To the Editor
In 2005, we reported a widespread and nonrandom distribution of DNA palindromes in cancer cells acting as a structural basis for subsequent gene amplification. These conclusions were based, in part, on the development of a new technique for the genome-wide analysis of palindrome formation (GAPF). This technique relies on the rapid intrastrand re-annealing of palindromic sequences under conditions that do not favor intermolecular annealing followed by S1 nuclease digestion of single-stranded DNA. Competitive hybridization of GAPF enriched DNA from control and cancer cells on an early generation spotted cDNA array resulted in cancer-specific GAPF-positive signals. Direct molecular analysis validated the fact that a subset of these GAPF-positive signals represented cancer-specific palindromes located at the boundary of gene amplicons1,2. However, in more recent studies using higher density tiling arrays that permit direct validation of a larger number of specific GAPF-positive signals, we have determined that a subset of GAPF-positive signals obtained using the original GAPF protocol are not associated with palindromes or other genome rearrangements. These GAPF-positive signals from non-palindromic regions correlate with regions of high CG content and high levels of CpG methylation3, both features known to increase the melting temperature of DNA. Therefore, the original GAPF technique enriches both for palindromes, through rapid intra-strand reannealing, and for regions of heavily methylated CpG islands that are selected because they do not fully denature at 100 degrees C in 100 mM NaCl.
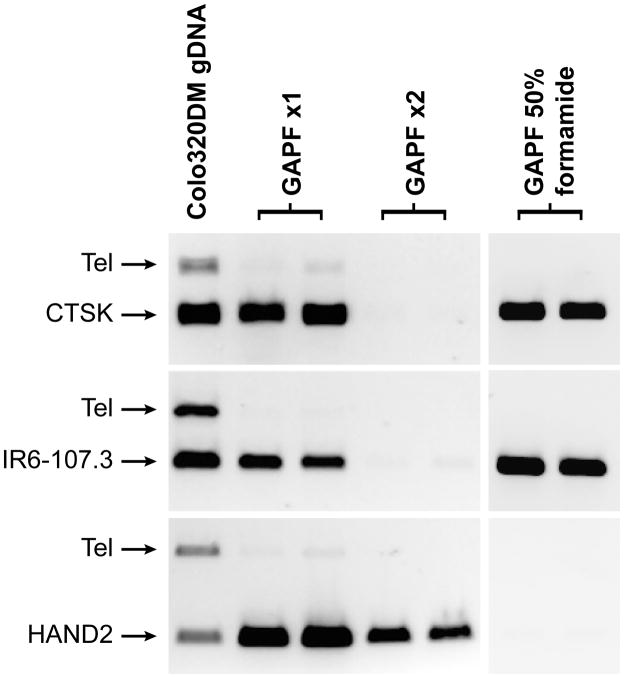
Increasing the stringency of the denaturation step in the GAPF assay by adding 50% formamide eliminates the signals from DNA methylation; and the signal from palindromes can be selectively eliminated by cycling through two rounds of the original GAPF procedure (Fig. 1). This is because the intra-molecular palindromic hairpin has a variably sized non-annealed loop at the center of the palindrome that is cleaved by the initial nuclease S1 digestion. Following cleavage of the loop, the second denaturation results in separation of the complementary strands. Therefore, the original GAPF procedure can be modified to either enrich for palindromes or to enrich for methylated CpG rich regions. We have recently used a modified GAPF procedure to identify differentially methylated regions in human medulloblastomas3.
Figure 1.
Specific enrichment for either CpG methylation or DNA palindromes using the GAPF assay.
Shown are results from Colo320DM cells of a PCR-based enrichment assay after one round of GAPF (GAPF ×1), two rounds of GAPF (GAPF ×2), or GAPF with 50% formamide added. The assay was performed in duplicate. PCR products obtained using unprocessed genomic DNA (gDNA) are included for comparison. As a negative control, the PCR product labeled as Tel amplifies a region on chromosome 1 that does not contain a DNA palindrome or CpG methylation, and primers to generate this fragment are added for multiplex PCR in each of the loci evaluated. A nonpalindromic GAPF-positive locus that is differentially CpG methylated (HAND2)3 is recalcitrant to a second round of GAPF. The DNA palindrome at the CTSK locus is enriched after one round of GAPF but does not survive the second round. This pattern is also recapitulated for a naturally occurring DNA palindrome on chromosome 6 (IR6-107.3)5. The addition of 50% formamide during denaturation optimizes GAPF for DNA palindromes3, as shown by the enrichment for the CTSK and IR6-107.3 DNA palindromes, while abolishing the signal from the methylated HAND2 locus.
In conclusion, the GAPF-positive signals in our original study represented both palindromes and CpG island methylation. Our published1,2,3 and ongoing studies directly confirm that GAPF identifies palindromes associated with gene amplicons in cancers; however, the initial publication over-estimated the frequency of palindrome formation and the frequency of palindromes occurring in similar locations among different cancers.
References
- 1.Tanaka H, Bergstrom DA, Yao M-C, Tapscott SJ. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nature Genetics. 2005;37:320–7. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka H, Cao Y, Bergstrom DA, Kooperberg C, Tapscott SJ, Yao M-C. Intrastrand annealing leads to the formation of a large DNA palindrome and determines the boundaries of genomic amplification in human cancer. Molec Cell Biol. 2007;27:1993–2002. doi: 10.1128/MCB.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diede SJ, Guenthoer J, Geng LN, Mahoney SE, Marotta M, Olson JM, Tanaka H, Tapscott SJ. Proc Natl Acad Sci USA. doi: 10.1073/pnas.0907606106. epub 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ. Nat Genet. 2010;42:361. doi: 10.1038/ng0410-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–9. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]