Summary
The entry of a water molecule into the distal heme pocket of pentacoordinate heme proteins such as myoglobin and the α,β chains of hemoglobin can be detected by time-resolved spectroscopy in the heme visible bands after photolysis of the CO complex. Reviewing the evidence from spectrokinetic studies of Mb variants, we find that this optical method measures the occupancy of non(heme)coordinated water in the distal pocket, nw, with high fidelity. This evidence further suggests that perturbation of the kinetic barrier presented by distal pocket water is often the dominant mechanism by which active site mutations affect the bimolecular rate constant for CO binding. Water entry into the heme pockets of isolated hemoglobin subunits was detected by optical methods. Internal hydration is higher in the native α chains than in the β chains, in agreement with previous crystallographic results for the subunits within Hb tetramers. The kinetic parameters obtained from modeling of the water entry and ligand rebinding in Mb mutants and native Hb chains are consistent with an inverse dependence of the bimolecular association rate constant on the water occupancy factor. This correlation suggests that water and ligand mutually exclude one another from the distal pockets of both types of hemoglobin chains and myoglobin..
Introduction
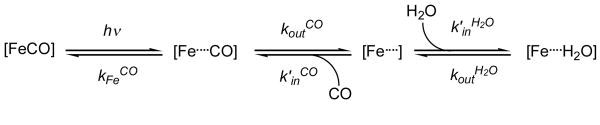
Water molecules buried within the internal cavities of proteins can modulate dynamical processes such as folding, catalysis, and proton transfer, as well as affect protein structural stability and rigidity.1–8 In the case of the oxygen storage protein myoglobin and other pentacoordinate heme proteins, including human hemoglobin A, a solvent water molecule is sequestered within the deoxygenated heme pocket by hydrogen bonding to the distal histidine (His at the E7 helical position) and exerts a controlling influence on the kinetics and thermodynamics of ligand binding during protein function.9–11 Displacement of this water from the distal pocket is required for ligand access to the active site in native sperm whale myoglobin and probably in human hemoglobin (Scheme 1). Thus, one important mechanism by which the structural and conformational perturbations induced by mutation and pH change influence the kinetics of ligand binding is through changes in the occupancy of water in the unliganded distal pocket.
Scheme 1.
We have recently discovered an optical signal for the entry of non-coordinated water into the distal heme pocket.12 These absorbance changes permit convenient monitoring of the dynamics of water entry, which has also been observed in atomic level detail by Schotte et al. by using picosecond time-resolved X-ray crystallography.13 The observation from both optical and X-ray time-resolved measurements that water entry is concurrent with ligand escape has confirmed the idea that the presence of ligand in the pocket prevents water entry and vice versa. This mutual exclusion has also been observed to occur between coordinated water and internal water molecules in molecular dynamics simulations of the distal pocket in met-myoglobin.14 The requirement to displace distal pocket water by apolar diatomic gases is the physical basis for the inhibition of ligand binding caused by increases in distal pocket polarity.11 The optical detection method provides additional and sometimes more reliable measurements of water occupancy than has been obtained by traditional structural methods such as X-ray crystallography and NMR spectroscopy, as discussed further bvelow. In combination, the structural and kinetic measurements provide a greater understanding of how distal pocket mutations and protein conformational changes, such as those observed at low pH, affect ligand binding in pentacoordinate heme proteins.
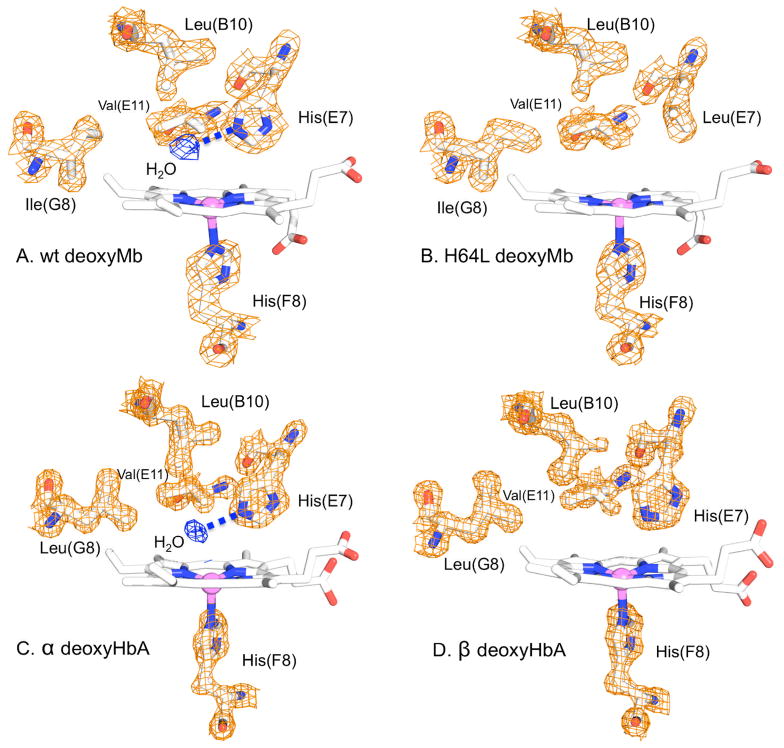
Previous crystallographic evidence indicated that the distal water occupancy is high, nw = 0.84, in native and wild type deoxymyoglobin and becomes effectively zero when the distal histidine residue (E7) is replaced with the apolar residue leucine (Figure 1).15 On the other hand, mutations from apolar to more polar residues in the distal pocket can increase distal water occupancy, as observed for the single V68F Mb mutant and for the double mutant H64L/V68N compared to H64L Mb.16, 17 The marked increase observed in the ligand binding rates of apolar His64 mutants relative to native myoglobin first led to the suggestion that the distal water molecule excluded small ligands like O2, CO and NO from the heme binding site.9 This interpretation suggests that there is a significant kinetic barrier to bimolecular ligand entry into the globin due to the requirement for water displacement, making the observed association rate constant proportional to the fraction of empty active sites (one minus the fraction of water occupancy, 1 − nW).11
Figure 1.
X-ray crystal structures of the distal heme pockets of (A) WT deoxymyoglobin (2MGL) and (B) H64L deoxyMb (2MGD) and (C) α and (D) β subunits within tetrameric deoxyHb (2DN1). A noncoordinated water molecule is H-bonded to the Nε atom of the distal histidine E7 in WT Mb (84% occupancy)15 and α subunits (100% occupancy) of deoxyHbA.41 Apolar mutations at the E7 position in Mb, e.g., leucine, reduce or eliminate distal water and there is no discrete water molecule in the heme pocket of unliganded β subunits.
Thus, the distal histidine can both inhibit ligand binding by stabilizing the water in the unliganded pocket and enhance affinity by donating a hydrogen bond to coordinated ligands. The latter electrostatic effect preferentially stabilizes bound O2 because of the large negative charge on the oxygen atoms of the Fe(II)-O-O complex. In contrast, the Fe-C-O complex is almost neutral and little affected by the presence of the His(E7) side chain. Thus, the net result of the distal histidine’s hydrogen bonding capacity is a decrease in the association rate constants for all ligands due to a reduction in the rate of entry into the active site caused by the presence of water and a marked, preferential decrease in the rate constant for O2 dissociation caused by stabilization of the FeO2 complex, with little or no effect on CO dissociation.11 The net effect of having a histidine at the E7 position in the active site is to enhance O2 affinity relative to that for CO. This ligand discrimination suppresses poisoning of the O2 storage and release function of Mb that might otherwise result from the production of endogenous CO during heme catabolism.18
The emerging recognition that internal water plays important roles in protein structure and function has motivated a need for more reliable methods for the detection of H2O molecules near the heme iron. The electron densities of buried water molecules may be obscured in X-ray diffraction studies by positional disorder, and NMR detection is often limited by orientational disorder and short residence times.19–22 Although computational methods can be of help in searching for the less ordered and more rapidly exchanging water molecules missed by traditional approaches, the extent to which water may be buried in the apolar cavities of proteins has remained an open question.8, 22
Optical absorbance changes due to alterations in polarity near heme groups appears to be unaffected by positional disorder, allowing dispersed buried water to be detected in Mb mutants wherein traditional structural methods have failed. Thus, the deoxy heme pockets of ligand-binding globins may offer model systems for investigating the effects of residue polarity and steric bulk on hydration in internal cavities. This information can, in turn, be used to guide the development of improved X-ray crystallographic, NMR, and computational approaches to detect internal water. The results presented here for myoglobin and hemoglobin chains also suggest that it may be possible to generalize optical methods for internal water detection in other chromophore-containing proteins.
Materials and methods
Preparation of Hb chains and variants
Hb α and β chains were isolated from the native human Hb tetramers by the method of Parkhurst and Parkhurst, as modified by Birukou et al.23, 24 Protein samples were prepared for laser photolysis experiments at a concentration of 75–100 μM in pH 7.3, 100 mM phosphate buffer under 1 atm CO and reduced with 1 mM sodium dithionite in sealed quartz cuvettes.
Time-resolved spectroscopy
Time-resolved absorption data were measured as described previously using a broadband xenon flashlamp probe, multichannel detection, and frequency-doubled Nd:YAG laser actinic pulses (8-ns, 40-mJ).25 Photolysis difference spectra were collected over the visible band spectral region (500–650 nm) at 41 time delays spaced logarithmically from 20 ns to 20 ms after the laser pulse. Each time-delayed spectrum represented the average of ~1000 photolysis measurements.
Spectrokinetic detection and modeling of water entry
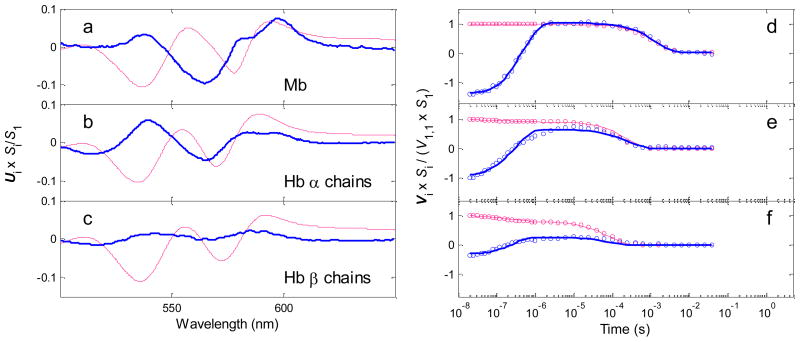
The procedure for kinetic modeling of water entry and ligand binding data described by Goldbeck et al. was applied to spectrokinetic data for the Hb chains.12 This model is based on the reaction sequence shown in Scheme 1.11 Singular value decomposition (SVD) was first applied to the matrix of time-resolved spectral data for each species to obtain matrices containing orthogonal spectral (U) and temporal (V) basis vectors and their respective weighted contributions, the singular values (S).26 The two largest SVD components were used as inputs to the modeling procedure, i.e., U1·S(1,1) ·V1′ and U2·S(2,2) · V2′. In general, SVD components may not necessarily correspond to particular physicochemical processes, although this correspondence is more likely when the processes are both spectrally and temporally distinct, as is the case for the myoglobin and hemoglobin visible band photolysis data. The first SVD component, which is very similar in spectral shape (U1) to the CO-unbound – CO-bound heme difference spectrum, approximately represents geminate and bimolecular CO rebinding, and the second component, which corresponds spectrally to a blue shift in the CO-unbound heme spectrum (U2), approximately represents water entry to the distal pocket (concurrent with geminate ligand escape) and exit (concurrent with bimolecular rebinding) (see Figure 6).12, 17
Figure 6.
First two SVD (singular value decomposition) components of CO photolysis TROA spectra for isolated human Hb chains (20 C, neutral pH): (a,d) Mb shown for comparison,27 (b,e) Hb α chains, (c,f) Hb β chains, The red traces correspond most closely to CO rebinding processes, the blue traces are most sensitive to water entry into the pocket. Panels a–c, spectral basis vectors; the second spectral vectors are scaled by their singular values and a factor of 20 for clarity. Panels d–f, temporal basis vectors; the second temporal basis vectors are scaled by their singular values and a factor of 25. The solid lines in panels d–f show fits to the Vi data points calculated from the kinetic hydration model of Goldbeck et al.12 with the parameter values (e.g., nw) shown in Table 1.
The fitting analysis provided values for all parameters in Scheme 1 using the time courses V1 and V2. The equilibrium water occupancy values were calculated from nw = kinH2O/(kinH2O + koutH2O). The water entry difference spectra were calculated as the spectral difference between the hydrated and anhydrous pockets, Scheme 1 species [Fe···H2O] and [Fe···], respectively. The water entry difference spectra for the native and variant Hb chains were constrained in the fitting procedure to be similar, when scaled to 100% occupancy, to the full-occupancy spectrum calculated previously for myoglobin.27 (The geminately dissociated and anhydrous deliganded species, [Fe···CO] and [Fe···], respectively, were assumed to have identical spectra in the modeling.) The geminate recombination yield was calculated from φg = kFeCO/(kFeCO + koutCO). Because the geminate rebinding time scale is much faster than bimolecular rebinding in hemoglobin and myoglobin (and the intrinsic rate for equilibration of water in the deoxy pocket, kinH2O + koutH2O, is very fast), Scheme 1 implies that the observed rate constant for bimolecular recombination, k′, will be given by the expression k′ = (1 − nw) ·φg· kinCO.
Results and discussion
Comparison of nw values from spectrokinetic and X-ray diffraction methods
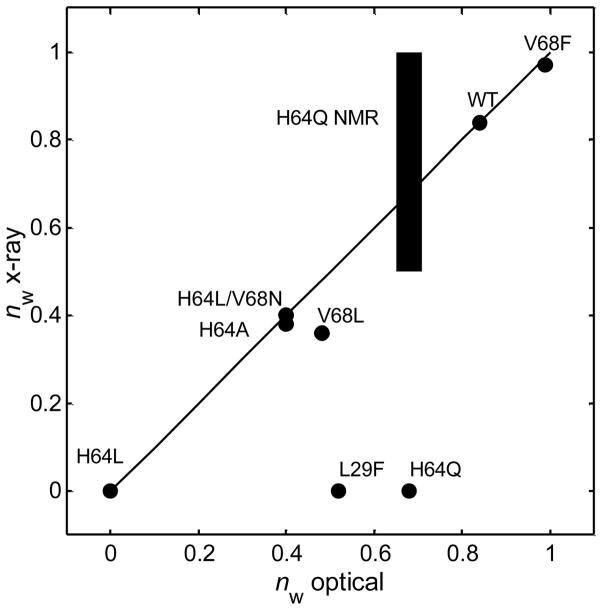
The spectrokinetic measurement of water entry after ligand photolysis permits determination of the kinetics and thermodynamics of heme hydration, information that can facilitate understanding of the influence of internal water on bimolecular ligand rebinding.12 The intrinsic rate of water entry into an empty distal pocket (kinH2O) is very fast, and thus the observed rate of solvent entry is limited to a great extent by the rate at which the CO ligand leaves the pocket (~2 μs−1 for native myoglobin). The correspondence between the spectrokinetically determined water occupancy values, nw, and the values determined by X-ray crystallography (where available) is excellent for WT Mb and most of the distal pocket Mb mutants studied so far, as shown in Figure 2. The two notable exceptions, L29F and H64Q Mb, contain distal pocket water that is detected spectrokinetically, but not observed by discrete electron density peaks in the crystal structures for these mutants, presumably because of positional disorder, as discussed further below.
Figure 2.
Distal pocket water occupancy values of deoxyMb variants determined by the optical spectrokinetic method correlate closely with values from X-ray crystallography. Paired values lie close to the line shown for perfect correlation, except for the mutants L29F and H64Q. NMR confirms the high distal water occupancy of H64Q (blue bar).37 High positional disorder accounts for the inability of X-ray diffraction to detect water in either the H64Q or L29F heme pockets (see text). Optical and X-ray diffraction derived nw values were taken from Goldbeck et al.12, 17
The kinetic mechanism for heme pocket hydration presented in Scheme 1 assumes that the entry of water from the bulk solvent into the distal portion of the heme pocket can be represented by a single, pseudo first order rate constant kinH2O[H2O]. This process could involve movement through the His(E7) gate or be a more complex process involving many distinct routes through the protein. No continuous open channels connecting the pocket to the solvent are obvious in the crystal structure, so the entry of small molecules like water or CO must depend on stochastic fluctuations in the close packing of protein residues. The parameter kinH2O thus represents an average of the distribution of second order rate constants corresponding to the various diffusional paths that water may take from the bulk solvent to the pocket.
We cannot rule out the possibility that small amounts of water enter the distal pocket more directly from nearby protein cavities, e.g. the four Xe cavities. However, the water occupancies of these apolar cavities are expected to be low, as suggested by the absence of electron density in these sites in crystal structures of Mb under 1 atm of gas pressure and by a recent molecular dynamics study of buried water in met-myoglobin.14 The distal pocket also becomes free of water when His64(E7) is replaced by Leu, as judged both by electron density maps of deoxy- and metMb crystals and by the spectrokinetic assay (Figure 1).16
The spectrokinetic water assay assumes that the extent to which the distal water dipole perturbs the observed deoxyMb visible band spectrum is proportional to the water occupancy. However, the spectral perturbation must depend on the water dipole orientation and distance relative to the heme ring. Thus, our emprical observation that the spectral assay reproduces the X-ray diffraction-determined water occupancies of a variety of distal pocket mutants with high fidelity suggests that the average distance, mobility, and orientation of the water dipoles with respect to the heme are similar among the Mb variants studied. This correlation may reflect the small size of the distal pocket, which constrains variation of the heme-water distance, and the fact that polar residues that stabilize internal water act as hydrogen bond acceptors, which would tend to consistently orient the negative end of the water dipole toward the heme.
Modulation of k′ by distal water occupancy
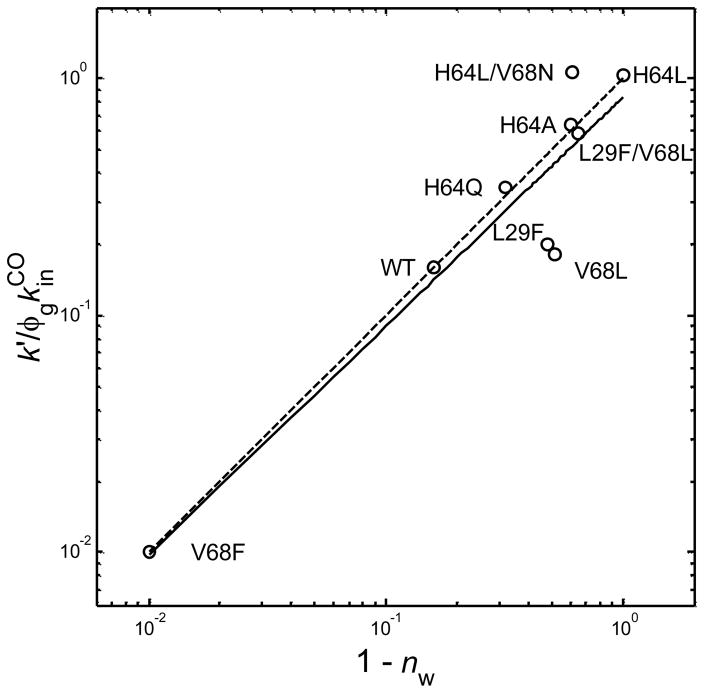
The observed bimolecular rate constant for CO rebinding, k′, depends on the fraction of empty active sites (1 − nw), which was suggested in the original mutagenesis studies,9,11 and is confirmed by the spectrokinetic results plotted in Figure 3 for a larger series of myoglobin variants. We have plotted k′/(φg·kinCO) vs. (1 − nw), where k′[CO] = τobs−1, τobs is the observed exponential time constant for ligand rebinding, [CO] = 1 mM, φg is the geminate recombination yield, and kinCO is the fitted microscopic rate constant for CO entry into an empty distal pocket. For the purpose of comparison, we assumed that the latter parameter is ~80 μM−1 s−1, which is near the diffusion controlled value for heme proteins and equal to the average of the kinCO values determined for Gly, Ala, Val, and Leu E7 mutants of sperm whale Mb. 28, 29 The linearity observed is consistent with a good fit to the expression k′ = (1 − nw) ·φg· kinCO. Note that the range of (1 − nw) values plotted in Figure 3 implies that the distal water occupancy factor modulates the observed ligand binding rates over two orders of magnitude. The fraction of empty deoxyMb active sites (1 − nw) ranges from 0.01 for V68F Mb (99% water occupancy) to the highest possible value, 1.0 for H64L Mb (0% water occupancy).
Figure 3.
Log-log plot of the observed bimolecular CO recombination rate constant normalized to the geminate recombination yield and the microscopic rate constant for CO entry into the pocket (assumed here to be 80 μM−1 s−1 for all species) vs. the distal water kinetic factor, (1 − nw), assayed spectrokinetically for myoglobin wild type and distal pocket mutants.12, 17 The solid line shows the linear least squares fit to the data and the dashed line represents a perfect fit to the expression k′ = (1 − nw)φgkinCO.
Comparison of modulation of k′ by nw, φg, and kinCO
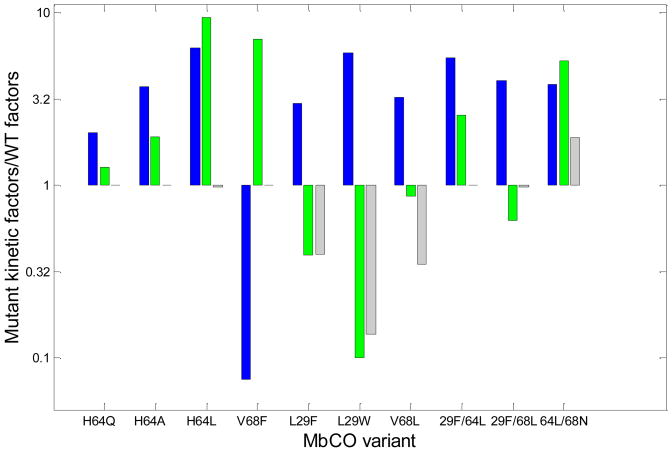
We have plotted the ratios of the values of nw, φg, and kinCO for all the Mb variants to those for wild-type myoglobin in Figure 4 to evaluate the individual contributions of these parameters to the overall bimolecular rate constant for CO binding. Surprisingly, the water occupancy factor, (1 − nw), often makes the largest contribution to changes in the overall bimolecular rate constant. Mutation-induced changes in water occupancy are typically the single most important factor affecting ligand association and frequently overshadow the direct effects of residue size and steric hindrance on kinCO and the fraction of geminate rebinding, φg.13
Figure 4.
Bar graph of the three factors contributing to k′ = (1 − nw) φg kinCO of myoglobin mutants relative to wild-type myoglobin: (blue) (1 − nw)mutant/(1-nw)wt, (green) φg,mutant/φg,wt, and (gray) kin,mutantCO/kin,wtCO. The values plotted were taken from references 12 and 17, with the exception of φg,mutant/φg,wt for L29W. Because φg was too small for direct measurement in L29W MbCO, the ratio observed in L29W MbO2 by Scott et al. was used: φg,mutant/φg,wt ≤ 0.1.43
Distal heme hydration is also an important factor governing the increase in CO binding rate and affinity observed in myoglobin at low pH in the range from 6 to 4.30–32 Protonation of the distal histidine promotes an “open” protein conformation in which the protonated distal histidine swings out into solvent, opening up the E7 channel and making the active site less polar and more anhydrous.33–35 Spectrokinetic results for nw measured at acidic pH values as low as 4.5 support the hypothesis that a decrease in occupancy of the distal water molecule accounts in part for the high ligand-binding rate attributed to the open conformation.27
Correlation between nw and hydrophobicity of distal pocket residues
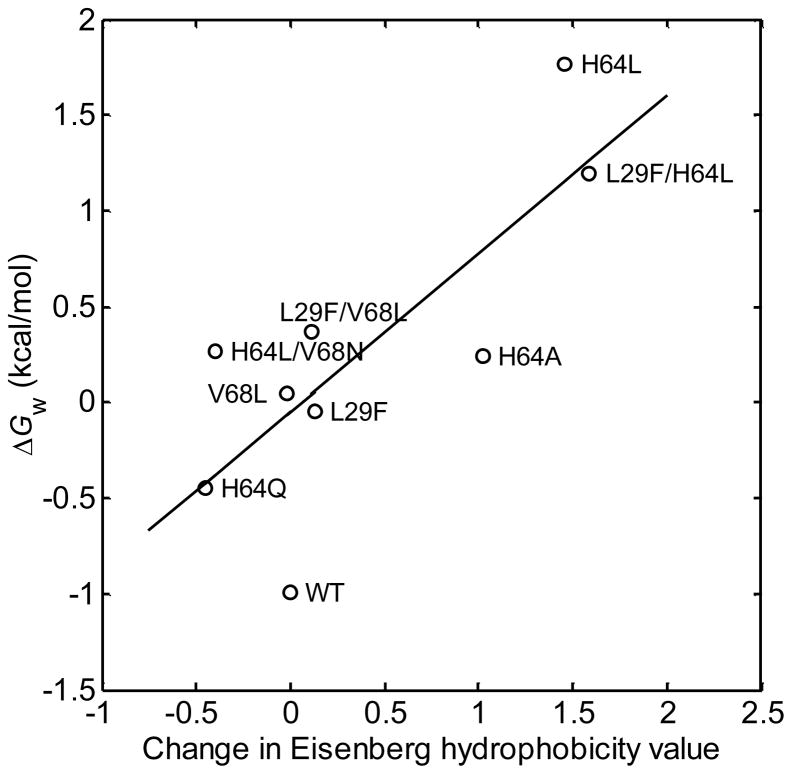
In general, mutations that increase the hydrophobicity of the pocket will decrease the distal water occupancy, as observed for apolar mutations of the distal histidine. To further examine this idea, a plot of the free energy of water occupancy in the active site of myoglobin, ΔGw = −RT ln[nw/(1 − nw)], vs. changes in active site hydrophobicity accompanying single and double mutations was constructed and is shown in Figure 5. The Mb mutant system appears to offer the first opportunity for a quantitative experimental test of the correlation between hydration free energies and residue hydrophobicities for an internal cavity in a protein. Consistent with expectation, there is a strong correlation, implying that polar side chain-water interactions are the main determinants of the free energies of water occupancy in Mb and probably more important than changes in residue size (which showed little correlation, results not shown). The near-unity slope of the correlation is also consistent with this interpretation, as hydrophobicity values such as those used here are designed to roughly represent the solvation free energies of residues in water vs. octanol.36
Figure 5.
Free energy of distal water occupancy vs. change in Eisenberg hydrophobicity values for single and double mutations of distal pocket residues in sperm whale myoglobin. ΔGw = −RT ln[nw/(1 − nw)], where nw values were measured using the optical spectrokinetic assay method.12, 17 Line is best linear least squares fit (r = 0.78).
Spectrokinetic detection of disordered water
The spectrokinetic approach overcomes several limitations of the structurally more specific X-ray crystallographic and NMR approaches in detecting disordered water in protein cavities. Our time-resolved optical spectroscopic studies have found evidence for the presence of water in the distal pockets of myoglobin mutants previously believed to be anhydrous from traditional structural methods. Both L29F and V68L deoxyMb were found to contain distal water molecules (~50% occupancy) that were not previously reported by X-ray diffraction studies.17 In our recent study, we reexamined the diffraction data for V68L Mb and found electron density in the pocket consistent with a water occupancy value of 0.36, in closer agreement with the optically determined value. However, a similar reexamination of the electron densities for L29F Mb failed to detect significant water occupancy, which we attributed to positional disorder of distal water molecules that exceeded the limit for crystallographic detection. The spectrokinetic method also detected a high water occupancy in H64Q Mb (nw = 0.68), a mutant in which no discrete water is seen by X-ray crystallography, although qualitative evidence from NMR spectroscopy indicated significant hydration of the heme pocket.12, 37
Water entry into the distal heme pockets of Hb chains
Native Hb α and βchains
We have recently examined water entry into hemoglobin after photolysis of the isolated α and β subunits. The α subunits of tetrameric deoxyHbA contain a well resolved noncoordinated water molecule in their heme pockets at room temperature, whereas, in contrast, the heme pockets of the β deoxyHb subunits have negligible electron density associated with distal water.38 We obtained a similar result from our spectrokinetic data for the isolated native hemoglobin chains shown in Figure 6. These measurements indicated that water enters the distal heme pocket of the α chains with a high fractional occupancy (nw > 0.5) after CO photodissociation. This process occurs on submicrosecond timescales similar to that for dissociated ligand escape from the pocket. The amplitude for water entry into β subunits is significantly smaller. The blue traces (2nd SVD component) for the Hb α chains (Figure 6, panels b and e) bear a strong similarity to the water entry signal observed previously in MbCO, both spectrally (cf. panel a), in time evolution, and in amplitude (cf. the trough to peak evolution of V2 at time delays ≤ ~1 μs, panel d). The corresponding signal is much smaller for the β chains (panels c and f), consistent with expectations from the crystal structure of the unliganded tetramer. We can get a rough initial estimate of the water occupancies in the Hb chains from the data in Figure 6 by assuming that the spectrokinetic water signal amplitudes for all species shown are proportional to nw by the same factor, i.e., that the spectral response of the heme chromophore to water entry is similar in myoglobin and the hemoglobin chains. Using myoglobin as a guide, we estimate that nw ≈ 0.6 for the α chains and ≈ 0.2 for the β chains (see Table 1 for more quantitative results from the spectrokinetic modeling). The differences between our results for isolated subunits and the crystallographic results for deoxyHbA tetramers could be real because of differences in quaternary structure; however the differences in hydration of the α and β subunits are apparent in both cases.
Table 1.
Kinetic parameters for MbCO and isolated HbCO α and β chains.a
| Variant | kFeCO (μs−1) | kinCO[CO]c (μs−1) | koutCO (μs−1) | kinH2O[H2O] (μs−1) | koutH2O (μs−1) | nw | X-ray Structure H2O Occupancy | φg | 1− nw | k′[CO]e (μs−1) | τobs−1 (μs−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mbb | 0.11 | (0.08) | 2.2 | 9.0 | 1.7 | 0.84 | 0.84 | 0.051 | 0.16 | 0.00064 | 0.00066 |
| α Native f | 3.4 | (0.08) | 21 | 6.0g | 3.4 | 0.64 | 1.00d | 0.14 | 0.36 | 0.0040 | 0.0057 |
| β Native f | 1.6 | (0.08) | 6.0 | 6.0 | 20 | 0.23 | 0.00d | 0.22 | 0.77 | 0.014 | 0.011 |
Rate constants and inverse lifetimes reported in μs−1; [CO] = 1.0 mM; [H2O] = 55.5 M.
Constrained during fitting procedure to a maximum (diffusion controlled) value of 0.08 μs−1.
Water occupancy values from high resolution crystal structure of deoxyHb tetramer.41
k′ ≡ (1 − nw)φgkinCO.
The φg values of the chains were fixed at the value measured by Birukou et al., as were kFeCO and koutCO.24
The α chain value for kinH2O[H2O] was fixed at the value obtained for the β chains.
Spectrokinetic modeling of theα and β subunit data
We applied the spectrokinetic modeling procedure described above to data for the native Hb chains to both (1) assess the applicability of Scheme 1 and the kinetic control of bimolecular ligand rebinding by the water factor (1 − nw) and (2) obtain more quantitative estimates of nw. We set the value kinCO[CO] for entry into an empty distal pocket to 0.08 μs−1, which is roughly equal to that for H64L Mb,12 and the value computed by Birukou et al.24 for various small apolar E7 mutants of both Hb subunits. We also set the geminate rebinding parameters to the values reported by Birukou et al.24 for native α and β subunits. The fitted parameters indicate that the differences between k′[CO] for the α and β subunits are due to a complex set of differences in iron ligand bond formation, ligand escape, and the extent of water hydration, (1 − nw) (see Table 1).
Conclusions
The spectrokinetic approach detects the presence of water with high fidelity in the distal heme pockets of a variety of myoglobin mutants, even those in which the water is apparently too disordered for detection by X-ray crystallography. The kinetic parameters obtained from this approach provide quantitative support for the idea that ligand association rates are proportional to the factor 1 − nw. The importance of this inverse dependence on water occupancy in modulating ligand binding rates was highlighted by the finding that this factor varied over two orders of magnitude in the Mb variants studied. It was also the major factor modulating ligand association rates in the majority of the variants studied. Water occupancy, along with the internal rate of iron-ligand bond formation and the rates of ligand escape and entry into vacant active site, are the three factors that govern the overall association rate constant. Variations in nw caused by distal residue mutations correlate well with concomitant changes in the overall hydrophobicity of the pocket.
The results presented here extend the optical approach for detecting water entry to the heme pockets of the isolated α and β chains of hemoglobin. There seems to be little difference between the ligand binding rates and affinities of the chains within the tetramer, despite their known structural differences.39, 40 This near equivalence of chain affinities is thought to be a result of evolutionary pressures to maximize the cooperative binding of O2 within the tetramer, and involves balancing factors such as bond formation and dissociation rates, the ligand escape rates from the pocket and the barrier to ligand entry that is presented by the distal pocket water molecules. In particular, the pressure to increase distal pocket polarity in order to increase affinity and reduce auto-oxidation by hydrogen bond donation to bound O2, must be balanced against a concomitant increase in water occupancy in the deoxygenated state, which lowers association rate constants and ligand affinity by restricting access to the heme. Our results show that the extent of hydration in isolated α subunits is 2 to 3 times larger than that in β subunits, in agreement with the room temperature, high resolution crystal structure of tetrameric deoxyHbA.38, 41 This agreement seems to be only little affected by the T-state quaternary constraint on heme tertiary structure that is present in the hemoglobin tetramer and absent in the isolated subunits, which exhibit R-state behavior.
The entry of water into the α hemoglobin deoxy pocket could also play a structural role in the protein conformational changes leading from the R to the T quaternary state. Density functional theory/molecular mechanics calculations for myoglobin have suggested that H-bonding of water to the distal histidine produces a steric contact between the water molecule and the Phe43 and Phe46 residues of the CD loop.42 Interestingly, this contact displaces the loop after deligation in a manner corresponding to the early steps along the R to T allosteric pathway observed in hemoglobin.
The results and conclusions presented here for myoglobin and the isolated α,β chains of hemoglobin help to illuminate the role that distal water plays in modulating the binding of ligands to pentacoordinate heme proteins. The knowledge gained from such studies may ultimately aid in the engineering of proteins for enhanced autooxidation resistance, O2 transport and CO discrimination.10
Acknowledgments
We would like to acknowledge stimulating discussions with Dr. Judith Klinman about hydrophobicity and buried water in proteins. This work was supported financially by NIH grants EB02056 (DSK), GM52588 (RME), MD000544 (RME), GM35649 (JSO), and HL47020 (JSO); Robert A. Welch Foundation Grant C-0612 (JSO) and Predoctoral Fellowship (IB); and a student fellowship from NIH grant GM08574 (ILP).
References
- 1.Park S, Saven JG. Proteins: Struct Funct Bioinf. 2005;60:450–463. doi: 10.1002/prot.20511. [DOI] [PubMed] [Google Scholar]
- 2.Takano K, Funahashi J, Yamagata Y, Fujii S, Yutani K. J Mol Biol. 1997;274:132–142. doi: 10.1006/jmbi.1997.1365. [DOI] [PubMed] [Google Scholar]
- 3.Mao Y, Ratner MA, Jarrold MF. J Am Chem Soc. 2000;122:2950–2951. [Google Scholar]
- 4.Schutz CN, Warshel A. Abstr Pap Am Chem Soc. 2002;223:C75–C75. [Google Scholar]
- 5.Cheung MS, Garcia AE, Onuchic JN. Proc Natl Acad Sci USA. 2002;99:685–690. doi: 10.1073/pnas.022387699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer E. Protein Sci. 1992;1:1543–1562. doi: 10.1002/pro.5560011203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofacker I, Schulten K. Proteins: Struct Funct Genet. 1998;30:100–107. [PubMed] [Google Scholar]
- 8.Damjanovic A, Schlessman JL, Fitch CA, Garcia AE, Garcia-Moreno B. Biophys J. 2007;93:2791–2804. doi: 10.1529/biophysj.107.104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohlfs RJ, Mathews AJ, Carver TE, Olson JS, Springer BA, Egeberg KD, Sligar SG. J Biol Chem. 1990;265:3168–3176. [PubMed] [Google Scholar]
- 10.Olson JS, Phillips GN., Jr J Biol Chem. 1996;271:17593–17596. doi: 10.1074/jbc.271.30.17593. [DOI] [PubMed] [Google Scholar]
- 11.Olson JS, Phillips GN. J Biol Inorg Chem. 1997;2:544–552. [Google Scholar]
- 12.Goldbeck RA, Bhaskaran S, Ortega C, Mendoza JL, Olson JS, Soman J, Kliger DS, Esquerra RM. Proc Natl Acad Sci USA. 2006;103:1254–1259. doi: 10.1073/pnas.0507840103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schotte F, Soman J, Olson JS, Wulff M, Anfinrud PA. J Struct Biol. 2004;147:235–246. doi: 10.1016/j.jsb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Scorciapino MA, Robertazzi A, Casu M, Ruggerone P, Ceccarelli M. J Am Chem Soc. 2010;132:5156–5163. doi: 10.1021/ja909822d. [DOI] [PubMed] [Google Scholar]
- 15.Quillin ML, Arduini RM, Olson JS, Phillips GN. J Mol Biol. 1993;234:140–155. doi: 10.1006/jmbi.1993.1569. [DOI] [PubMed] [Google Scholar]
- 16.Quillin ML, Li T, Olson JS, Phillips GN, Jr, Dou Y, Ikeda-Saito M, Regan R, Carlson M, Gibson QH, Li H, et al. J Mol Biol. 1995;245:416–436. doi: 10.1006/jmbi.1994.0034. [DOI] [PubMed] [Google Scholar]
- 17.Goldbeck RA, Pillsbury ML, Lintner BW, Mendoza JL, Nguyen RL, Olson JS, Soman J, Kliger DS, Esquerra RM. J Am Chem Soc. 2009;131:12265–12272. doi: 10.1021/ja903409j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piantadosi CA. Antioxid Redox Signal. 2002;4:259–270. doi: 10.1089/152308602753666316. [DOI] [PubMed] [Google Scholar]
- 19.Phillips GN, Pettitt BM. Protein Sci. 1995;4:149–158. doi: 10.1002/pro.5560040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XJ, Matthews BW. Protein Sci. 1994;3:1031–1039. doi: 10.1002/pro.5560030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denisov VP, Halle B. J Am Chem Soc. 1994;116:10324–10325. [Google Scholar]
- 22.Imai T, Hiraoka R, Kovalenko A, Hirata F. Proteins: Struct Funct Bioinf. 2007;66:804–813. doi: 10.1002/prot.21311. [DOI] [PubMed] [Google Scholar]
- 23.Parkhurst KM, Parkhurst LJ. Int J Biochem. 1992;24:993–998. doi: 10.1016/0020-711x(92)90109-e. [DOI] [PubMed] [Google Scholar]
- 24.Birukou I, Schweers RL, Olson JS. J Biol Chem. 2010;285:8840–8854. doi: 10.1074/jbc.M109.053934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esquerra RM, Goldbeck RA, Kim-Shapiro DB, Kliger DS. Biochemistry. 1998;37:17527–17536. doi: 10.1021/bi9814437. [DOI] [PubMed] [Google Scholar]
- 26.Henry ER, Hofrichter J. Methods Enzymol. 1992;210:129–192. [Google Scholar]
- 27.Esquerra RM, Jensen RA, Bhaskaran S, Pillsbury ML, Mendoza JL, Lintner BW, Kliger DS, Goldbeck RA. J Biol Chem. 2008;283:14165–14175. doi: 10.1074/jbc.M709710200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao W, Christian JF, Champion PM, Rosca F, Sage JT. Biochemistry. 2001;40:5728–5737. doi: 10.1021/bi010067e. [DOI] [PubMed] [Google Scholar]
- 29.Barboy N, Feitelson J. Biochemistry. 1987;26:3240–3244. doi: 10.1021/bi00385a046. [DOI] [PubMed] [Google Scholar]
- 30.Giacometti GM, Traylor TG, Ascenzi P, Brunori M, Antonini E. J Biol Chem. 1977;252:7447–7448. [PubMed] [Google Scholar]
- 31.Coletta M, Ascenzi P, Traylor TG, Brunori M. J Biol Chem. 1985;260:4151–4155. [PubMed] [Google Scholar]
- 32.Sage JT, Morikis D, Champion PM. Biochemistry. 1991;30:1227–1237. doi: 10.1021/bi00219a010. [DOI] [PubMed] [Google Scholar]
- 33.Morikis D, Champion PM, Springer BA, Sligar SG. Biochemistry. 1989;28:4791–4800. doi: 10.1021/bi00437a041. [DOI] [PubMed] [Google Scholar]
- 34.Tian WD, Sage JT, Champion PM. J Mol Biol. 1993;233:155–166. doi: 10.1006/jmbi.1993.1491. [DOI] [PubMed] [Google Scholar]
- 35.Yang F, Phillips GN. J Mol Biol. 1996;256:762–774. doi: 10.1006/jmbi.1996.0123. [DOI] [PubMed] [Google Scholar]
- 36.Eisenberg D, Weiss RM, Terwilliger TC, Wilcox W. Faraday Symp Chem Soc. 1982:109–120. [Google Scholar]
- 37.La Mar GN, Dalichow F, Zhao X, Dou Y, Ikeda-Saito M, Chiu ML, Sligar SG. J Biol Chem. 1994;269:29629–29635. [PubMed] [Google Scholar]
- 38.Fermi G, Perutz MF, Shaanan B, Fourme R. J Mol Biol. 1984;175:159–174. doi: 10.1016/0022-2836(84)90472-8. [DOI] [PubMed] [Google Scholar]
- 39.Perutz MF. Nature. 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 40.Balakrishnan G, Ibrahim M, Mak PJ, Hata J, Kincaid JR, Spiro TG. J Biol Inorg Chem. 2009;14:741–750. doi: 10.1007/s00775-009-0487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SY, Yokoyama T, Shibayama N, Shiro Y, Tame JRH. J Mol Biol. 2006;360:690–701. doi: 10.1016/j.jmb.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 42.Guallar V, Jarzecki AA, Friesner RA, Spiro TG. J Am Chem Soc. 2006;128:5427–5435. doi: 10.1021/ja057318h. [DOI] [PubMed] [Google Scholar]
- 43.Scott EE, Gibson QH, Olson JS. J Biol Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]