Abstract
The identities of the regulators that mediate commitment of hematopoietic precursors to the T lymphocyte lineage have been unknown. The last stage of T lineage commitment in vivo involves mechanisms to suppress natural killer cell potential, to suppress myeloid and dendritic cell potential, and to silence the stem cell or progenitor cell regulatory functions that initially provide T cell receptor–independent self-renewal capability. The zinc finger transcription factor Bcl11b is T cell–specific in expression among hematopoietic cell types and is first expressed in precursors immediately before T lineage commitment. We found that Bcl11b is necessary for T lineage commitment in mice and is specifically required both to repress natural killer cell–associated genes and to down-regulate a battery of stem cell or progenitor cell genes at the pivotal stage of commitment.
T lymphocyte precursors begin the T lineage differentiation program in the thymus, before they fully lose alternative differentiation potentials inherited from their stem cell precursors (1). Through a sequence of distinct stages coupled with proliferation, termed DN1 to DN3, these cells shed B cell, myeloid cell, dendritic cell, and natural killer (NK) cell options while turning on successive batteries of T cell genes. A necessary, although not sufficient, driver of this process is triggering of Notch1 through interaction with Delta-like (DL) ligands in the thymic microenvironment, and many transcription factors must collaborate with Notch to establish T cell identity. A pivotal stage in the process is the DN2 stage. This is when the first fully committed cells emerge, and their commitment is marked by the concerted, permanent down-regulation of a group of stem cell– or progenitor cell–associated regulatory genes (2). Repression of these genes is crucial for completing normal T lineage specification; at least three of them—Tal1, Lyl1, and Sfpi1 (PU.1)—can each block T cell development or cause T cell leukemia if sustained beyond this point (3–6). Correct silencing of such genes can be directly implicated in the extinction of certain alternative lineage potentials [for a review, see (7–9)] and probably also underlies the loss of broader stem cell properties. A major challenge for the field has been to identify the rate-limiting regulator that triggers this complex of silencing events (10).
We have used the expression patterns of multiple transcription factors in T cell precursors as clues to identify candidates that might provide a rate-limiting function to trigger commitment. Bcl11b is highly unusual both in its T lineage specificity and for its steep onset of expression in the early DN2 stage, just preceding commitment (11, 12). Expression is then sustained throughout T cell development, placing Bcl11b expression at the right places and the right times to contribute to T lineage commitment. Previous reports have shown that loss of Bcl11b blocks development of T cell receptor (TCR) αβ T cells after commitment, at the first TCR-dependent selection event in the thymus (13, 14); later, conditional deletion of this gene causes impaired survival and profound abnormalities of CD4+ CD8+ TCRαβ T cells (15). Genetic evidence also suggests that Bcl11b acts as a tumor suppressor for thymocytes (16, 17). Although abnormalities in precommitment development without Bcl11b have not been described previously, early stages when Bcl11b is normally first expressed were not dissected in detail in those reports, and cohorts of differentiating cells were not followed kinetically. Therefore, to examine the earliest roles Bcl11b might play in T cell development, we deleted Bcl11b from floxed Bcl11bL2/L2 conditional knockout prethymic precursor cells (18) using retroviral transduction of Cre recombinase (fig. S1). To track their development through the T cell specification process, we then cultured these cells and Cre-transduced wild-type cells in vitro with OP9 stromal cells that express Delta-like 1 (DL1) and cytokines Flt3L (FMS-like tyrosine kinase 3 ligand) and interleukin-7 (IL-7) (19), conditions that induce T cell differentiation in vitro (19) (Fig. 1A).
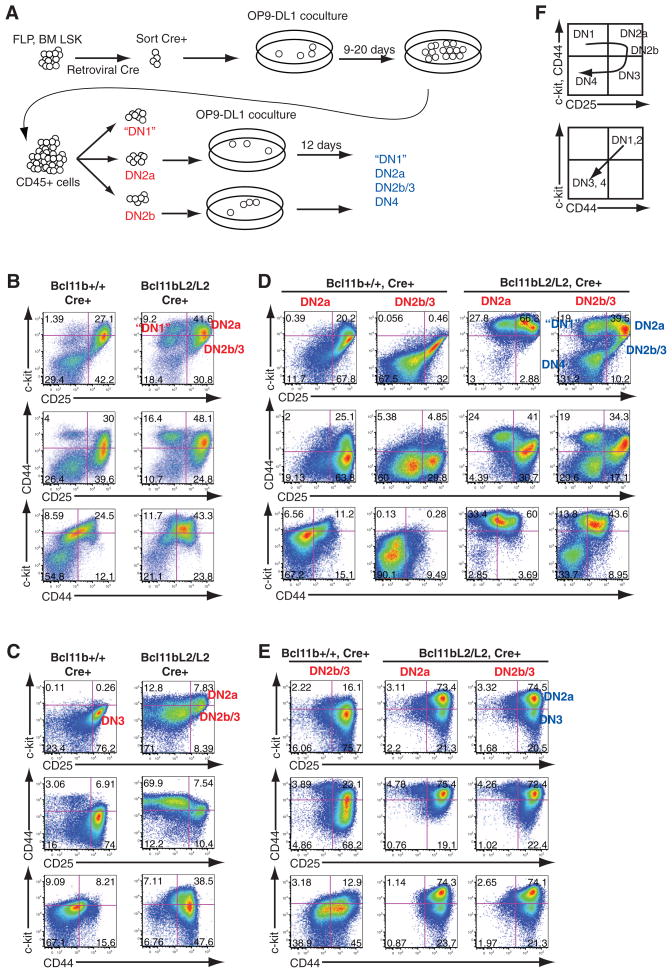
Fig. 1.
Generation and developmental arrest of Bcl11b-deficient T cell precursors. (A) Experimental scheme. Fetal liver precursors (Lin−c-kit+ CD27+; FLP) from embryonic day 13.5 embryos or bone marrow hematopoietic stem cells (BM LSK) were isolated from Bcl11bL2/L2 or wild-type mice, transduced with hNGFR-Cre retrovirus, and sorted 48 hours later and cocultured with OP9-DL1 cells. After 9 days of culture, DN subsets of cells were sorted (red populations). A fraction of each subset was returned to OP9-DL1 coculture for later analysis (blue populations). The remaining cells were analyzed for RNA expression. (B) Flow cytometric analysis of transduced fetal liver–derived precursors after 9 days of OP9-DL1 coculture. (C) As in (B), for bone marrow–derived precursors after 14 days of OP9-DL1 coculture. (D) Flow cytometric analysis of sorted DN2a and DN2b cells from fetal liver precursors after an additional 12 days of OP9-DL1 coculture. (E) As in (D), but for sorted DN2a and DN2b cells from bone marrow precursors. (F) Schematic of normal differentiation.
Definitive T lineage initiation is marked by the appearance of Kit+ CD44+ CD25+ DN2 cells; these normally give rise to Kitlow CD44low CD25+ DN3 cells after commitment. Both control and Bcl11b-deleted progenitor cells from fetal liver or adult bone marrow generated early Kithigh DN2 cells (DN2a) (Fig. 1, B and C). Bcl11b-deleted cells differentiated poorly to DN3 cells, however, arresting instead at DN2 and producing an abnormally large population of CD44+ Kit+ CD25− DN1-like cells. Both of these types of descendants from Bcl11b-deleted precursors expressed distinctively high amounts of the progenitor cell growth factor receptor Kit, especially when derived from fetal precursors (Fig. 1B). The most striking feature of Bcl11b-deficient cells, however, was that differentiation was blocked despite proliferation at least as strong as in controls (fig. S2, A to D). When Bcl11b-deficient arrested DN2a cells were purified by fluorescence-activated cell sorting and replated for further differentiation, they consistently generated more DN2 cells and more DN1-like cells for at least another 12 days but did not progress to a later stage (table S1); under the same conditions, control cells progressed through DN3 stage and beyond (Fig. 1, D and E, and figs. S2E and S3), to the CD4+ CD8+ stage (fig. S3D). Bcl11b-deficient cells grew as well as controls in reduced concentrations of IL-7, but, whereas controls progressed faster to DN3 in these conditions, the Bcl11b-deleted cells did not (fig. S2, E and F). Cells that did leak through this block (Fig. 1, B and C; “DN2b” in Bcl11bL2/L2) were highly enriched for retention of at least one Bcl11b allele (fig. S4). However, loss of Bcl11b provided a survival advantage for DN2a and DN1-like cells. In experiments where initial Bcl11b deletion was inefficient, cells with a more “advanced” DN2b or DN3 phenotype were generated (Fig. 1B and fig. S4A). After purification and replating, however, these populations yielded “retrograde” DN1 and DN2a descendants (Fig. 1D and table S1) that were highly enriched for complete loss of Bcl11b (fig. S4, B and C) The robustness and self-renewal capacity of both DN1-like and DN2 cells without Bcl11b contrast with the reported poor viability of later T lineage cells that are deprived of Bcl11b (15).
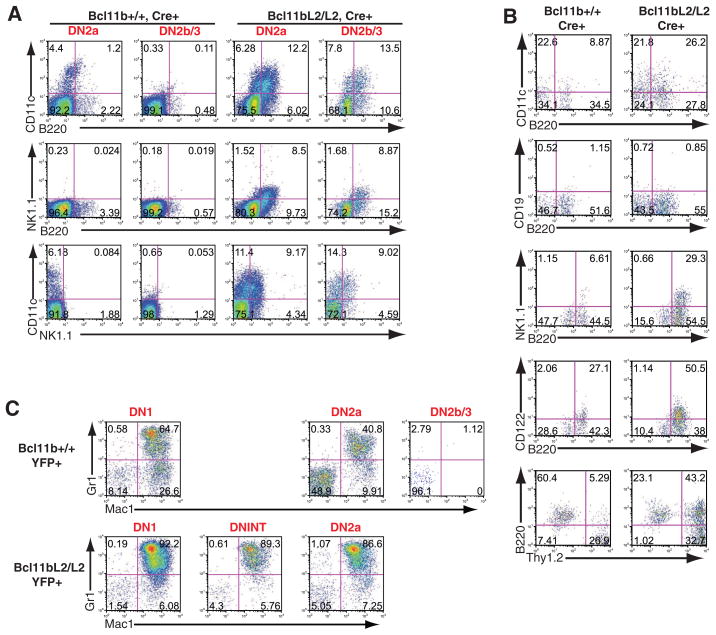
Whereas normal DN2a cells generate few non-T progeny and normal DN2b and DN3 cells are fully committed (2), both DN2a- and DN2b-like cells from Bcl11b-deficient precursors generated cells that expressed non-T markers (Fig. 2A). The DN1-like cells generated in cultures of Bcl11b-deficient precursors were not early T cell precursors. Most of them expressed B220 together with CD11c, as well as moderate to high amounts of Kit and substantial Thy1 (Fig. 2A). A substantial fraction of DN1-like cells also expressed the NK cell marker NK1.1 (fig. S5). Cells with these non-T features were generated efficiently from purified Bcl11b-deficient DN2 precursors when Notch-DL1 signaling was reduced by transfer to OP9 cells without DL1 (Fig. 2B) or by adding γ-secretase inhibitor (fig. S5), and with the support of Flt3 ligand and IL-7 they expanded in culture on OP9 cells without DL1 (fig. S6).
Fig. 2.
Non–T lineage differentiation of Bcl11b-deleted DN2 and DN1-like cells. (A) Flow cytometric analysis of B220, CD11c, and NK1.1 coexpression on progeny of DN2a and DN2b precursors in OP9-DL1 secondary culture. (B) Flow cytometric analysis of cells derived from DN2 precursors transferred to OP9 control secondary culture. (C) Myeloid developmental competence of T cell precursors from control and Bcl11b-deficient precursors. DN1, DN2a, and DN2b cells were generated from bone marrow precursors in primary culture with OP9-DL1 (see fig. S3), sorted after ~20 days, and transferred to OP9 control secondary cultures supplemented with macrophage colony-stimulating factor only. Flow cytometric analyses of Mac1 and Gr-1 expression are shown. Results in each panel are from different independent, single experiments, each representative of three or four experiments.
Although expression of B220, CD11c, and NK1.1 is usually associated with disparate lineages (20–25), gene expression analysis implied many DN1-like cells to be Kit+ precursors of interferon-producing killer dendritic cells (IKDCs), proposed to be a subtype of NK cell (20, 21) (Fig. 3). Although they were B220+, they expressed no other B cell genes tested (fig. S7; Cd19, Ebf1). Their modest expression levels of Mac1 (Itgam), PU.1 (Sfpi1), SpiB, and RORγt (Rorct) made them unlikely to be myeloid cells, conventional or plasmacytoid dendritic cells, or lymphoid tissue inducer cells (Fig. 3 and figs. S7 to S9). They were not precursors because they had lost expression of canonical stem cell or progenitor cell genes (see below). Instead, these “DN1-like” cells expressed high amounts of the NK genes Id2, Il2rb, Tbet, and Eomes, along with Il7r and Gata3 expression typical of thymus-derived NK cells (26). The transcription factor genes Plzf (Zbtb16) and Nfil3 (E4bp4), also implicated in NK cell development (27, 28), were also strongly expressed (Fig. 3 and figs. S7 to S9). Thus, after the DN2 stage, expression of Bcl11b may limit T cell access to this NK-like program.
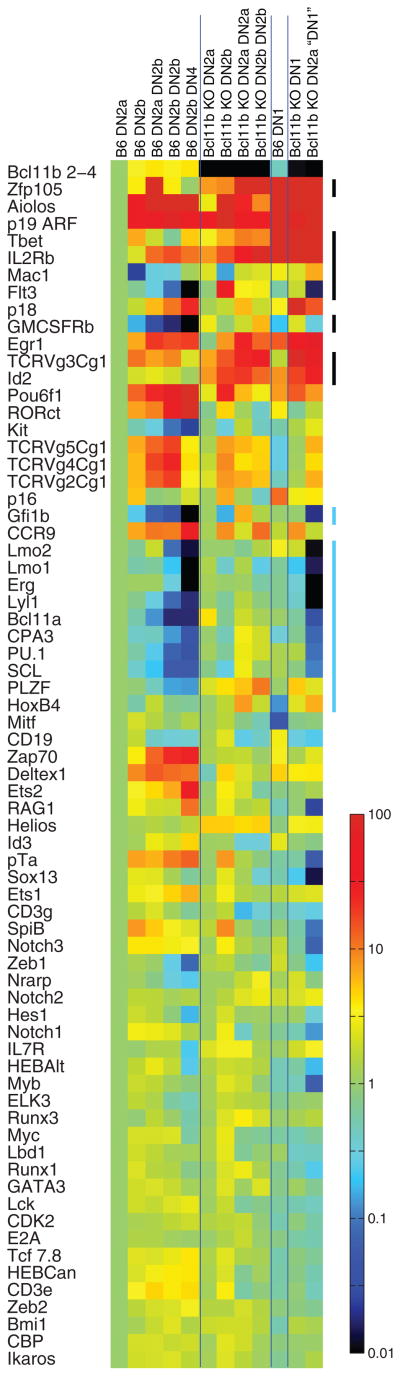
Fig. 3.
Gene expression comparison between normal and Bcl11b-deleted cells in early T cell development in vitro. Subsets of control and Bcl11b-deleted cells were sorted both after primary 9-day culture and after secondary culture as indicated. For sorting gates and quantitation of expression levels in fixed units, see fig. S7. “DN4” cells generated by controls in secondary culture may include later stages. Real-time quantitative polymerase chain reaction data are shown as a heat map normalized to levels of each gene in control DN2a cells. Color scale units are log10 for a 104 overall range. Vertical bars on right highlight gene groups of interest. Results are from one of four representative experiments (independent experiments are shown in figs. S8 and S9).
Although access to the NK-like option was most favored under standard OP9 culture conditions, it was not the only one enhanced by loss of Bcl11b. Normally, competence to make myeloid progeny is also reduced between the early T cell precursor DN1 stage and DN2a, then extinguished at DN2b (2). Deletion of Bcl11b enabled DN2 cells to remain fully competent to generate Mac1+ Gr-1+ myeloid cells if transferred to myeloid supportive conditions (Fig. 2C). Bcl11b-deficient cells therefore retain access to at least two different non–T lineage alternatives.
For normal thymocytes, DN2a is the last stage not only to preserve alternative developmental potentials but also to maintain an extensive complement of stem cell and progenitor cell regulatory gene expression (2). Several highly T cell–specific genes are normally up-regulated at DN2a, including Bcl11b, but full T lineage–specific gene expression awaits the DN3 stage (1, 2). The profound shift in regulatory state after the DN2a stage has not been explained by changes in expression of previously studied T lineage transcription factors (10). We therefore asked whether Bcl11b might also have a specific role either in triggering T lineage gene expression or in repression of stem cell and progenitor cell genes after the DN2a stage, by comparing gene expression in Bcl11b-deficient DN2 cells with that in DN2- and DN3-stage controls (three independent experiments shown in Fig. 3 and figs. S7 to S9).
Bcl11b-deficient precursors began T cell specification in a manner similar to controls, as indicated by the up-regulation of Cd3e, Cd3g, Ptcra, and Rag1 between the DN1 and DN2a stages in both Bcl11b-deficient and control cells (1, 2) (Fig. 3). Expression of these genes was sustained in replated progeny of Bcl11b-deficient precursors that retain a DN2a-like phenotype, resembling wild-type DN2a cells (fig. S9). These cells also fully expressed regulatory genes known to be essential for T cell specification (e.g., Gata3, Tcf7, Runx1, Ikzf1, Myb, Ets1, Tcfe2a, Tcf12, the lymphoid growth factor receptor Il7r, and Notch targets Hes1, Notch1 itself, and Myc, which supports DN3 cell viability) (1, 29). Unlike control cells, Bcl11b-deficient DN2 cells in primary culture simultaneously expressed elevated amounts of NK-promoting genes: Id2, Il2rb, Nfil3 (E4bp4), and Plzf (Zbtb16) (27, 28). These genes were further up-regulated in progeny that became NK-like, but continued even in cells maintaining a long-term DN2 phenotype (Fig. 3 and figs. S7 to S9). Thus, loss of Bcl11b may permit early T cell specification to occur in parallel with regulatory “priming” for an innate-like differentiation alternative.
In the context of gene networks that control T cell differentiation, the Bcl11b-deficient DN2a cells also failed in another aspect of repression (10). A “legacy” of progenitor cell regulatory genes (11, 12), including several proto-oncogenes (Tal1, Sfpi1, Lyl1, Gfi1b, Erg, and Bcl11a), may normally help to sustain proliferation to the DN2a stage. During commitment, the cells normally shift their survival requirements, becoming Notch-dependent, and the progenitor cell genes are repressed in this process (2, 30). Although in vitro T cell differentiation has some abnormal features, all the subsets of cells generated in these cultures from control fetal liver precursors correctly shut off stem cell and progenitor cell genes during secondary culture. When replated, Bcl11b-deficient DN2a cells consistently did not do so (Fig. 3 and figs. S7 and S8). Initially, Bcl11b-deficient cells expressed similar amounts of stem cell and progenitor cell transcription factors as control cells, although often with slightly higher amounts of the progenitor cell cytokine receptor Flt3. With sustained culture, Bcl11b-deficient cells continued to resemble the decreasing proportion of control cells that retained a DN1-like or DN2a-like phenotype (fig. S9). However, as the majority of control cells generally progressed to more mature T cell development stages, Bcl11b-deficient cells as a whole diverged from controls. They not only failed to silence but sometimes further up-regulated progenitor cell genes. Alternative lineage markers such as the mast cell gene Cpa3 and the myeloid growth factor receptor gene Csfr2b were also abnormally sustained. The inability of the cells to differentiate further in the T lineage may thus be explained not by a failure to initiate T cell program activation, but by a failure to mobilize repression, under conditions of Notch and cytokine signaling, if Bcl11b is absent (fig. S10).
Germline Bcl11b knockouts can generate some TCRγδ cells despite a block in TCRαβ development (13). Bcl11b-deficient cells could also generate surface TCRγδ+ progeny in vitro (fig. S11). Do these cells not require the same commitment mechanism? Normally, Rag1 expression is not fully up-regulated until the DN3 stage (1, 2), and this would limit TCRγδ gene rearrangement until after commitment. Even so, DN2a cells can normally generate larger yields of TCRγδ descendants per precursor than DN3 cells (2, 31), which suggests that some TCRγδ emergence may indeed precede mainstream commitment events. Consistent with the strong expression of Rag1 in Bcl11b-deficient DN2a cells, mutant cells within both DN1 and DN2a samples expressed Vγ-Jγ joined transcripts (Fig. 3 and figs. S7 to S9). Frequently, Bcl11b-deficient cells expressed the fetal-specific Vγ3-Jγ1Cγ1 transcript more than did controls. Fetal-type TCRγδ cells, like NK cells, are programmed to develop in a unique context of high Id2 expression, which is less permissive for other TCR rearrangements (32). Other “innate” lineages of TCRγδ cells require high levels of Plzf, which here are promoted by Bcl11b loss (33). This may indicate that particular TCRγδ sublineages sharing NK-like features may escape an absolute requirement for Bcl11b. Nonetheless, the absolute yield of TCRγδ+ cells from any cohort of Bcl11b-deleted precursors was always substantially reduced relative to controls (20 to 30%) (fig. S11). Thus, although in vivo such cells might appear enriched against a more profound TCRαβ deficiency, Bcl11b loss still inhibits the events in T cell development that require passage from DN2a to DN3 stages.
Through dysregulation of progenitor genes, the absence of Bcl11b directly or indirectly disables a crucial component of T lineage–specific commitment machinery. This profound underlying defect may explain why the DN3-like surface phenotype reported for Bcl11b-deficient cells in neonatal germline knockout mice does not confer competence to undergo β-selection (14). The conditions of in vitro culture that reveal the latent self-renewal potential in Bcl11b-deficient DN2a cells undoubtedly sustain greater expansion of DN2-stage cells than the conditions that predominate in the normal thymus. Similar conditions could occur in some intrathymic niches, however, and in these sites loss of Bcl11b could be “rewarded,” lowering a threshold to leukemogenesis. Most important, these results show that Bcl11b’s role distinguishes initial responses of T cell precursors to Notch signaling from a second set of regulatory events that are required for T lineage commitment (fig. S10). Bcl11b may also dissect “adaptive” from “innate” modes of T cell development at an unexpectedly early stage. Our results thus establish this factor as a crucial contributor to both the quality control and the repression of stemness in T cell development.
Supplementary Material
Acknowledgments
We thank P. Liu, H. Kawamoto, P. Li, T. Ikawa, D. Scripture-Adams, and members of the Rothenberg lab for sharing unpublished results and helpful discussion; D. Metzger and J.-M. Bornert for help with generation of the floxed Bcl11b mice; H.-Y. Kueh for guidance in data analysis; T. Taghon and F. Costantini for vectors and mice; D. Perez and R. Diamond for flow cytometry support; and R. Butler and S. Washburn for animal care and breeding supervision. Supported by a California Institute for Regenerative Medicine fellowship (L.L.), NIH grants R33 HL089123 and RC2 CA148278 (E.V.R.), NIH grant R01 GM60852 (M.L.), the Caltech–City of Hope Biomedical Initiative, the Louis Garfinkle Memorial Laboratory Fund, the Al Sherman Foundation, and the A. B. Ruddock Professorship.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/329/5987/89/DC1
Materials and Methods
SOM Text
References
References and Notes
- 1.Rothenberg EV, Moore JE, Yui MA. Nat Rev Immunol. 2008;8:9. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yui MA, Feng N, Rothenberg EV. J Immunol. 2010 doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Neil J, Shank J, Cusson N, Murre C, Kelliher M. Cancer Cell. 2004;5:587. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 4.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. Nat Immunol. 2000;1:138. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 5.Zhong Y, Jiang L, Hiai H, Toyokuni S, Yamada Y. Oncogene. 2007;26:6937. doi: 10.1038/sj.onc.1210494. [DOI] [PubMed] [Google Scholar]
- 6.Rosenbauer F, et al. Nat Genet. 2006;38:27. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 7.Dias S, Xu W, McGregor S, Kee B. Curr Opin Genet Dev. 2008;18:441. doi: 10.1016/j.gde.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye M, Graf T. Curr Opin Immunol. 2007;19:123. doi: 10.1016/j.coi.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Rothenberg EV. Immunity. 2007;26:690. doi: 10.1016/j.immuni.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Georgescu C, et al. Proc Natl Acad Sci USA. 2008;105:20100. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tydell CC, et al. J Immunol. 2007;179:421. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- 12.David-Fung ES, et al. Dev Biol. 2009;325:444. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakabayashi Y, et al. Nat Immunol. 2003;4:533. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 14.Inoue J, et al. J Immunol. 2006;176:5871. doi: 10.4049/jimmunol.176.10.5871. [DOI] [PubMed] [Google Scholar]
- 15.Albu DI, et al. J Exp Med. 2007;204:3003. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamimura K, et al. Biochem Biophys Res Commun. 2007;355:538. doi: 10.1016/j.bbrc.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Przybylski GK, et al. Leukemia. 2005;19:201. doi: 10.1038/sj.leu.2403619. [DOI] [PubMed] [Google Scholar]
- 18.Golonzhka O, et al. J Invest Dermatol. 2009;129:1459. doi: 10.1038/jid.2008.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.See supporting material on Science Online.
- 20.Welner RS, et al. Blood. 2007;109:4825. doi: 10.1182/blood-2006-08-043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vosshenrich CA, et al. J Exp Med. 2007;204:2569. doi: 10.1084/jem.20071451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dontje W, et al. Blood. 2006;107:2446. doi: 10.1182/blood-2005-05-2090. [DOI] [PubMed] [Google Scholar]
- 23.Shigematsu H, et al. Immunity. 2004;21:43. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Martin CH, et al. Nat Immunol. 2003;4:866. doi: 10.1038/ni965. [DOI] [PubMed] [Google Scholar]
- 25.Takatori H, et al. J Exp Med. 2009;206:35. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vosshenrich CA, et al. Nat Immunol. 2006;7:1217. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 27.Gascoyne DM, et al. Nat Immunol. 2009;10:1118. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 28.Doulatov S, et al. Genes Dev. 2009;23:2076. doi: 10.1101/gad.1788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weng AP, et al. Genes Dev. 2006;20:2096. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciofani M, Zúñiga-Pflücker JC. Immunol Res. 2006;34:117. doi: 10.1385/IR:34:2:117. [DOI] [PubMed] [Google Scholar]
- 31.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zúñiga-Pflücker JC. Immunity. 2006;25:105. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Bain G, Romanow WJ, Albers K, Havran WL, Murre C. J Exp Med. 1999;189:289. doi: 10.1084/jem.189.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreslavsky T, et al. Proc Natl Acad Sci USA. 2009;106:12453. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.