40:347–360. doi: 10.1046/j.1365-2958.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.