Abstract
The human cytomegalovirus UL97 kinase, an important target of antiviral therapy, has an impact on at least two distinct phases of viral replication. Compared with wild-type virus, the UL97 deletion mutant exhibits an early replication defect that reduces DNA accumulation by 4- to 6-fold, as well as a late capsid maturation defect responsible for most of the observed 100- to 1000-fold reduction in replication. Block-release experiments with the antiviral 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)-benzimidazole revealed an important role for UL97 kinase in capsid assembly. Although cleavage of concatemeric DNA intermediates to unit-length genomes remained unaffected, progeny mutant virus maturation was delayed, with accumulation of progeny at significantly reduced levels compared with wild type after release of this block. Transmission electron microscopy confirmed the aberrant accumulation of empty A-like capsids containing neither viral DNA nor an internal scaffold structure, consistent with a failure to stably package DNA in mutant virus-infected cells. The function of UL97 in DNA synthesis as well as capsid assembly suggests that protein phosphorylation mediated by this herpesvirus-conserved kinase increases the efficiency of these two distinct phases of virus replication.
Human cytomegalovirus (CMV), a betaherpesvirus, is a major cause of disease in immunocompromised individuals, including AIDS patients and recipients of blood or bone marrow allografts or of solid organ transplants (1). CMV is also a common cause of congenital infection leading to neurological damage and hearing loss (1).
CMV DNA synthesis proceeds in a manner similar to that of other herpesviruses (2), with a set of six viral DNA synthesis proteins, including the DNA polymerase (UL54), directing the production of concatemeric DNA intermediates that are subsequently cleaved to genome-length units and packaged into preformed capsids (2). The precise molecular events of CMV capsid assembly and DNA packaging are not yet understood, although these processes are thought to occur in a stepwise fashion analogous to these processes in double-stranded DNA bacteriophages (3–5). Three capsid types, termed A, B, and C, are found in CMV-infected cells (2, 4, 5). Type B capsids represent maturation intermediates consisting of a capsid shell with an internal scaffold core. DNA packaging into these precursors displaces the scaffold structure to yield C capsids that mature by budding through nuclear membranes. Type A capsids, lacking both DNA and a scaffold core, are aberrant maturation products resulting from failure to stably package viral DNA (4, 5).
All antiviral drugs currently used for the treatment of systemic CMV infection, including ganciclovir, foscarnet, and cidofovir, ultimately target the CMV DNA polymerase (6). Of these, ganciclovir is the most widely used for treatment and prophylaxis of CMV disease (6, 7). Ganciclovir must be phosphorylated to a triphosphate form to inhibit the viral DNA polymerase (7). Initial phosphorylation of ganciclovir is carried out by the viral UL97 kinase (8, 9) such that mutations in the UL97 kinase domain lead to ganciclovir resistance (7, 9–11). Recently, an interesting class of potent, orally bioavailable nonnucleosidic antiviral compounds has been developed, which includes benzimidazole- and sulfonamide-based derivatives (12).§ Based on genetic mapping of resistance mutations, these compounds inhibit viral replication independently of viral DNA polymerase or DNA synthesis. 2-Bromo-5,6-dichloro-1-(β-d-ribofuranosyl)-benzimidazole (BDCRB) and the related compound 2,5,6-trichloro-1-(β-d-ribofuranosyl)benzimidazole block the viral DNA encapsidation process (13, 14). Both benzimidizole derivatives are chemically related to 5,6-dichloro-2-(isopropylamino)-1-l-ribofuranosyl-1H-benzimidazole (also called 1263W94), an antiviral compound that inhibits replication by directly affecting a UL97-dependent step in replication.¶
The UL97 gene product is a member of a family of serine/threonine protein kinases conserved in all mammalian herpesviruses (15) especially with regard to domains conserved in protein kinases. Alphaherpesvirus homologs of UL97, such as UL13 of herpes simplex virus (HSV) and ORF47 of varicella zoster virus (VZV), have been shown to phosphorylate viral immediate early proteins and cellular proteins (16–22) and have been implicated in both regulation of viral gene expression (16–22) and tissue tropism (23). From this work, the CMV UL97 kinase is predicted to phosphorylate protein substrates, and, although autophosphorylation by recombinant UL97 protein has been reported (24, 25), this kinase retains the distinction of being the member of this family that is best known for its ability to phosphorylate nucleoside analogs (8, 9, 26). The alphaherpesvirus UL97 homologs are nonessential for replication in cell culture (17, 27), yet the HSV UL13 shows a cell type-dependent impact on the levels of viral replication and late protein expression (16). The stage of replication that UL97 kinase controls has not been established. A UL97 deletion mutant (28) exhibits a severe growth defect, with a 100- to 1000-fold reduction in virus yield. This growth phenotype, along with the documented importance of the UL97 gene product as a unique target for novel antiviral therapy, prompted us to assess the nature of the block to replication imposed by the deletion. These studies reveal a biphasic phenotype, including an inhibition during the early phase of viral DNA synthesis followed by a block in the late phase of infection affecting capsid maturation, that establishes a role for this kinase at distinct steps in viral replication.
Materials and Methods
Cells, Viruses, and Drug Inhibition-Release Experiments.
Human foreskin fibroblasts (HFs) were cultured and used to propagate CMV strain AD169 (American Type Culture Collection, ATCC) and UL97 deletion mutants RCΔ97.08 and RCΔ97.19 (28) as previously described (29). Where appropriate, phosphonoformate (PFA) (300 μg/ml; Sigma) or BDCRB (20 μM, from L. B. Townsend, University of Michigan) was added after a 1-h period of virus adsorption to cells that were incubated for 96 h postinfection (hpi). At that time cultures were washed five times with PBS and then incubated in drug-free medium. Duplicate samples were used for all drug block-release studies. Growth curves were carried out in freshly plated HFs infected at a specified multiplicity of infection (moi). Samples of infected-cell supernatant were removed at designated time points and stored at −80°C before titration by plaque assay on HFs.
Analysis of Viral DNA.
Total cellular DNA was isolated at specified times from virus-infected (moi of 0.1–1) and mock-infected cell monolayers. For total cellular DNA, cells were collected in 1 ml of 1 mM EDTA, 10 mM Tris⋅HCl (pH 7.4), 0.5% SDS, and 0.5 mg/ml proteinase K (lysis buffer), incubated at 65°C for 2 h, and subjected to extraction with phenol and chloroform before ethanol precipitation. For isolation of encapsidated (DNase-resistant) DNA, cells were scraped into 1 ml of 10 mM Tris⋅HCl (pH 7.4), 10 mM KCl, 1.5 mM MgCl2 and sonicated before the addition of DNase I (Promega) at a concentration of 50 μg/ml. After incubation at 37°C for 2 h, SDS (to 0.5%), EDTA (to 1 mM), and Proteinase K (to 0.5 mg/ml) were all added before incubation at 65°C, followed by DNA extraction as described above. To follow the accumulation of viral DNA, total cellular DNA was digested with BamHI, separated by electrophoresis on a 0.8% agarose gel, transferred to nitrocellulose, and hybridized with radiolabeled BamHI fragment probe (AD169 nucleotides 122,699–124,902) prepared by random primed incorporation of [α-32P]dCTP (Amersham Pharmacia) as described (29). After hybridization, a PhosphorImager with imagequant software was used to calculate CMV DNA levels. For measurements of small DNA copy numbers, quantitative competitive PCR (QC-PCR) was performed as previously described (30). To detect the presence of viral genomic termini in DNA samples, DNA was digested with EcoRI, separated as described above, and hybridized with EcoRI terminal fragment W probe, pON227 (31).
Analysis of Viral Proteins.
Rabbit polyclonal antiserum (RαIE1-IE2) raised against glutathione S-transferase fusion proteins containing UL122 and UL123 was used to detect IE1 p72, IE2 p86, and the late IE2 p40 proteins, and mouse monoclonal antibodies 1202 and 1205B (Goodwin Institute, Plantation, FL) were used to detect ppUL44 and pp65, respectively. Horseradish peroxidase-conjugated rabbit anti-mouse IgG (Dako) or goat anti rabbit IgG (Vector Laboratories) was used as a secondary antibody. Actin was detected with the use of goat polyclonal anti-actin and rabbit anti-goat IgG (Santa Cruz Biotechnology). For protein analyses, total proteins from infected (moi of 1) or mock-infected cells were harvested at the indicated times and stored in sample buffer at −20°C until analyzed by immunoblot (24) that was visualized by Enhanced Chemiluminescence (Amersham Pharmacia).
Generation of UL97-Expressing Cell Line.
U373-MG glioblastoma astrocytoma cells (ATCC) were transfected with plasmid pLXSN-UL97, containing the entire UL97 gene (CMV nucleotides 140,460–142,716) from plasmid pMAL97 (24) inserted into EcoRI and XhoI sites in pLXSN (32) and the neo resistance gene. Cells resistant to 400 μg of G418 per ml were isolated, and individual colonies were amplified and screened for their ability to confer ganciclovir sensitivity on TK-mutant HSV-1 (33). Two colonies, denoted UL97–9 and UL97–21, were selected for use.
Transmission Electron Microscopy.
Cells grown on coverslips in 24-well plates were infected at an moi of 0.1. When appropriate, cells were fixed in 2% glutaraldehyde in phosphate buffer, postfixed with 1% aqueous uranyl acetate, dehydrated through graded ethanol steps, and embedded in Poly/Bed (Polysciences). Thin sections (70 nm) were then prepared and stained with 1% uranyl acetate and 1% lead citrate for electron microscopy on a Phillips CM12 microscope.
Results
UL97 Role in DNA Synthesis.
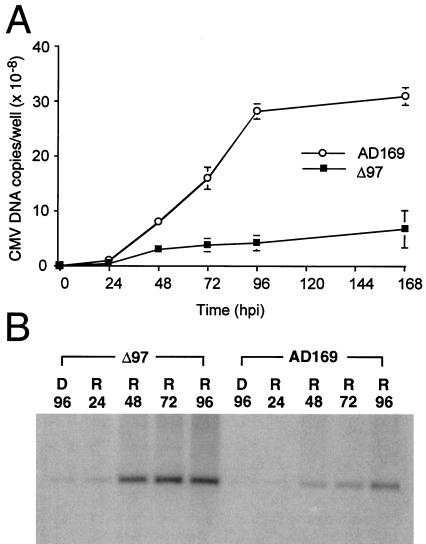
Total infected cell DNA was collected from cells infected with UL97 mutants RCΔ97.08 and RCΔ97.19 (28) or parental CMV strain AD169 to assess the accumulation of viral DNA. Low quantities of DNA before 48 hpi necessitated the use of QC-PCR up to that time point. Blot hybridization was used for later time points. Mutant viruses displayed 4- to 6-fold less viral DNA accumulation at late times compared with wild type (Fig. 1A shows data for RCΔ97.08). To determine whether a preelongation step was involved in this difference, the reversible antiviral compound PFA was used to block the elongation step of wild-type and mutant virus DNA synthesis. The input viral DNA present during 96 h of PFA block remained barely detectable by blot hybridization (Fig. 1B, D96 lanes). Between 24 and 48 h after drug release, DNA accumulation resumed and increased through 96 h (Fig. 1B, R24 through R96 lanes). Analysis of the hybridization signal showed that mutant virus DNA accumulation was similar to wild type at 96 h, although it appeared to be elevated above wild type at earlier times after drug release. These levels were also influenced by a slightly higher level of input mutant virus DNA at 24 hpi as measured by QC-PCR (data not shown). Similar results were obtained with mutant virus RCΔ97.19 (data not shown). This finding was consistent with a role for UL97-mediated protein phosphorylation events in the preelongation phase of viral DNA synthesis.
Figure 1.
Accumulation of viral DNA in cells infected with UL97 mutant RCΔ97.08 (Δ97) and parental strain AD169. (A) DNA accumulation curves of DNA copies per well from QC-PCR (0, 24, 48 hpi) and blot hybridization (72, 96, 168 hpi) after infection of HFs with an moi of 1. ○, AD169; ■, RCΔ97.08. (B) DNA blot hybridization to detect levels of viral DNA after a 96-h PFA-induced DNA synthesis block (D) and release (R) for a subsequent 24, 48, 72, or 96 h. Total cellular DNA was digested with BamHI and blot hybridized to detect a 2.3-kb viral BamHI fragment.
Viral Protein Accumulation.
Alphaherpesvirus UL97 homologs such as HSV-1 UL13 mediate phosphorylation of regulatory proteins that affect viral protein synthesis (16–21). To determine whether putative protein modifications or the reduced level of DNA synthesis in UL97 mutants were accompanied by reduced or delayed protein expression, we examined the levels of representative early and late viral proteins after infection with mutant RCΔ97.19 or wild-type virus (Fig. 2). No differences in the accumulation of α proteins IE1491aa and IE2579aa were detected at 24, 48, 72, and 96 hpi in cells infected with either virus. Importantly, IE2 p40 protein (γIE2331aa), a γ2 (true late) protein, was readily detected in mutant infected cells at 96 hpi. The β2 (early-late) protein ppUL44 (DNA polymerase processivity factor) and the γ1 (early-late) protein pp65 both accumulated in mutant infected cells to levels slightly less than those in wild type at late times (96 hpi); however, ppUL44 appeared to be more readily detected in mutant virus infected cells at earlier time points. Similar results were obtained with mutant RCΔ97.08 (data not shown). Further analysis of 35S- or [33P]orthophosphate-labeled total infected cell proteins at 24, 48,72, and 96 hpi did not reveal significant differences between mutant and wild type (data not shown). These observations demonstrate that synthesis of viral proteins, including replication proteins, is not appreciably altered by the disruption of UL97 under conditions where replication levels are significantly reduced.
Figure 2.
Immunoblot analysis of proteins expressed during RCΔ97.19 (Δ97) or AD169 infection of HFs (moi of 1). The four panels (from Top to Bottom) show detection of IE1 and IE2 antigens with the use of RαIE1-IE2, ppUL44 antigen with the use of monoclonal antibody 1202, pp65 (ppUL83), antigen with the use of monoclonal antibody 1205B, and cellular actin loading control with the use of goat polyclonal antiserum. Size markers (in kDa) are indicated on the right. M, mock-infected.
UL97 Role in the Late Phase of Viral Replication.
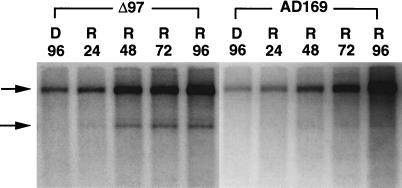
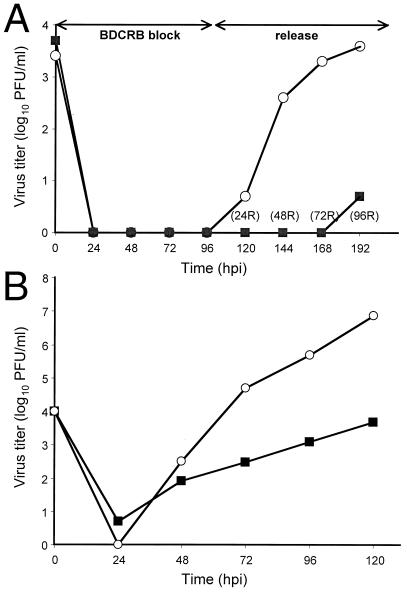
The differences in DNA synthesis or protein expression did not account for the poor replication of the UL97 mutant. Consequently, we investigated the behavior of mutant virus during the late phase of viral replication by employing the reversible encapsidation inhibitor BDCRB. BDCRB is known to inhibit cleavage of concatemeric viral DNA molecules as well as packaging of progeny DNA into nucleocapsids (13, 14). Accumulation and cleavage of viral DNA during BDCRB block release were evaluated by DNA blot hybridization by following the appearance of L-S junction and terminal fragments after digestion with EcoRI. Hybridization with EcoRI W probe revealed both terminal and junction fragments in cleaved monomeric progeny DNA but only junction fragments in concatemeric DNA intermediates. During BDCRB block (96 hpi), only noncleaved DNA intermediates accumulated, as reflected by the presence of junction but not terminal fragments in EcoRI digests (Fig. 3, D96 lanes). After drug release, terminal fragments were detectable in mutant infected cells within 24 h, indicating that cleavage proceeded soon after BDCRB was removed, and the amount of viral DNA continued to increase, indicating ongoing DNA synthesis. We observed a significant increase in the appearance of junction fragment in both viruses up to 72 h after reversal, with a continued increase in wild type to 96 h after reversal. The ratio of terminal to junction fragment bands was difficult to compare, although this ratio appeared to be greater in the mutant DNA samples. The band representing the terminal fragment was sharper in mutant virus DNA than in wild type because of less a sequence heterogeneity. Parental wild-type AD169 virus strain is highly heterogeneous, which makes direct comparison difficult (34) and contributes to the apparent difference in the ratio of junction and terminal fragments. Whereas the DNA accumulation/cleavage of wild type and mutant were synchronized by the BCDRB-induced block, the yield of mutant virus remained reduced by three orders of magnitude after BDCRB release (Fig. 4A). Thus, the difference between mutant and wild-type virus yield after BDCRB synchronization was similar to the difference observed when drug was not included (Fig. 4B and ref. 28). The discrepancy between DNA accumulation/cleavage synchronization and virus yield suggested an important second role for UL97 in maturation.
Figure 3.
DNA blot hybridization showing accumulation and cleavage of viral DNA in cells infected with UL97 mutant RCΔ97.08 (Δ97) and parental strain AD169 (moi of 0.1) after BDCRB drug block (D) and release (R) at the indicated times. Total cellular DNA was digested with EcoRI and subjected to blot analysis to detect L-S junction (upper arrow) and L terminal (lower arrow) fragments, with the use of radiolabeled probe pON227.
Figure 4.
Growth curves for RCΔ97.08 and AD169. (A) Growth curves during BDCRB block-release. HFs were infected (moi of 0.1), and supernatant progeny virus were harvested at the indicated times after infection or after drug release (R) and titered on HF monolayers. (B) Growth curves for RCΔ97.08 and AD169. HFs were infected (moi of 1), and supernatant progeny virus was harvested at the indicated times after infection and titered on HF monolayers. ■, RCΔ97.08; ○, AD169.
UL97 Role in Viral DNA Encapsidation.
To determine whether the DNA accumulating in mutant RCΔ97.08-infected cells was being packaged, total infected cell DNA was harvested either directly from cells (total DNA) or after a DNase I digestion step (DNase-resistant) to degrade nonencapsidated viral DNA. When mutant virus-infected cells were subjected to DNase treatment at 72 or 96 hpi, junction and terminal fragments were not detected (Fig. 5A; compare Lane 10 to Lane 7, and Lane 11 to Lane 8), indicating that viral DNA was not encapsidated at these times during mutant virus infection. In contrast, the junction fragment was reduced, but the terminal fragment was less affected when wild-type virus-infected cells were evaluated, consistent with expected packaging of mature virion DNA (Fig. 5A; compare Lane 5 to Lane 2, and Lane 6 to Lane 3). Aliquots of the same DNA samples were subjected to QC-PCR to compare the total and DNase-resistant (encapsidated) viral DNA (Fig. 5B). Given the detection sensitivity of 1–10 DNA copies per well, quantitation was possible on samples containing DNA at levels below the detection limits of blot hybridization. Based on QC-PCR, the ratios of total DNA to encapsidated DNA in mutant RCΔ97.08- or RCΔ97.19-infected cells was 200:1–300:1 at 72 or 96 hpi, whereas the ratio in wild-type AD169 virus-infected cells was 5:1–10:1. These results indicate a severe encapsidation defect in the mutant virus.
Figure 5.
Analysis of total and encapsidated viral DNA in AD169-, RCΔ97.08-, and RCΔ97.19-infected cells. (A) DNA blot hybridization showing accumulation of total and DNase-resistant viral DNA in cells infected with mutant RCΔ97.08 (Δ97) and parental strain (moi of 1). Total infected cell DNA was isolated before or after DNase digestion (+DNase) at the indicated times (hpi). Viral DNA digestion and hybridization were carried out to identify junction and terminal EcoRI fragments (arrows) as indicated for Fig. 3. M, mock-infected. (B) Quantitative analysis of total and DNase-resistant (+DNase) viral DNA in AD169-, RCΔ97.08-, and RCΔ97.19- (Δ97) infected cells at the times indicated. Quantitation was carried out with the use of blot hybridization for all samples, except for the DNase-resistant mutant DNA, and QC-PCR was used for all samples, yielding values similar to blots at higher DNA levels. The mean DNA copy number values, derived from three independent experiments, are displayed. Empty bars, total AD169 DNA; stippled bars, DNase-resistant AD169 DNA; black bars, total RCΔ97 strains DNA; hatched bars, DNase-resistant RCΔ97 strains DNA.
UL97 Role in Capsid Maturation.
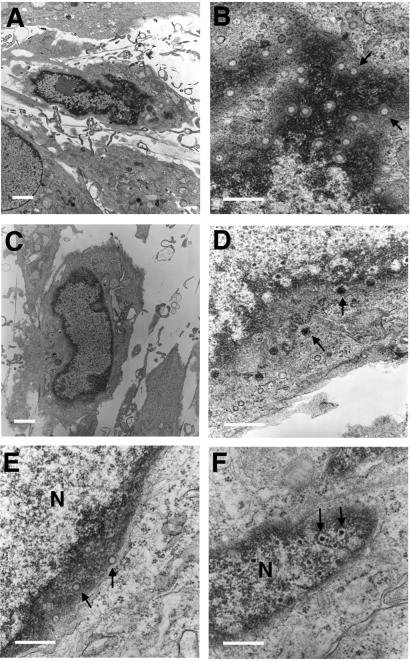
The presence and distribution of capsid assembly intermediates were evaluated by transmission electron microscopy in thin sections of HFs infected with mutant RCΔ97.08 or wild-type virus (Fig. 6). Empty capsids accumulated in the nuclei of mutant virus-infected cells at late times after infection that were morphologically similar to A capsids (Fig. 6 A and B). These capsids contained neither viral DNA nor internal core structures (Fig. 6B). The mean ratio of mutant RCΔ97.08 A:C capsids was 9:1, based on counts of representative fields from several different sections. Similar results were obtained with mutant RCΔ97.19 (data not shown). In contrast, the A:C capsid ratio in wild-type virus infected cells was 1:3, because of a marked increase in the numbers of C capsids as well as a significantly reduced number of A capsids. In addition, wild-type virus-infected cells contained all three expected nuclear-localized capsid forms (A, B, and C capsids) and had DNA-containing virions in the cytoplasm (Fig. 6D). DNA-containing virions were not observed in the cytoplasm of mutant virus-infected cells. To determine whether the presence of UL97 would compensate for the aberrant accumulation of A capsids, stably transfected U373 cells constitutively expressing functional UL97 were established (UL97–9 and UL97–21) and evaluated after infection with mutant and wild-type viruses. G418-resistant U373 cells carrying an empty vector construct (U373-vec) were prepared as a control. Nuclear accumulation of A capsids was observed at late times (Fig. 6E, data shown for RCΔ97.08) after infection of U373-vec cells with either RCΔ97.08 or RCΔ97.19. These A-type capsid forms that accumulated in the nuclear periphery similar to those observed in HFs. When UL97–9 or UL97–21 cells were infected with mutant virus, DNA was clearly incorporated into nuclear-localized capsid forms (Fig. 6F) at levels similar to those seen with AD169 infection of U373-vec, UL97–9, or UL97–21 cells (data not shown); however, the maturation of CMV was not as efficient in this cell type. Mature virions were only rarely observed in the cytoplasm of U373 cells, even when wild-type virus was used. These results showed transcomplementation of the DNA packaging defect exhibited by the UL97 mutant virus in UL97-expressing cells, confirming the role of UL97 in the efficient formation of DNA-containing capsids.
Figure 6.
Transmission electron micrographs of virus-infected cells at 72 hpi. (A and B) HFs infected with mutant RCΔ97.08. (C and D) HFs infected with strain AD169. (E) U373-vec cells infected with mutant RCΔ97.08. (F) UL97–9 cells infected with mutant RCΔ97.08. Arrows in B and E point to empty A capsids. Arrows in D and F point to DNA-containing capsids. [Magnification: A and C, ×3,800; B, D–F, ×35,000. Size bars: A and C, 5 μm; B, D–F, 1 μm.]
Discussion
This study has defined a role for the UL97 kinase, a herpesvirus-conserved protein kinase, in two distinct phases of the viral replication cycle, DNA replication and capsid assembly. A preelongation step in DNA synthesis and a late step in genome encapsidation are both less efficient in the absence of this kinase. UL97 did not appear to influence viral protein expression levels in the manner reported for UL97 homologs HSV UL13 and varicella-zoster virus ORF 47, which are known to phosphorylate viral immediate early regulatory proteins and to exert an impact on viral gene expression (16–23). Even though the activities of this herpesvirus-conserved kinase are not as well established in CMV as in other herpesviruses (26, 35), the impact of UL97 on DNA synthesis and capsid assembly is best ascribed to its role as a protein kinase. This phenotype, which has been readily demonstrated in cultured HFs with the CMV UL97 mutant, is much more striking than that observed with kinase mutants of HSV-1 and varicella-zoster virus, where a critical requirement for the viral kinase, when recognized, has been tissue specific (16, 23). The partial replication defect of the CMV mutant and the lack of a phenotype of either HSV-1 or varicella-zoster virus kinase mutants in cell culture suggests that viral kinase function is complemented by host cell kinases as a result of cell cycle or differentiation state. The kinases related to UL97 carried by all mammalian herpesviruses may therefore be predicted to have roles similar to those described here.
DNA accumulation was consistently reduced in the UL97 deletion mutant compared with wild type, which, together with the ability of PFA block-release to reverse the synthesis defect, suggested that UL97-dependent protein phosphorylation events precede replication fork assembly. These events may be linked to origin recognition or protein–protein interactions that precede active replication. Such a role for this viral enzyme is supported by a preliminary report that UL97 controls the phosphorylation state of the DNA polymerase processivity factor ppUL44.‖ Furthermore, treatment with 1263W94, which directly blocks UL97 kinase activity, is associated with disruption of DNA replication compartments.** Interestingly, 1263W94 has also been shown to inhibit the synthesis of Epstein–Barr virus DNA and the phosphorylation of the Epstein–Barr virus DNA processivity factor (36), suggesting that a gammaherpesvirus UL97 homolog retains a function similar to that of UL97. Regardless of the replication functions that may be targeted by UL97-mediated phosphorylation, the overall effect on DNA synthesis is modest, suggesting that host cell kinases compensate for any critical UL97 role at this replication step.
A more substantial role for UL97 was observed during virion maturation and was revealed by a release of a BDCRB block to synchronize mutant and wild-type virus DNA encapsidation at capsid assembly. This process remains relatively poorly understood in CMV replication but was the most dramatically affected in UL97 mutant-infected cells and correlated best with the severely reduced mutant virus growth properties. The striking accumulation of A capsids, along with the absence of detectable packaged DNA, revealed a role for this kinase in genome encapsidation or capsid assembly. By analogy with the double-stranded DNA bacteriophages, herpesvirus capsid maturation may proceed in a stepwise fashion and requires the concerted activities of cleavage-packaging proteins that function to cleave concatemeric DNA into unit-length genomes as well as to insert DNA into capsids (3, 4). Cleavage-packaging machinery controls docking and insertion of DNA into capsids, cleavage of genome ends, and maintenance of encapsidated DNA within capsids through the final stages of virion morphogenesis (3, 4). In HSV-1, at least seven gene products have been shown to provide critical DNA packaging functions, and mutations in most of them have been shown to result in the concomitant accumulation of uncleaved concatemeric DNA and scaffold-containing B capsids (4, 37–44). In contrast, the UL97 deletion mutant demonstrated a unique phenotype associated with concomitant production of cleaved unpackaged DNA and accumulation of A capsids. Type A capsids are regarded as aberrant end products of the capsid assembly process, resulting from either incomplete insertion of DNA into capsids or the premature release of DNA from capsids (4, 5). The viral replication cycle demands that DNA be stably inserted into capsids before the completion of virion maturation, but that it be readily released from those same capsids during entry in a subsequent round of infection. It is quite possible that virion-associated kinases and phosphatases provide the modifications that throw this switch, either alone or in conjunction with cellular kinases and phosphatases soon after entry. Most virion maturation models (5) predict that DNA cleavage and packaging are coupled events, suggesting that UL97 phosphorylation events regulate the efficiency of capsid assembly after the insertion of DNA into capsids. The phenotype of the UL97 deletion mutant appears similar in both the pattern of A capsid predominance and the accumulation of cleaved, nonencapsidated viral DNA to those displayed by a mutant in the HSV gene UL25 (44), which encodes a minor herpesvirus-conserved capsid component that stabilizes the capsid during packaging.
The phenotype of the UL97 mutant is also similar to an HSV-1 alkaline nuclease mutant that exhibits a DNA packaging defect resulting from failure to resolve DNA intermediates that arise during replication (45, 46). Thus we cannot exclude an additional impact of UL97-mediated phosphorylation on processing of DNA intermediates to create packageable forms. At the current level of analysis, any viral protein involved in packaging may be the critical target of UL97 kinase. A role for this kinase in cleavage–packaging is further supported by the chemical similarity of 1263W94, which inhibits UL97 directly, to the benzimidazole analog BDCRB, which targets cleavage–packaging proteins (12–14, 36).
The behavior of UL97 as a ganciclovir kinase (8, 9) at first glance seems to distinguish this enzyme from homologs encoded by alphaherpesvirus; however, its role in viral replication is consistent with that of a protein kinase. Although the differences observed here may reflect viral UL97 function as well as the impact of host cell factors induced during CMV infection, it is notable that UL97 was previously shown to compensate for HSV-1 UL13 in restoring efficient growth while only partially restoring protein modification and without conferring ganciclovir sensitivity (26, 35). Regardless of the differences we and other investigators have observed in the behavior of these kinase homologs, two common functions retained by these enzymes may be regulation of DNA replication and encapsidation. These data are consistent with a common origin as well as some divergence of the function during coevolution of this gene within each respective biologically distinct herpesvirus. Further studies of this enzyme and its targets will provide insight into the regulation of CMV DNA packaging and capsid assembly and possibly reveal connections between distinct phases of viral replication.
Acknowledgments
We thank Nafisa Ghori for her excellent assistance with electron microscopy and A. Louise McCormick for comments on the manuscript. This work was supported by U.S. Public Health Service Grant RO1 AI20211 (to E.S.M.).
Abbreviations
- CMV
cytomegalovirus
- HSV
herpes simplex virus
- BDCRB
2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidazole
- PFA
phosphonoformate
- HF human foreskin fibroblast
hpi, hours postinfection
- moi
multiplicity of infection
- QC-PCR
quantitative competitive PCR
Footnotes
Hallenberger, S., Trappe, J., Buerger, I., Bender, W., Eckenberg, P., Goldman, S., Haerter, M., Reefschlaeger, J. & Weber, O. Interscience Conference on Antimicrobial Agents and Chemotherapy, September 26–29, 1999, San Francisco, abstr. 203.
Davis, M. G., Talarico, C. C., Underwood, M. R., Baldanti, F. & Biron, K. K., 23rd International Herpesvirus Workshop, August 1–7, 1998, York, United Kingdom.
Krosky, P. M., Baek, M. C., Barrera, I., Harvey, R. J., Biron, K. K., Coen, D. M., & Sethna, P. B., 24th International Herpesvirus Workshop, July 17–23, 1999, Boston, MA, abstr. 12.016.
Harvey, R. J., Chamberlain, S., Davis, M., Koszalka, G. W., Smith, A. & Biron, K. K., 22nd International Herpesvirus Workshop, August 2–8, 1997, San Diego, CA, abstr. 245.
References
- 1.Alford C A, Britt W J. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. New York: Lippincott-Raven; 1996. pp. 2493–2534. [Google Scholar]
- 2.Mocarski E S, Jr, Tan Courcelle C. In: Fields Virology. Knipe D M, Howley P M, editors. Philadelphia: Lippincott Williams & Wilkens; 2001. , in press. [Google Scholar]
- 3.Black L W. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 4.Steven A C, Spear P G. In: Structural Biology of Viruses. Chiu W, Burnett R M, Garcea R L, editors. Oxford: Oxford Univ. Press; 1997. pp. 312–351. [Google Scholar]
- 5.Gibson W. Intervirology. 1996;39:389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- 6.Drew W L, Lalezari J P. Curr Clin Top Infect Dis. 1999;19:16–29. [PubMed] [Google Scholar]
- 7.Crumpacker C S. N Engl J Med. 1996;335:721–729. doi: 10.1056/NEJM199609053351007. [DOI] [PubMed] [Google Scholar]
- 8.Littler E, Stuart A D, Chee M S. Nature (London) 1992;358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. Nature (London) 1992;358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- 10.Wolf D G, Smith I L, Lee D J, Freeman W R, Flores-Aguilar M, Spector S A. J Clin Invest. 1995;95:257–263. doi: 10.1172/JCI117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou S, Erice A, Jordan M C, Vercellotti G M, Michels K R, Talarico C L, Stanat S C, Biron K K. J Infect Dis. 1995;171:576–583. doi: 10.1093/infdis/171.3.576. [DOI] [PubMed] [Google Scholar]
- 12.Townsend L B, Devivar R V, Turk S R, Nassiri M R, Drach J C. J Med Chem. 1995;38:4098–4105. doi: 10.1021/jm00020a025. [DOI] [PubMed] [Google Scholar]
- 13.Krosky P M, Underwood M R, Turk S R, Feng K W, Jain R K, Ptak R G, Westerman A C, Biron K K, Townsend L B, Drach J C. J Virol. 1998;72:4721–4728. doi: 10.1128/jvi.72.6.4721-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Underwood M R, Harvey R J, Stanat S C, Hemphill M L, Miller T, Drach J C, Townsend L B, Biron K K. J Virol. 1998;72:717–725. doi: 10.1128/jvi.72.1.717-725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chee M S, Lawrence G L, Barrell B G. J Gen Virol. 1989;70:1151–1160. doi: 10.1099/0022-1317-70-5-1151. [DOI] [PubMed] [Google Scholar]
- 16.Purves F C, Ogle W O, Roizman B. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coulter L J, Moss H W, Lang J, McGeoch D J. J Gen Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- 18.Ng T I, Keenan L, Kinchington P R, Grose C. Virology. 1994;68:1350–1359. doi: 10.1128/jvi.68.3.1350-1359.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogle W O, Ng T I, Carter K L, Roizman B. Virology. 1997;235:406–413. doi: 10.1006/viro.1997.8710. [DOI] [PubMed] [Google Scholar]
- 20.Kawaguchi Y, Matsumura T, Roizman B, Hirai K. J Virol. 1999;73:4456–4460. doi: 10.1128/jvi.73.5.4456-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long M C, Leong V, Schaffer P A, Spencer C A, Rice S A. J Virol. 1999;73:5593–5604. doi: 10.1128/jvi.73.7.5593-5604.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Advani S J, Brandimarti R, Weichselbaum R R, Roizman B. J Virol. 2000;74:8–15. [PMC free article] [PubMed] [Google Scholar]
- 23.Moffat J F, Zerboni L, Sommer M H, Heineman T C, Cohen J I, Kaneshima H, Arvin A M. Proc Natl Acad Sci USA. 1998;95:11969–11974. doi: 10.1073/pnas.95.20.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Z, He Y S, Kim Y, Chu L, Ohmstede C, Biron K K, Coen D M. J Virol. 1997;71:405–411. doi: 10.1128/jvi.71.1.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel D, Kramer S, Hahn S, Schaarschmidt P, Wunderlich K, Mertens T. J Virol. 1999;73:8898–8901. doi: 10.1128/jvi.73.10.8898-8901.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talarico C L, Burnette T C, Miller W H, Smith S L, Davis M G, Stanat S C, Ng T I, He Z, Coen D M, Roizman B, et al. Antimicrob Agents Chemother. 1999;43:1941–1946. doi: 10.1128/aac.43.8.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heineman T C, Cohen J I. J Virol. 1995;69:7367–7370. doi: 10.1128/jvi.69.11.7367-7370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prichard M N, Gao N, Jairath S, Mulambia G, Krosky P, Coen D M, Parker B O, Pari G S. J Virol. 1999;73:5663–5670. doi: 10.1128/jvi.73.7.5663-5670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greaves R F, Brown J M, Vieira J, Mocarski E S. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 30.Slobedman B, Mocarski E S. J Virol. 1999;73:4806–4481. doi: 10.1128/jvi.73.6.4806-4812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McVoy M A, Adler S P. J Virol. 1994;68:1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller A D, Rosman G J. BioTechniques. 1989;7:980–982. [PMC free article] [PubMed] [Google Scholar]
- 33.Van Zeijl M, Fairhurst J, Baum E Z, Sun L, Jones T R. Virology. 1997;231:72–80. doi: 10.1006/viro.1997.8523. [DOI] [PubMed] [Google Scholar]
- 34.Tamashiro J C, Spector D H. J Virol. 1986;59:591–604. doi: 10.1128/jvi.59.3.591-604.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng T I, Talarico C, Burnette T C, Biron K, Roizman B. Virology. 1996;225:347–358. doi: 10.1006/viro.1996.0609. [DOI] [PubMed] [Google Scholar]
- 36.Zacny V L, Gershburg E, Davis M G, Biron K K, Pagano J S. J Virol. 1999;73:7271–7277. doi: 10.1128/jvi.73.9.7271-7277.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Addison C, Rixon F J, Preston V G. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 38.Al-Kobashi M F, Rixon F J, McDougall I, Preston V G. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 39.Patel A H, Rixon F J, Cunningham C, Davison A J. Virology. 1996;217:111–123. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- 40.Poon A P W, Roizman B. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salmon B, Cunningham C, Davison A J, Harris W J, Baines J D. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherman G, Bachenheimer S L. Virology. 1987;158:427–430. doi: 10.1016/0042-6822(87)90214-5. [DOI] [PubMed] [Google Scholar]
- 43.Yu D, Sheaffer A K, Tenney D J, Weller S K. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao L, Rapp L M, Weller S K. Virology. 1993;196:146–162. doi: 10.1006/viro.1993.1463. [DOI] [PubMed] [Google Scholar]
- 46.Martinez R, Sarisky R T, Weber P C, Weller S K. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]