Abstract
Mitosis is a highly regulated process in which errors can lead to genomic instability, a hallmark of cancer. During this phase of the cell cycle, transcription is silent and RNA translation is inhibited. Thus, mitosis is largely driven by posttranslational modification of proteins, including phosphorylation, methylation, ubiquitination and sumoylation. Here, we show that protein acetylation is prevalent during mitosis. To identify proteins that are acetylated, we synchronized HeLa cells in early prometaphase and immunoprecipitated lysine-acetylated proteins with anti-acetyl-lysine antibody. The immunoprecipitated proteins were identified by LC-ESI-MS/MS analysis. These include proteins involved in RNA translation, RNA processing, cell cycle regulation, transcription, chaperone function, DNA damage repair, metabolism, immune response and cell structure. Immunoprecipitation followed by Western blot analyses confirmed that two RNA processing proteins, eIF4G and RNA helicase A, and several cell cycle proteins, including APC1, anillin and NudC, were acetylated in mitosis. We further showed that acetylation of APC1 and NudC was enhanced by apicidin treatment, suggesting that their acetylation was regulated by histone deacetylase. Moreover, treating mitotic cells with apicidin or trichostatin A induced spindle abnormalities and cytokinesis failure. These studies suggest that protein acetylation/deacetylation is likely an important regulatory mechanism in mitosis.
Keywords: acetylation, mitosis, RNA processing, cell cycle, histone deacetylase inhibitor
Introduction
Mitosis is largely driven by posttranslational modification of proteins. During this phase of the cell cycle, transcription is silent and RNA translation is globally inhibited1. The posttranslational modification mechanisms, including protein phosphorylation2,3, methylation4, ubiquitination5 and sumoylation6, have been shown to regulate the activity and subcellular localization of proteins in various phases of mitosis. However, protein modification by acetylation in mitosis is largely unexplored.
We recently discovered that a histone deacetylase, HDAC3, is localized on the mitotic spindle and knockdown of HDAC3 by siRNA or reconstitution with a deacetylase dead mutant HDAC3 resulted in a collapsed spindle and aberrant mitosis7. These observations suggest that acetylation/deacetylation of proteins is likely to be involved in regulating protein functions during mitosis. However, proteins that are acetylated/deacetylated during the mitotic phase of the cell cycle are not known.
To delineate the proteins that are acetylated, we synchronized HeLa cells in mitosis and immunoprecipitated lysine-acetylated proteins with anti-acetyl-lysine antibody. Using LC-ESI-MS/MS analysis8,9, we identified 51 unique nonhistone proteins that were immunoprecipitated by the anti-acetyl-lysine antibody. We found that the acetylation status of some of these proteins is regulated by histone deacetylase while others are not, suggesting a differential regulatory mechanism. Treatment of mitotic cells with histone deacetylase inhibitors led to mitotic defects. Uncovering acetylation as a posttranslational modification in mitosis is likely to reveal new paradigms for mitotic regulation.
Materials and Methods
Antibodies and Histone Deacetylase Inhibitors
The following antibodies were used for immunoprecipitation (IP) and immunoblotting (IB, dilutions shown): acetyl-lysine (Millipore, Woburn, MA, USA, rabbit, 0.5 μl/mg lysate for IP, 1:2000), acetyl-lysine (Millipore, mouse, 1 μg/mg lysate for IP for this and all other antibodies), acetyl-lysine (Cell Signaling, Danvers, MA, USA, mouse, 1:1000), anillin (Bethyl, Montgomery, TX, USA, rabbit, 1:2000), NudC (our laboratory, rabbit R2 against NudC C terminus 15 aa peptide, 1:3000)10, NudC (our laboratory, goat G1 against NudC C terminus 15 aa peptide, 1:2000) (under submission), APC1 (Bethyl, rabbit, 1:1000), eIF4G (gift of Dr. Richard Lloyd, Baylor College of Medicine, rabbit 583, 1:5000)11, RNA helicase A (Abcam, Cambridge, MA, USA, rabbit, 1:1000), α-tubulin (GeneTex, San Antonio, TX, USA, rabbit, 1:2000), α-tubulin (Sigma, St. Louis, MO, USA, mouse), histone H3 (Abcam, rabbit, ChIP grade, 1:2000), β-tubulin(tub2.1) (Sigma, mouse, 1:1000), and CREST serum (gift of Dr. Bill Brinkley, Baylor College of Medicine, human, 1:10,000 for immunofluorescence). Histone deacetylase inhibitors trichostatin A (TSA), apicidin and sodium butyrate (NaB) were purchased from Sigma.
Cell Culture and Synchronization
HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA). To enrich for mitotic cells, cells were synchronized by incubation with 2 mM thymidine for 15 h, released for 10.5 h, followed by another thymidine block for 14 h (double thymidine block, DTB). Cells were then either released into fresh medium for 8 h to reach the peak of mitosis, or released for 8 h during which 50 ng/ml nocodazole, a microtubule depolymerizer, was added for the last 3.5 h to further enrich for cells in early prometaphase. Using this protocol, more than 80% of cells exhibited a rounded morphology that is characteristic of mitotic cells. Mitotic cells were harvested by lightly pipeting with Beral® transfer pipets (Samco Scientific, San Fernando, CA, USA) to wash the round cells off the culture dish. In other experiments, a histone deacetylase (HDAC) inhibitor, trichostatin A (TSA, 660 nM) or apicidin (100 nM or 500 nM), was added for the last 3.5 h of the second thymidine release to enhance acetylation of proteins only during mitosis.
Flow Cytometry
Randomly cycling HeLa cells were collected by trypsinization while mitotic cells were collected by light pipeting as described above. Cells were fixed and stained as follows12. Briefly, 1 × 106 to 1 × 107 cells were resuspended in 0.5 ml of 4°C phosphate-buffered saline (PBS), fixed in 4.5 ml 70% ethanol at 4°C for 2 h, and washed twice in PBS by centrifugation at 500 × g for 5 min. Fixed cells were incubated in 1 ml of propidium iodide staining solution (0.1% Triton X-100, 20 μg/μl DNase-free RNase A [Sigma], 20 μg/ml propidium iodide in PBS) for 15 min at 37°C. DNA content frequency was acquired using a FACS Canto II benchtop cytometer (BD Biosciences, Franklin Lake, NJ, USA). Cell doublets were excluded from the FACS data using doublet discrimination gating: FSC-H/FSC-W gate followed by an SSC-H/SSC-W gate and then a PE-H/PE-A gate. Cell cycle distribution was analyzed using FlowJo (Tree Star, Inc., Ashland, OR, USA) and the percentage of cells in each cell cycle phase was determined by fitting the DNA content histogram to the Watson Pragmatic Model13.
Immunoprecipitation for Mass Spectrometry
Mitotic HeLa cells were needle sheared and lysed for 20 min on ice in lysis buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 5 mM EGTA, 1.5 mM EDTA, 0.1% Triton X-100, 5% glycerol) supplemented with 1 mM PMSF, mammalian protease-inhibitor cocktail, 5 mM Na3VO4, 5 mM NaF, serine-threonine and tyrosine phosphatase inhibitor cocktails, and 10 mM NaB (all from Sigma). Protein concentrations were determined by Bradford assay (Bio-Rad, Hercules, CA, USA). Mitotic cell lysates (9 – 15 mg) were incubated with polyclonal anti-acetyl-lysine antibody (4.5 – 7.5 μg) at 4°C overnight and washed five times in lysis buffer supplemented with 10 mM NaB. Immunoprecipitated proteins were incubated at 95°C for 5 min, resolved by 4 – 12% SDS-PAGE, and stained by GelCode Blue (Thermo Fisher Scientific, Rockford, IL, USA).
In-gel Protein Digestion
24 gel slices were excised from three independent samples, and subjected to trypsin digestion as described8,9. Briefly, the gel slices were washed two times in a mixture of 1:1 v/v analytical-grade water: 0.1 M NH4HCO3 in twice the gel volume for 15 min with agitation. After removing the wash solution completely, the gel pieces were covered with acetonitrile (ACN), left to shrink and become stuck together, removed from ACN, rehydrated in 0.1M NH4HCO3 for 10 min, incubated in an equal volume of ACN for another 10 min, and drained of all liquids. The gel pieces were dried down in a vacuum centrifuge, and the proteins were reduced in 10 mM dithiothreitol, alkylated in 55 mM iodoacetamide in 0.1 M NH4HCO3, washed as above, and digested with trypsin for 24 h at 37 °C. The peptides were extracted from the gel pieces by the addition of 10 μl 25 mM NH4HCO3, 5 μl 5% formic acid and 5 μl ACN, dried down, and dissolved in a mixture containing formic acid:water:ACN:trifluoroacetic acid (0.1:95:5:0.01) for LC-MS/MS analysis.
Identification of Proteins with LC-ESI-MS/MS
After immunoprecipitation, the bands separated by SDS-PAGE were excised and digested with trypsin. Tryptic digests were separated with a reverse-phase column (C-18 PepMap 100, LC Packings/Dionex, Sunnyvale, CA)8,9. The column eluate was directly introduced onto a QSTAR XL mass spectrometer (Applied Biosystems, Foster City, CA, USA and Sciex, Concord, ONT, Canada) via ESI. Candidate ion selection and data collection were performed as described previously8,9. Half second MS scans (300 – 1500 Thompson) were used to identify candidates for fragmentation during MS/MS scans. Up to five 1.5s MS/MS scans (65 – 1500 Thompson) were collected after each scan. An ion was assigned a charge in the range of +2 to +4. The dynamic exclusion was 40. Protein identifications were completed with ProteinPilot (versions 1.0 and 2.0, Applied Biosystems and Sciex), using a setting with 1.5 Da mass tolerance for both MS and MS/MS and the human “RefSeq” databases from NCBI (http:www.ncbi.nlm.nih.gov/RefSeq). ProteinPilot is the successor to ProID and ProGroup, and uses the same peptide and protein scoring method. Briefly, given a protein score, S, the likelihood that the protein assignment is incorrect is 10-S. Scores above 2.0 require that at least two sequence-independent (unique) peptides will be identified8,9.
Immunoprecipitation and Immunoblotting
To confirm acetylation of proteins in mitosis, two approaches were employed. Mitotic HeLa cell lysates (2 mg) were immunoprecipitated with a second anti-acetyl-lysine antibody (2 μg monoclonal acetyl-lysine antibody from Millipore) in the presence of 10 mM NaB as described above. For the reciprocal immunoprecipitation, mitotic HeLa cells were lysed in a reducing buffer (10 mM dithiothreitol, 1% SDS, 5 mM EDTA) for 5 min on ice. Cell lysates were diluted 10-fold with RIPA buffer (150 mM NaCl, 25 mM Tris pH 7.5, 1 mM EDTA, 0.5% deoxycholate, 1% NP40) supplemented with 1 mM PMSF, mammalian protease-inhibitor cocktail, 5 mM Na3VO4, 5 mM NaF, serine-threonine and tyrosine phosphatase inhibitor cocktails, 10 mM NaB (all from Sigma) and 15 U/ml DNase1 (Roche, Branford, CN, USA), needle sheared, and precleared with normal rabbit serum bound protein G sepharose beads at 4 °C for 1 h. Antibodies against each specific protein of interest were used to immunoprecipitate 1 mg mitotic cell lysate, followed by immunoblotting with a third anti-acetyl-lysine antibody (monoclonal acetyl-lysine antibody from Cell Signaling, 1:1000) and reblotting with the specific antibodies in the presence of ReliaBLOT® (Bethyl) to reduce background signals. After SDS-PAGE, proteins were transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked for 1 h at room temperature with blocking buffer (5% BSA in TBST [Tris-buffered saline with 0.2% Tween-20]) or ReliaBLOT® Block (Bethyl), incubated with primary antibodies overnight at 4°C, washed three times with TBST, incubated for 1 – 2 h at room temperature with horseradish-peroxidase linked secondary antibodies (Vector Laboratories, Burlingame, CA, USA) or ReliaBLOT® HRP Conjugate (Bethyl), washed three times with TBST, and developed using chemiluminescence SuperSignal West Pico (Thermo Scientific).
Immunofluorescence Microscopy
HeLa cells were cultured on coverslips and synchronized by DTB without nocodazole treatment, rinsed twice with 37°C PHEM (60 mM K-PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgSO4, pH6.9 with KOH), fixed with 4% paraformaldehyde in PHEM at 4°C for 20 min, rinsed twice with 4°C PBS and permeabilized with 0.5% Triton X-100 in PBS at room temperature for 15 min. The fixed cells were incubated at room temperature for 30 min with antibody blocking solution (0.1 M K-PIPES, 1 mM MgSO4, 1 mM EGTA, 1.83% L-lysine, 1% BSA, 0.1% NaN3, pH7.2 with KOH, pre-saturated with 2% nonfat milk at 4°C), then incubated overnight at 4°C with primary antibody, washed three times with cold PBS, incubated in secondary antibody for 3 h at 4°C, and washed three times with cold PBS. Coverslips were mounted in ProLong Gold antifade reagent with DAPI (Molecular Probes, Eugene, OR, USA). Images were acquired using a Nikon TE2000 widefield microscope system (Nikon Instruments, Lewisville, TX, USA) and a 40× oil/1.40 NA objective.
Results
Identification of Proteins that are Acetylated During Mitosis
To identify proteins that are acetylated during mitosis, we immunoprecipitated acetylated proteins from mitotic cells. We employed a well-established cell synchrony protocol10 to enrich for cells in mitosis. HeLa cells were synchronized by double thymidine block and released for 8 h to allow cells to enter mitosis (Fig. 1A). Cells were further incubated for the last 3.5 h with 50 ng/ml nocodazole, a drug that depolymerizes microtubule, to enrich for cells in early prometaphase. FACS analysis showed that the randomly cycling cell population contained about 12% cells in the G2/M phase (Fig. 1A i) while the double thymidine block and release cell synchrony protocol produced over 80% of cells in the G2/M phase (Fig. 1A ii). Mitotic cells with the characteristic round morphology (Fig. 1A ii) were collected by a gentle rinse of the plate and the cells were used for immunoprecipitation with anti-acetyl-lysine antibody. Protein samples from two independent experiments were resolved on an SDS-PAGE gel, and protein bands were cut into 24 gel slices, digested with trypsin, and subjected to LC-ESI-MS/MS analysis (Fig. 1B). In addition to histones, which are known to be acetylated14, 51 unique nonhistone proteins were identified (Table 1). Interestingly, proteins involved in RNA translation and RNA processing were highly represented in these samples. Proteins involved in cell cycle regulation during mitosis and cytokinesis were also identified. Other proteins identified are involved in gene transcription, DNA damage repair, chaperone functions, metabolism, immune response, and cell structure.
Figure 1.
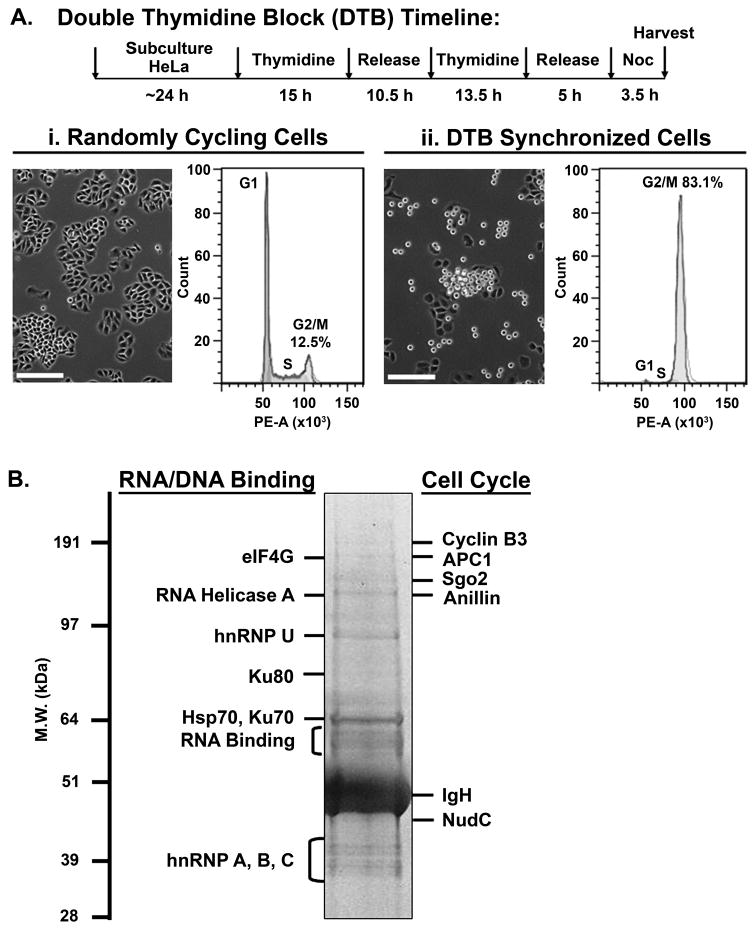
Isolation of acetylated proteins from cells in mitosis. (A) Cell synchronization protocol. HeLa cells were synchronized in mitosis by double thymidine block (DTB) and release into 50 ng/ml nocodazole to enrich for mitotic cells. Phase contrast images show the mitotic population as rounded cells. Images were acquired using a Nikon TE2000 microscope system. Bars, 200 μm. Flow cytometry was performed to determine cell cycle distribution of (i) randomly cycling cells versus (ii) DTB synchronized mitotic cells. Percentage of cells in each cell cycle phase was determined by fitting the DNA content histogram to the Watson Pragmatic model13 using FlowJo with a root mean squared value of 6.98 and 21.29 for (i) and (ii), respectively. Randomly cycling cells: G1, 53.4%; S, 29.1%; G2/M, 12.5%. DTB synchronized cells: G1, 0.04%; S, 3.03%; G2/M, 83.1%. (B) A representative mitotic sample used for acetyl-lysine immunoprecipitation followed by mass spectrometry. Three samples prepared as in (A) (9 – 15 mg each) from two independent experiments were immunoprecipitated with anti-acetyl lysine polyclonal antibody (Millipore), resolved on SDS-PAGE, and stained with GelCode Blue. 24 gel slices were processed for LC-ESI-MS/MS analysis. HC, IgG heavy chain.
Table 1.
Proteins identified by acetyl-lysine immunoprecipitation and LC-ESI-MS/MS in mitotic HeLa cells.
| Protein names | gi | Biological Process | Molecular Function | *Known Ac status |
|---|---|---|---|---|
| Histones | ||||
| Histone H2A.1 | 31980 | Chromosome organization | DNA binding | 51,52 |
| Histone H3 | 31982 | Chromosome organization | DNA binding | 51,52 |
| Histone H4 | 31995 | Chromosome organization | DNA binding | 51,52 |
| Translation | ||||
| 40S ribosomal protein S25 | 51338648 | Translation | Catalyzes protein synthesis | 51 |
| 40S ribosomal protein S26 | 266970 | Translation | Catalyzes protein synthesis | 51 |
| Eukaryotic initiation factor 4 gamma (eIF4G) | 219613 | Translation | Initiates translation | 51 |
| Polyadenylate binding protein | 35570 | Translation | Initiates translation | 51 |
| Polyadenylate binding protein II | 74706522 | Translation | Initiates translation | 51 |
| RNA binding | ||||
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | 57209229 | mRNA processing | Unwinds RNA | 51 |
| Heterogeneous nuclear ribonucleoprotein AO | 773644 | mRNA processing/transport | RNA binding | |
| Heterogeneous nuclear ribonucleoprotein A1 | 296650 | mRNA processing/transport | RNA binding | 51,52 |
| Heterogeneous nuclear ribonucleoprotein A/B | 13528732 | mRNA processing/transport | RNA binding | |
| Heterogeneous nuclear ribonucleoproteins A2/B1 | 133257 | mRNA processing/transport | RNA binding | 51 |
| Heterogeneous nuclear ribonucleoproteins C1/C2 | 108935845 | mRNA processing/transport | RNA binding | 51 |
| Heterogeneous nuclear ribonucleoproteins C3 | 3334899 | mRNA processing/transport | RNA binding | |
| Heterogeneous nuclear ribonucleoprotein D0 | 13124489 | mRNA processing/transport | RNA binding | 51 |
| Heterogeneous nuclear ribonucleoprotein D-like | 39644771 | mRNA processing/transport | RNA binding | |
| Heterogeneous nuclear ribonucleoprotein R | 2697103 | mRNA processing/transport | RNA binding | 51 |
| Heterogeneous nuclear ribonucleoproteins U | 32358 | mRNA processing/transport | RNA binding | 51 |
| Heterogeneous nuclear ribonucleoproteins U-like | 21536326 | mRNA processing/transport | RNA binding | 51 |
| mRNA-binding protein CRDBP | 7141072 | RNA metabolism | RNA binding | |
| RNA helicase A | 307383 | RNA processing | Unwinds dsDNA and dsRNA | 51 |
| Translocation in Liposarcoma (TLS) | 386157 | mRNA processing/transport | RNA binding | |
| Cell cycle | ||||
| Actin binding protein anillin | 8489881 | Cytokinesis | Acto-myosin ring assembly | 51 |
| APC1 (Tsg 24 protein) | 11967711 | Cell cycle | Anaphase promotion | |
| Cip 1- interacting zinc finger protein (Ciz1) | 6136800 | Cell cycle | Inhibits cdks | |
| Cyclin B3 | 14275558 | Meiotic prophase I | Interacts with cdk2 | |
| NudC | 619907 | Cell cycle | Mitotic progression | 51 |
| Tripin/Shugoshin 2 | 23986276 | Cell cycle | Protects centromeres | |
| Zizimin-3 (Dedicator of cytokinesis, protein 10) | 32469767 | Cytokinesis | Guanine exchange factor | |
| Transcription | ||||
| ATP-dependent helicase SMARCA4 | 116242792 | Transcription | Transcriptional coactivator | |
| DNA damage | ||||
| Ku70 | 125729 | DNA damage response | Non-recombinational repair | 51 |
| Ku80 | 35038 | DNA damage response | Non-recombinational repair | 51 |
| NF45 protein | 532313 | DNA damage response | Non-recombinational repair | |
| Mitochondiral single stranded DNA binding protein | 188856 | DNA damage response | Protects ssDNA | |
| Chaperones | ||||
| Stress-70 protein, mitochondrial, precursor | 21264428 | Stress/metabolic response | Protein folding | 51,52 |
| 71 kDa heat shock cognate protein | 32467 | Stress/metabolic response | Protein folding | 51 |
| Heat shock 70 kDa protein 5, precursor | 14916999 | Stress/metabolic response | Protein folding | 51 |
| Metabolism | ||||
| Placental alkaline phosphatase 1, precursor | 130737 | Basic phosphatase | Hydrolase enzyme | |
| Carbamoyl-phosphate synthase I, mitochondria | 4033707 | Urea cycle | Degrades ammonia | 52 |
| Carbamoyl-phosphate synthetase II, cytosol | 1228049 | Pyrimidine biosynthesis | Degrades glutamine | 51 |
| Immune response | ||||
| Bone marrow stromal cell antigen 2 | 506861 | Humoral immune response | B cell growth and development | |
| Complement component 3, precursor | 119370332 | Inflammation | Complement activation | |
| Fibrinogen alpha chain, precursor | 1706799 | Blood clotting | Platelet aggregation cofactor | |
| Migration inhibitory factor related protein 14 (MRP14) | 34771 | Inflammation | Calcium binding | |
| Structural | ||||
| Desmoplakin I | 1147813 | Intercellular junction | Binds desmosomes | |
| Ifapsoriasin (filaggrin 2) | 59939295 | Structural | Binds keratin | |
| Keratin Type II | 34069 | Structural | Intermediate filament | |
| Myosin, heavy chain 9, non-muscle | 3169000 | Structural | Contractile protein | 51 |
| Miscellaneous | ||||
| Annexin A2 | 113950 | Cell growth; Signal transduction | Phospholipid binding | 51 |
| F-box only protein 11 isoform 1 | 30089926 | Protein Ubiquitination | Interacts with Skp1 | |
| Granulin | 31193 | Cell growth | Secreted, glycosylated peptide | |
| FLJ00195 | 18676594 | N/A | N/A | |
| Hypothetical protein | 21739818 | N/A | N/A |
Acetylation of Structural Proteins in Mitosis
Next, we sought to confirm that these proteins are acetylated in mitosis. We first analyzed proteins that are generally abundant. Mitotic cell lysates were prepared from HeLa cells in the same manner as was performed for the mass spectrometry analysis in Fig. 1A. To demonstrate specificity, we used a different anti-acetyl-lysine monoclonal antibody (Millipore), instead of the original polyclonal anti-acetyl-lysine antibody, for immunoprecipitation. The immunoprecipitated acetylated proteins were then immunoblotted for the individual proteins of interest. Using this approach, we confirmed that histone H3 (17 kDa) (Table 1) was acetylated in mitotic cells (Fig. 2A, left).
Figure 2.

Acetylation of structural proteins in mitosis. Mitotic cell lysates (2 mg), prepared as in Fig. 1, were immunoprecipitated with a second anti-acetyl-lysine monoclonal antibody (Millipore) (Ac-K IP) and immunoblotted with specific antibodies (arrowhead) as shown. (A, left) Histone H3. (B, left) α-tubulin. For confirmation, reciprocal protein-specific immunoprecipitation of H3 (A, right) and α-tubulin (B, right) was performed. Mitotic cell lysates (2 mg) were prepared in a reducing condition as described in Materials and Methods, immunoprecipitated with anti-peptide antibodies against the protein of interest, immunoblotted with a third anti-acetyl-lysine monoclonal antibody (Cell Signaling) to show acetylation (arrowhead), and re-immunoblotted with the same anti-peptide antibodies to show efficiency of immunoprecipitation and protein loading. The data are reproducible in three independent experiments. Input lanes, 20 μg total mitotic cell lysates. Antibody alone was included as an IgG control. *, non-specific band; HC, IgG heavy chain; LC, IgG light chain.
As further confirmation, we performed the reciprocal immunoprecipitation by immunoprecipitating with anti-histone H3 antibody and immunoblotted with a third anti-acetyl-lysine monoclonal antibody (Cell Signaling). Reciprocal protein-specific immunoprecipitation showed that histone H3 was acetylated in mitotic cells (Fig. 2A, right). In parallel, we examined the acetylation of α-tubulin, a cytoskeletal protein previously shown to be acetylated in mitosis7. Using both acetyl-lysine immunoprecipitation and reciprocal immunoprecipitation, we showed that α-tubulin (51 kDa) was indeed acetylated in the same mitotic lysates (Fig. 2B).
Acetylation of RNA Processing Proteins in Mitosis
Next, we examined two proteins involved in RNA processing, that is, the RNA translation initiation factor eIF4G1,11,15 and RNA helicase A16,17 (Table1). Using the anti-acetyl-lysine immunoprecipitaton approach as in Fig. 2, we showed that eIF4G (220 kDa) (Fig. 3A, left) and RNA helicase A (140 kDa) (Fig. 3B, left) were both acetylated during mitosis. Reciprocal immunoprecipitation further confirmed that eFI4G (Fig. 3A, right) and RNA helicase A (Fig. 3B, right) were acetylated in mitosis.
Figure 3.

Acetylation of RNA processing proteins in mitosis. The acetylation of RNA processing proteins eIF4G (A) and RNA helicase A (RHA) (B) was demonstrated by acetyl-lysine immunoprecipitation (Ac-K IP) (left panels) and by reciprocal protein-specific immunoprecipitation (right panels) as described in Fig. 2. The data are reproducible in three to six independent experiments. Input lanes, 20 μg total mitotic cell lysates. Antibody alone was included as an IgG control. *, non-specific band(s); HC, IgG heavy chain.
Acetylation of Cell Cycle Proteins in Mitosis
For cell cycle proteins, we selected for analysis proteins that have well-characterized functions in mitosis and cytokinesis. These include the scaffold protein Anaphase Promoting Complex 1 (APC1)18-20 in the E3 ubiquitin ligase APC/C, the cleavage furrow protein anillin21,22, and the dynein/dynactin associated and Polo-like kinase 1 (Plk1)-interacting protein NudC10,23,24 (Table 1). Using the anti-acetyl-lysine immunoprecipitation approach as in Fig. 2, we showed that APC1 (215 kDa) (Fig. 4A, left), anillin (124 kDa) (Fig. 4B, left), and NudC (42 kDa) (Fig. 4C, left) were acetylated in mitotic cells. The right panels in Fig. 4 confirmed that APC1 (Fig. 4A, right), anillin (Fig. 4B, right) and NudC (Fig. 4C, right) were indeed acetylated in mitotic cells by reciprocal immunoprecipitation.
Figure 4.
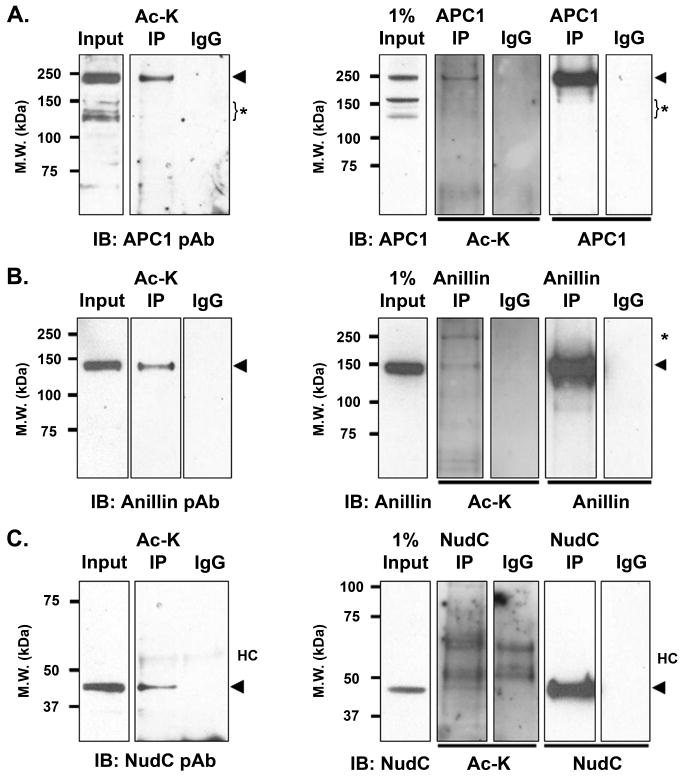
Acetylation of cell cycle proteins in mitosis. The acetylation of cell cycle proteins APC1 (A), anillin (B) and NudC (C) was demonstrated by acetyl-lysine immunoprecipitation (Ac-K IP) (left panels) and by reciprocal protein-specific immunoprecipitation (right panels) as described in Fig. 2. The data are reproducible in three to six independent experiments. Input lanes, 20 μg total mitotic cell lysates. Antibody alone was included as an IgG control. *, non-specific band(s); HC, IgG heavy chain.
Histone Deacetylase and Protein Acetylation/Deacetylation in Mitosis
Previous studies showed that the deacetylase HDAC3 is localized on the mitotic spindle and its activity is involved in mitotic progression as the expression of an inactive HDAC3 led to aberrant mitosis7. To determine whether any of the acetylated mitotic proteins might be a target of HDAC3 deactylation, we treated the cells with apicidin25, an HDAC inhibitor that preferentially inhibits HDAC2 and HDAC3 of the Class I family of HDACs26. Mitotic cells were enriched by a double thymidicine block, released for 5 h, and treated with apicidin for 3.5 h during the peak of mitosis. Under this condition, acetylation of APC1 and NudC was found to be increased with apicidin treatment (Fig. 5A). This observation suggests that the acetylation status of APC1 and NudC is regulated by HDAC2/3 in mitosis, and explains in part why NudC acetylation in mitotic cells was difficult to detect in the absence of apicidin treatment (Fig. 4C and Fig. 5A). Histone H3, known to be deacetylated by both HDAC227 and HDAC328 in vitro, showed an increase in acetylation with apicidin treatment (Fig. 5A). On the other hand, treatment of mitotic cells with apicidin did not affect the acetylation of eIF4G, RNA helicase A and anillin (Fig. 5B). Acetylation of α-tubulin, which is known to be deacetylated by HDAC629, was not altered by apicidin treatment (Fig. 5B). These results suggest that some of the acetylated proteins in mitotic cells are targets of HDAC2/3 regulation.
Figure 5.
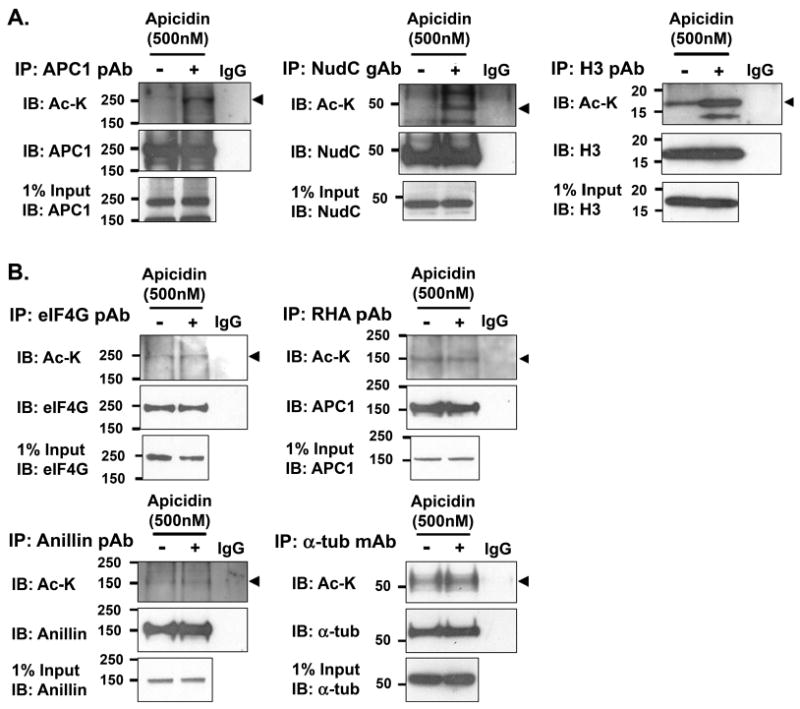
Mitotic proteins are regulated by acetylation and deacetylation. Mitotic HeLa cells were synchronized as in Fig. 1 and treated with or without Apicidin (500 nM) for 3.5 h prior to harvest to block protein deacetylation during the peak of mitosis. Cell lysates were prepared as in Fig. 2B, immunoprecipitated with antibodies against each specific protein of interest, immunoblotted with a third anti-acetyl-lysine monoclonal antibody (Cell Signaling) (arrowhead), and re-immunoblotted with protein-specific antibodies to show efficiency of immunoprecipitation and protein loading. (A) Proteins whose acetylation is affected by apicidin treatment during mitosis. (B) Proteins whose acetylation is not affected by apicidin treatment during mitosis. The data are reproducible in three independent experiments.
Acetylation Regulates Mitosis and Cytokinesis
We further used HDAC inhibitors to examine the effects of acetylation on mitotic progression. Apicidin treatment at 100 nM is specific for HDAC3 inhibition, while 500 nM apicidin inhibits both HDAC2 and HDAC3 deacetylase activity but not that of other HDACs26. Trichostatin A (TSA) is a pan-HDAC inhibitor. Mitotic cells were enriched by a double thymidine block, released for 5 h, and treated with HDAC inhibitors for 3.5 h during the peak of mitosis. Cells were stained with CREST, a human autoserum that marks the inner centromere, tubulin to visualize the mitotic spindle, and DAPI to visualize DNA. While control cells showed a tight band of chromosomes aligned in between the mitotic spindle at the metaphase plate (Fig. 6A), cells treated with 100 nM apicidin exhibited a range of mitotic phenotypes (Fig. 6B). These include a collapsed mitotic spindle (i)7, lopsided spindle poles where the larger pole was surrounded by a ring of chromosomes (ii), and misaligned chromosomes lying laterally on or external to the spindle pole (i). Similar collapsed (iii, v) or lopsided spindles (iv) as well as misaligned chromosomes (iii, v) were observed when the cells were treated with 500 nM apicidin (Fig. 6C) or with 660 nM TSA (Fig. 6D). Further, a more severe phenotype, such as a loss of chromosome attachment from the mitotic spindle (Fig. 6D, vi), was also observed in TSA-treated cells. The collapsed spindle phenotype and dome-like chromosome configuration were found to be statistically significant in both the 100 nM apicidin treated and 660 nM TSA treated cells as compared to control cells (Fig. 6G).
Figure 6.
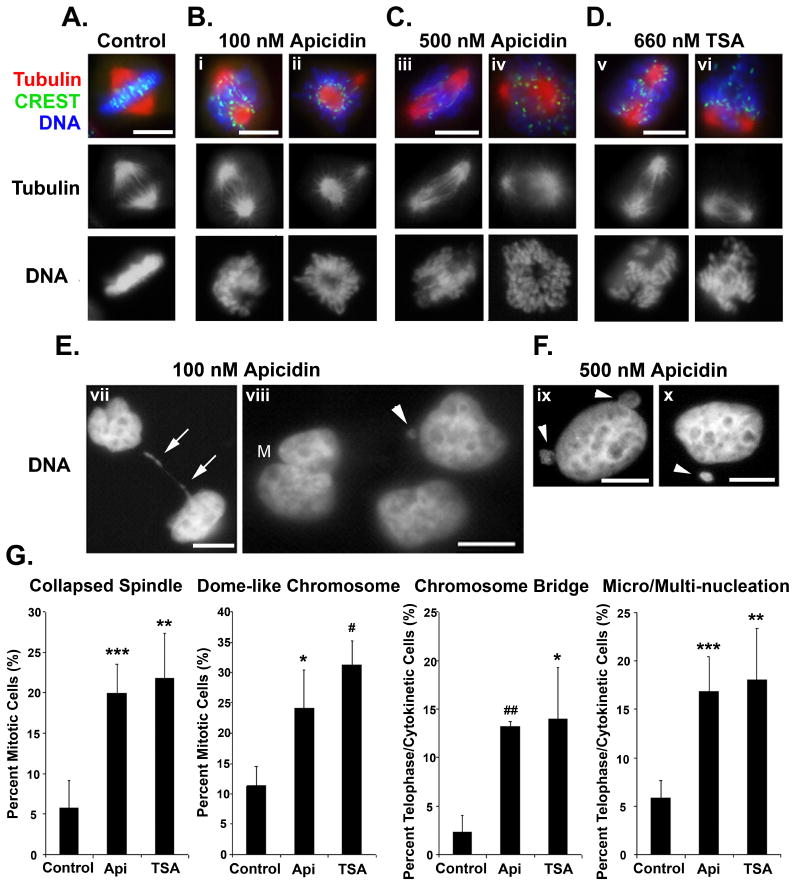
Protein acetylation plays a role in mitosis and cytokinesis. HeLa cells were synchronized by a double thymidine block and released to enter into mitosis as described in Fig. 1. At 5 h after release at around the G2/M transition, cells were treated with isopropanol solvent control (A), 100 nM apicidin (B, E), 500 nM apicidin (C, F), or 660 nM TSA (D) for an additional 3.5 h during mitosis. Cells were stained with CREST antiserum (green) to label the inner centromere, anti-tubulin antibody (red) to label the mitotic spindle, and counterstained with DAPI (blue) to label the DNA. Tubulin and DNA staining are also shown in black and white for contrast. (E, F) show only DNA in black and white for contrast. Arrows, chromatin bridge in between two divided cells; M, multinucleation; arrowheads, micronucleation. Images were acquired using a Nikon TE2000 microscope system. Bars, 10 μm. (G) Early and late mitotic phenotypes observed in (A – F) were quantified. About 200 cells were counted for each condition for the early mitotic phenotypes (A – D) and 200 cells were counted for each condition for the late mitotic phenotypes (E, F). Api, 100 nM apicidin; TSA, 660 nM TSA. The graph shows the average (± SD) from three independent experiments. *, p < 0.025; **, p < 0.01; ***, p < 0.005; #, p < 0.0025; ##, p < 0.00025.
In addition to early mitotic abnormalities, a late mitotic phenotype was observed in both 100 nM (Figs. 6E) and 500 nM (Fig. 6F) apicidin-treated cells that have progressed beyond mitosis. Some cells exhibited chromatin bridges in between divided daughter cells (vii, arrows), micronucleation (viii – x, arrowheads), and multinucleation (viii, M). The formation of chromosome bridges, micronucleation and multinucleation were found to be statistically significant in both the 100 nM apicidin treated and 660 nM TSA treated cells as compared to control cells (Fig. 6G). These results suggest that enhanced acetylation due to inhibition of HDAC3 during mitosis resulted in aberrant mitosis and cytokinesis. These observations suggest that acetylation/deacetylation plays a critical role in mitosis and cytokinesis.
Discussion
We identified 51 unique nonhistone proteins that are acetylated in mitosis by mass spectrometry. These include proteins involved in RNA translation, RNA processing, cell cycle regulation, transcription, chaperone function, DNA damage repair, metabolism, immune response and cell structure. Our findings suggest that acetylation is likely a form of posttranslational modification in mitosis that is more prevalent than previously appreciated. Acetylation is known to neutralize the positive charge on basic lysine residues in proteins, and may thus change protein conformation, protein-protein interaction, protein localization and protein function4. Moreover, protein acetylation may affect other posttranslational modifications, that is, activate or inhibit phosphorylation4,30,31, ubiquitination5 or sumoylation6, to regulate the activities and/or localization of multi-protein complexes4,30. The acetylated proteins identified in this study will provide a framework from which to understand how acetylation regulates cell cycle progression.
We found that many RNA binding and processing proteins were acetylated in mitotic cells. The high representation of proteins involved in RNA processing and translation is interesting because RNA translation is largely inhibited during mitosis1,15,32. However, how acetylation affects RNA translation and processing is unknown. eIF4G is a translation initiation factor that acts as a scaffold protein to recruit other factors as well as the 40S ribosomal subunit to form the preinitiation complex at the 5′ end of mRNA32-34. During mitosis, eIF4G is dissociated from the 5′ end of mRNA1,32. Stalling of RNA translation on polysomes protects mRNAs during mitosis and allows cells to rapidly resume protein synthesis upon entry into G135. RNAse helicase A (DDX9) is a member of the DExD/H protein family36. RNA helicases bind ATP and RNA, exhibit RNA unwinding activities, and are involved in all levels of RNA metabolism, from transcription, splicing, transport, translation, to RNA decay16,17. The acetylation of two key RNA processing proteins in mitotic cells opens the way to investigate how acetylation may be involved in various RNA activities during mitosis.
We also found that many cell cycle regulating proteins were acetylated. However, how acetylation affects the function of APC1, anillin and NudC during mitotic progression is unknown. APC1 is the largest subunit of the 1.5 MDa E3 ubiquitin ligase APC/C37-39 that is involved in the degradation of cyclins and anaphase inhibitors at the kinetochore19,38,40. APC/C thus regulates sister chromatid separation and the timing of metaphase-to-anaphase transition. Anillin is an actin binding protein that is localized at the cleavage furrow22. Anillin acts as a scaffold to recruit other proteins to the cleavage furrow, to stimulate the formation of the actomyosin contractile ring and cleavage furrow ingression21,22,41. NudC is a dynein-dynactin motor associated factor42. In early mitosis, NudC is involved in recruiting Plk1 to the kinetochores and kinetochore-microtubule attachment10. In late mitosis, NudC is localized to the cleavage furrow and midzone/midbody23,24,43 and is required for stable midzone organization and the completion of cytokinesis. These proteins are either associated with (NudC) or exist in multi-protein complexes (APC1, anillin) in distinct mitotic structures. Thus, it is very likely that their acetylation status may affect protein-protein interactions and protein functions in mitosis and/or cytokinesis.
Protein acetylation and deacetylation is a dynamic process regulated by histone acetyltransferase (HAT) and histone deacetylase (HDAC) proteins in a spatial and temporal-dependent manner. Our study found that the acetylation of APC1, NudC, and histone H3 was increased after apicidin treatment, while the acetylation of eIF4G, RNA helicase A, anillin, and α-tubulin was not affected. Apicidin treatment at 500 nM inhibits both HDAC2 and HDAC3 deacetylase activity but not that of other HDACs. These observations suggest that protein acetylation/deacetylation, like other types of posttranslational modifications, is specific and differentially regulated.
The significance of acetylation/deacetylation in mitosis was revealed when persistent acetylation with HDAC inhibitor treatment during mitosis resulted in a range of mitotic and cytokinetic phenotypes. These include a collapsed spindle, unequal spindle pole formation, aberrant kinetochore-microtubule attachment, and loss of chromosome attachment to the spindles in early mitosis. Cells also showed chromatin bridge formation, micronucleation, and multinucleation, which likely resulted from errors in chromosome segregation and a failed cytokinesis. These defects demonstrate that acetylation/deacetylation plays a critical role in regulating mitosis and cytokinesis. In agreement with our studies, recent reports have suggested that nonhistone proteins may also be targets of HDAC inhibitors, leading to disorganized kinetochore assembly and organization44-48. Further, acetylation of Cyclin A49 and the checkpoint protein BubR150 have been shown to be important for mitotic progression. Our study showed that many more proteins are acetylated in mitosis and provided the biochemical basis to assess mitotic phenotypes due to their hyperacetylation. We predict that aberrant acetylation of APC1 and NudC in response to HDAC inhibitor treatment may account for some of the observed mitotic and cytokinetic phenotypes, and this possibility will be examined in future studies.
Recent technological advances in proteomics analysis revealed that protein acetylation is almost as prevalent as protein phosphorylation51,52. These studies examined acetylated proteins after treating randomly cycling cells with pan-HDAC inhibitors over a 24-h period51,52. In contrast, our study focused on posttranslational protein modification that occurred during the mitotic phase of the cell cycle. We predict that there are more acetylated proteins in mitosis than those identified in the current study. Thus, further proteomics analysis of acetylated proteins in mitosis is warranted.
HDAC inhibitors for transcriptional regulation are being tested for cancer treatments53. However, the role of HDAC inhibitors on posttranslational modification of proteins has not been considered. Our study suggests that acetylation is a novel biochemical control during mitotic progression that may have significant clinical implication. Understanding the role of acetylation and deacetylation in mitotic progression is likely to open up new pathways and drug targets for tackling human diseases and cancers.
Acknowledgments
This work was supported by NIH training grant T32 DK07696 (C.C.), and grants from the NIH (CA111479, CA090270 for S.-H.L.; SR10RR025623, RR-P20-RR17695 for D.J.; DK053176, AI071130 for L.-y.Y.-L.) and the Dan L. Duncan Cancer Center (L.-y.Y.-L. and S.-H.L.).
References
- 1.Le Breton M, Cormier P, Belle R, Mulner-Lorillon O, Morales J. Translational control during mitosis. Biochimie. 2005;87:805–811. doi: 10.1016/j.biochi.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malik R, Lenobel R, Santamaria A, Ries A, Nigg EA, Korner R. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J Proteome Res. 2009;10:4553–4563. doi: 10.1021/pr9003773. [DOI] [PubMed] [Google Scholar]
- 4.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modification. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merbl Y, Kirschner MW. Large scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc Natl Acad Sci. 2008;106:2543–2548. doi: 10.1073/pnas.0812892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh ETH. SUMOylation and de-SUMOylation: wrestling with life's processes. J Biol Chem. 2009;248:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishii S, Kurasawa Y, Wong J, Yu-Lee L. Histone deacetylase 3 localizes to the mitotic spindle and is required for kinetochore-microtubule attachment. Proc Natl Acad Sci. 2008;105:4179–4184. doi: 10.1073/pnas.0710140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang F, Clifton J, Yang X, Rosenquist T, Hixon DC, Kovacs S, Josic D. SELDI-TOF as a method for biomarker discovery in the urine of aristolochic-acid-treated mice. Electrophoresis. 2009;30:1168–1174. doi: 10.1002/elps.200800548. [DOI] [PubMed] [Google Scholar]
- 9.Clifton JG, Huang F, Kovac S, Yang X, Hixson DC, Josic D. Proteomic characterization of plasma-derived clotting factor VIII-von Willebrand factor concentrates. Electrophoresis. 2009;30:3636–3646. doi: 10.1002/elps.200900270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishino M, Kurasawa Y, Evans R, Lin SH, Brinkley BR, Yu-Lee L. NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr Biol. 2006;16:1414–1421. doi: 10.1016/j.cub.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 11.Byrd MP, Zamora M, Lloyd RE. Translation of eukaryotic translation initiation factor 4G1 (eIF4G1) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J Biol Chem. 2005;19:18610–18622. doi: 10.1074/jbc.M414014200. [DOI] [PubMed] [Google Scholar]
- 12.Darzynkiewicz Z, Juan G, Bedner E. Determining cell cycle stages by flow cytometry. Curr Protoc Cell Biol. 2001;Chapter 8(Unit 8.4):1–18. doi: 10.1002/0471143030.cb0804s01. [DOI] [PubMed] [Google Scholar]
- 13.Watson JV, Chambers SH, Smith PJ. A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry. 1987;8:1–8. doi: 10.1002/cyto.990080101. [DOI] [PubMed] [Google Scholar]
- 14.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Sivan G, Elroy-Stein O. Regulation of mRNA translation during cellular division. Cell Cycle. 2008;7:741–744. doi: 10.4161/cc.7.6.5596. [DOI] [PubMed] [Google Scholar]
- 16.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 17.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen PM, Graslund S, Betz R, Stahl S, Larson C, Hoog C. Characterization of the human APC1, the largest subunit of the anaphase promoting complex. Gene. 2001;262:51–59. doi: 10.1016/s0378-1119(00)00511-4. [DOI] [PubMed] [Google Scholar]
- 19.Lindon C. Control of mitotic exit and cytokinesis by the APC/C. Biochem Soc Trans. 2008;36:405–410. doi: 10.1042/BST0360405. [DOI] [PubMed] [Google Scholar]
- 20.Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao WM, Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J Biol Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
- 22.Hickson GRX, O'Farrell PH. Anillin: a pivotal organizer of the cytokinetic machinery. Biochem Soc Trans. 2008;36:439–441. doi: 10.1042/BST0360439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aumais JP, Williams SN, Luo W, Nishino M, Caldwell KA, Caldwell GA, Lin SH, Yu-Lee L. A role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J Cell Sci. 2003;116:1991–2003. doi: 10.1242/jcs.00412. [DOI] [PubMed] [Google Scholar]
- 24.Zhou T, Aumais JP, Liu X, Yu-Lee L, Erikson RL. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev Cell. 2003;5:127–138. doi: 10.1016/s1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 25.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discovery. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 26.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramstsov N, Qian X, Mills E, Berghs SC, Carey N, Finn PW, Collins LS, Rumber A, Ritchie JW, Jensen PB, Lichenstein HS, Sehested M. Determination of the class and isoform selectivity of small molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Rodas G, Brosch G, Golderer G, Lindner H, Grobner P, Loidl P. Enzymes involved in the dynamic equilibrium of core histone acetylation of Physarum polycephalum. FEBS Lett. 1992;296:82–86. doi: 10.1016/0014-5793(92)80408-9. [DOI] [PubMed] [Google Scholar]
- 28.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang X, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 30.Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24:1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 31.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signaling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 32.Groppo R, Richter JD. Translation control from head to tail. Curr Opin Cell Biol. 2009;21:444–451. doi: 10.1016/j.ceb.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marintchev A, Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: Implications for the structure and function of eIF4G. Biochemistry. 2005;44:12265–12272. doi: 10.1021/bi051271v. [DOI] [PubMed] [Google Scholar]
- 34.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sivan G, Kedersha N, Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol. 2007;27:6639–6646. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Grosse F. Domain structure of human nuclear DNA helicase II (RNA helicase A) J Biol Chem. 1997;272:11487–11494. doi: 10.1074/jbc.272.17.11487. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen PM, Brundell E, Starborg M, Hoog C. A subunit of the anaphase promoting complex is a centromere-associated protein in mammalian cells. Mol Cell Biol. 1998;18:468–476. doi: 10.1128/mcb.18.1.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 39.van Leuken R, Clijsters L, Wolthuis R. To cell cycle, swing the APC/C. Biochim Biophys Acta. 2008;1786:49–59. doi: 10.1016/j.bbcan.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Baker DJ, Dawlaty MM, Galardy P, van Deursen JM. Mitotic regulation of the anaphase promoting complex. Cell Mol Life Sci. 2007;64:589–600. doi: 10.1007/s00018-007-6443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Avino PP. How to scaffold the contractile ring for a safe cytokinesis - lessons from anillin-related proteins. J Cell Sci. 2009;233:1071–1079. doi: 10.1242/jcs.034785. [DOI] [PubMed] [Google Scholar]
- 42.Aumais JP, Tunstead JR, McNeil R, Schaar B, McConnell SK, Lin SH, Clark GD, Yu-Lee L. NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J Neurosci. 2001;21:RC187–RC193. doi: 10.1523/JNEUROSCI.21-24-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreau N, Aumais JP, Prudhomme C, Morris SM, Yu-Lee L. NudC expression during amphibian development. Int J Dev Biol. 2001;45:839–843. [PubMed] [Google Scholar]
- 44.Robbins AR, Jablonski SA, Yen TJ, Yoda K, Robey R, Bates SE, Sackett DL. Inhibitors of histone deacetylases alter kinetochore assembly by disrupting pericentromeric heterochromatin. Cell Cycle. 2005;4:717–726. doi: 10.4161/cc.4.5.1690. [DOI] [PubMed] [Google Scholar]
- 45.Dowling M, Voong KR, Kim M, Keutmann MK, Harris E, Kao GD. Mitotic spindle checkpoint inactivation by trichostatin A defines a mechanism for increasing cancer cell killing by microtubule-disrupting agents. Cancer Biol Ther. 2005;4:197–206. [PubMed] [Google Scholar]
- 46.Stevens FE, Beamish H, Warrener R, Gabrielli B. Histone deacetylase inhibitors induce mitotic slippage. Oncogene. 2007;27:1345–1354. doi: 10.1038/sj.onc.1210779. [DOI] [PubMed] [Google Scholar]
- 47.Magnaghi-Jaulin L, Eot-Houllier G, Fulcrand G, Jualin C. Histone deacetylase inhibitors induce premature sister chromatid separation and override the mitotic spindle assembly checkpoint. Cancer Res. 2007;67:6360–6367. doi: 10.1158/0008-5472.CAN-06-3012. [DOI] [PubMed] [Google Scholar]
- 48.Ma Y, Cai S, Cai S, Lu Q, Lu X, Jiang Q, Zhou J, Zhang C. Inhibition of protein deacetylation by TSA impairs microtubule-kinteochore attachment. Cell Mol Life Sci. 2008;65:3100–3109. doi: 10.1007/s00018-008-8237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mateo F, Vidal-Laliena M, Canela N, Busino L, Martinez-Balbas MA, Pagano M, Agell N, Bachs O. Degradation of cyclin A is regulated by acetylation. Oncogene. 2009;28:2654–2666. doi: 10.1038/onc.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi E, Choe H, Min J, Choi JY, Kim J, Lee H. BubR1 acetylation at prometaphase is required for modulating APC/C activity and timing of mitosis. EMBO J. 2009;28:2077–2089. doi: 10.1038/emboj.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choudhary C, Kumar C, Gnad F, Nielsen M, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 52.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 53.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]


