Abstract
Plasma of the striped bass (Morone saxatilis) contains a fucose-specific lectin (MsaFBP32) that consists of two F-type carbohydrate recognition domains (CRDs) in tandem. The crystal structure of the complex of MsaFBP32 with l-fucose reported here shows a cylindrically-shaped 81 Å-long and 60 Å-wide trimer divided into two globular halves, one containing N-terminal CRDs and the other C-terminal CRDs. The resulting binding surfaces at the opposite ends of the cylindrical trimer have the potential to cross-link cell surface or humoral carbohydrate ligands. The N- and C-CRDs of MsaFBP32 exhibit significant structural differences, suggesting that they recognize different glycans. An analysis of the carbohydrate binding sites by computer modeling provides the structural basis for the observed specificity pattern of MsaFBP32 for simple carbohydrates, and suggests that the N-CRD recognizes more complex fucosylated-oligosaccharides and with a relatively higher avidity than the C-CRD. The modeling of MsaFBP32 complexed with fucosylated-glycans that are widely distributed in prokaryotes and eukaryotes rationalizes the observation that binary tandem CRD F-lectins function as opsonins by cross-linking “non-self” and “self” carbohydrate ligands, such as sugar structures displayed by microbial pathogens and glycans on the surface of phagocytic cells from the host.
Keywords: Animal lectins, Fucolectins, Fucose, Lewis groups, Carbohydrate Binding, Modeling, Crystal Structure
INTRODUCTION
Molecular recognition of glycans by lectins plays an important role in diverse biological processes1. In the immune system, lectins mediate cell-cell and host-pathogen recognition2; 3; 4; 5 as well as key regulatory functions5. Members from several lectin families (including C-type, I-type, and galectins) exhibiting distinct protein folds have been well characterized in this regard6.
F-type lectins (F-lectins) constitute a newly identified family of fucosyl-binding lectins7; 8 with members initially described in the Japanese horseshoe crab9, and later as “fucolectins” in fish8; 10; 11; 12. Interestingly, the F-lectin family appears so far to be restricted to phyla that either lack an immunoglobulin-based adaptive immune system, or are highly reliant on innate immune mechanisms for defense against infectious challenges8; 9; 10. The single carbohydrate recognition domain (CRD) of the eel F-lectin (Anguilla anguilla agglutinin, AAA) folds with a jelly-roll β-barrel topology7. AAA recognizes the l-αFucose in a shallow depression, where the axial 4-OH group is strongly recognized by the residues of the fucose binding motif (HX26RX5-6R) that characterizes these lectins7. The AAA physiological unit forms a homotrimer with all binding sites oriented facing the same plane. Similarly folded domains are also observed in carbohydrate binding modules (CBMs) of bacterial enzymes13; 14; 15, in the C1 and C2 domains coagulation Factors V and VIII, and in a variety of other proteins without significant sequence similarity broadly involved in membrane adhesion, suggesting functional convergence in the evolution of the F-fold7. Although most F-lectins appear to be secreted, some otherwise homologous proteins predicted to be membrane receptors10 probably lack specificity for fucosyl ligands, as suggested by the absence of the canonical F-lectin fucose binding motif.
Individual lectin-carbohydrate interactions are weak as compared to those mediated by other molecules involved in immune recognition, such as immunoglobulins. Simultaneous recognition by multiple CRDs, either arranged in oligomeric associations of lectin subunits or in a single lectin polypeptide, generates an increased avidity towards clustered carbohydrates despite of a relatively weak affinity for the isolated carbohydrate16; 17. For example, collectins mediate complement activation upon binding the pathogen by a “bouquet” arrangement of C-type CRDs18. Several F-lectins display a tandem arrangement of CRDs in their polypeptide sequence8. F-Lectins having multiple CRDs have been identified in bacteria7, fish and amphibians8; 10 and have the potential for greater structural and functional complexity than single-CRD F-lectins such as AAA. Clearly, the crystal structure of AAA does not fully illustrate the diversity of potential binding site interactions or the diversity of quaternary structures that F-lectins may assume.
MsaFBP32, an F-lectin present in striped bass (Morone saxatilis) plasma, is an example of a protein with two tandemly arranged domains8. MsaFBP32 contains an N-terminal CRD (N-CRD) that resembles the eel lectin, whereas the C-terminal CRD (C-CRD) is significantly different. The similarity between the MsaFBP32 CRDs and AAA suggests that the bass lectin is likely trimeric, although the hydrodynamic data and cross-linking experiments suggest monomeric behavior8.
The Streptococcus pneumoniae virulence factor spGH98 contains three F-type CRDs in tandem, each CRD sequence being approximately 25 % identical to that of AAA7. The crystal structures of isolated spGH98 CRDs bound to fucose and fucosylated glycans were determined15. The spGH98’s F-lectin fold and its affinity for fucose confirm an earlier prediction based on to the presence of the F-lectin sequence motif7. Unfortunately, the crystals of the isolated spGH98’s CRDs do not provide information about the quaternary structure of CRDs in the full-length protein.
The crystal structure of the slime mold (Dictyostelium discoideum) discoidin II (DiscII)19, which is expressed on the cell surface of the amoeba stage, shows a two-CRD chimeric structure, with an N-terminal domain that folds as an F-lectin and a C-terminal domain resembling the Helix pomatia agglutinin (HPA) trimer. Interestingly, DiscII shows a trimeric arrangement of molecules similar to that of the AAA homotrimer19.
To further characterize the structural/functional relations of this unique lectin family we have determined the crystal structure of MsaFBP32 bound to l-fucose at 2.3 Å resolution. Interestingly, under crystallization conditions the striped bass lectin forms a homotrimer of parallel monomers with tandem CRDs oriented tail-to-tail with their three N- and C-terminal binding sites facing in opposite directions (each triplet of CRDs resembles the AAA trimer). This arrangement of binding sites revealed by the crystal structure suggests a potential for cross-linking different humoral or cell-associated fucosylated glycoconjugates.
To analyze the effects of differences between the CRDs in a tandem F-lectin subunit might have on carbohydrate recognition, well characterized oligosaccharides displaying non-reducing terminal Fucα(1-2,3,4) such as Lewis and H blood antigens were modeled in both MsaFBP32’s recognition sites. The Fucα(1-2,3,4) moiety is present not only on glycans from eukaryotes but also on their microbial pathogens20. The models suggested that MsaFBP32 tightly recognizes the terminal fucose in both Lewis and H blood antigens via its fucose recognition motif residues, while their other carbohydrate moieties only contact the CRD through a few additional residues from the surrounding loops (cdr1, 2 and 4; extended site). These few interactions provide a rationale for the observed pattern of carbohydrate specificity of MsaFPB328, and suggest that it has a broader binding spectrum than that of AAA. Remarkably, the differences between the landscapes of the two recognition sites of MsaFBP32 point to their distinct fine specificity, and support the notion that this lectin mediates innate immune functions as an opsonin, by cross-linking microbial pathogens to the phagocytic host cells.
RESULTS
Structure determination
Crystals of the MsaFBP32 complex with fucose were obtained as described in the Experimental Procedures section. The crystal structure of the complex was determined at 2.3 Å resolution (Figure 1). Non protein electron densities in each binding site were interpreted as bound l-fucose molecules (Fig. 1c). The final model of the crystal asymmetric unit, containing a trimeric association of MsaFBP32 monomers (amino acids 1 to 293) with two fucose molecules and two Ca2+ ions bound to each monomer plus 418 waters molecules and three Cl− ions, was refined to an R value of 0.18 and R-free value of 0.24 (Table 1).
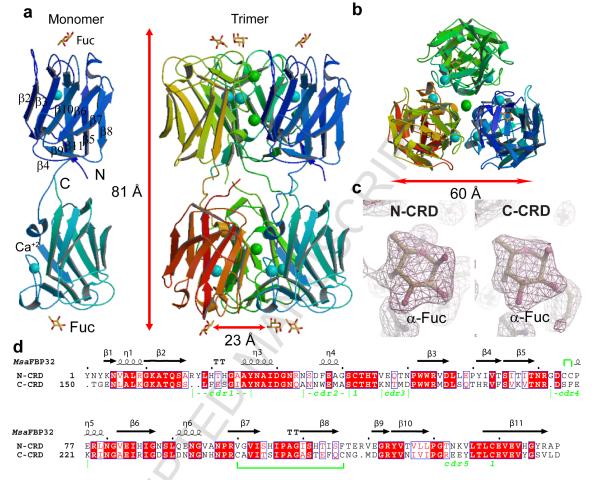
Figure 1. Structure of MsaFPB32.
a) View of MsaFPB32 asymmetric unit of the crystal (trimer) and of an isolated monomer. b) top view of the ASU trimer (N-CRD side). Monomers are colored with different hues. l-Fucose atoms are colored yellow for carbon atoms, and red for oxygen atoms. Ca+2 ions are colored cyan and Cl− ions are colored green. c) 2mFo-DFc sigmaA-weigthed electronic density at the CRDs binding sites. The magenta map is contoured at 0.35 e/Å3. d) Sequence alignment of the two MsaFPB32 CRDs resulting in a 48.6% and a 51% of identity for the N- and the C-CRD respectively. The first line indicates the observed secondary structure elements of each CRD. Coils indicate 3-10 helices, T indicates turns and arrows indicate β-sheets strands. Red solid boxes over the sequences indicate identity and boxed red characters indicate amino acids with similar polarity. The last line shows the positions of the different cdrs. Disulfide bridges are indicated either by number “1” under the connected cysteines or a green line between them when are within one CRD.
Table 1.
Data collection and refinement statistics
| MsaFPB32/Fuc | |
|---|---|
| Data collection | |
| Space group | P43212 |
| Cell dimensions | |
| a, b, c (Å) | 88.1, 88.1, 230.2 |
| Resolution (Last shell) (Å) | 41.1-2.3 (2.4-2.3) |
| R sym | 0.11 (0.47) |
| I/σ(I) | 26.6 (4.2) |
| Completeness (%) | 96.7 (76.5) |
| Redundancy | 4.1 (3.6) |
| No of Unique Refl. | 38891(4612) |
| Β-factor Wilson plot estimated (Å2) | 26.7 |
| Refinement | |
| Resolution (Å) | 41.1-2.3 |
| No. reflections | 38891 |
| Rwork/ Rfree | 0.18, 0.24 |
| N° atoms | |
| Protein | 6822 |
| Ligand/ions | 75 |
| Waters | 418 |
| B-factors (Å2) | |
| Protein | 28.0 |
| Ligand/ions | 30.1 |
| Waters | 29.3 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.4 |
| Ramachandram | |
| Favored regions (%) | 94.2 |
| Allowed regions (%) | 99.4 |
Description of the monomer
In the complex, the MsaFBP32 monomer folds as an extended tail-to-tail arrangement of the two F-type CRDs (total length 81 Å) such that the carbohydrate recognition sites are at opposite ends of the elongated structure (Fig. 1a). The N-terminal domain spans residues 1 to 147 and the C-terminal domain residues 153 to 293. The N- and C-termini are in close proximity in the middle of the molecule but do not interact extensively. Residues 148 to 152 of the linker that connects the two CRDs are in an extended conformation such that there are only weak interactions between the two domains: the area buried at the domains’ interface is only 910 Å2 (7% of the total area) per monomer.
The overall structure of the N-CRD is highly similar to that of the C-CRD (rms of 1.06 Å for 144 Cα atoms). However, small but significant differences exist between the binding sites of the two domains (see below). Each domain folds as the AAA F-lectin, a β-barrel with a jelly-roll topology, consisting of eight major antiparallel β-strands arranged in two β-sheets, one of five strands (β2, β3, β10, β6, and β7) and one of three strands (β8, β5 and β11) (Fig. 1). In each CRD, five loops, the carbohydrate complementarity determining residues (cdr1-5), connect the two sheets and encircle a hollow that binds the fucose. These loops span the following residues: 16 to 27 (N-cdr1), 33 to 42 (N-cdr2), 45 to 50 (N-cdr3), 71 to 78 (N-cdr4) and 130 to 134 (N-cdr5) in the N-CRD, and, the following residues: 165 to 174 (C-cdr1), 180 to 188 (C-cdr2); 193 to 196 (C-cdr3), 217 to 223 (C-cdr4) and 273 to 279 (C-cdr5) in the C-CRD. Each CRD, as in the AAA F-lectin, bind a Ca+2 atom that contributes to maintain the conformation of cdr1 and cdr27. In addition, a similar combination of disulfide bridges and salt bridges holds the CRDs structure together.
Description of the trimer
The three MsaFPB32 molecules in the asymmetric unit of the crystal are related by a local 3-fold axis of symmetry (average rms difference between any two monomers is 0.4 Å) and form a cylindrical trimer 83 Å long and 60 Å in diameter (Fig 1). The trimer is divided into two halves, one containing the three N-CRDs (N-trimer), the other three C-CRDs (C-trimer). The top and bottom ends of the cylindrical trimer show a three-fold arrangement of carbohydrate binding sites. All six CRDs contain a bound Ca2+ ion. Three Cl− ions present on the 3-fold axis (two in the N-trimer and one on the C-trimer) are coordinated by positively charged residues of 3-fold related symmetry mates: in the N-CRD the three Lys 11 coordinates Cl− (1), the three Arg 17 residues coordinates Cl− (2) and in C-CRD the three Lys 159 residues coordinates Cl− (3). Similar features are present in the AAA crystal structure. The large accessible surface area buried upon trimer formation: 7702 Å2 (3916 Å2 in the N-CRD half and 3786 Å2 in the C-CRD half) that correspond to 20% of the total area of three separated monomers, suggests that the MsaFBP32 trimer is the physiological state.
Carbohydrate binding sites
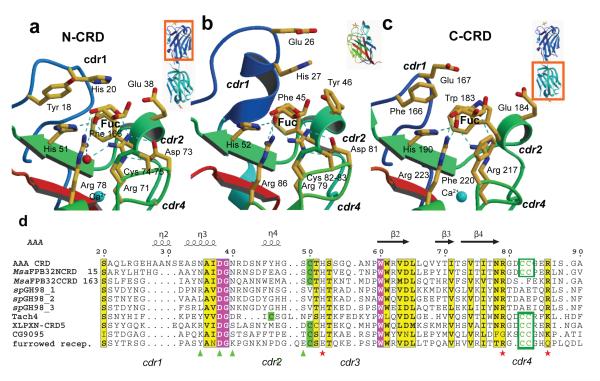
All six carbohydrate binding sites in the asymmetric unit of the crystal are occupied, with a single fucose per binding site (Fig. 1). The full range of interactions that characterize the fucose-recognition by AAA F-lectins is observed in each CRD of striped bass: polar interactions of the fucose axial 4-OH with His 44, Arg 71, and Arg 78 of the motif HX26RX5-6R (and corresponding residues His 190, Arg 217, and Arg 223 in C-CRD), and apolar interactions of C6 and the face B of the carbohydrate with the protein (Fig. 2a,c). Arg 71 also makes an H-bond to O5 and Arg 78 to the 3-OH group of the sugar. However, there are significant sequence and structure differences between the recognition sites of the N- and C-CRD. It is evident that the N-CRD binding site resembles the AAA site more closely than the C-CRD site (Fig. 2). Although, C6 group pockets of MsaFPB32 are more solvent accessible than the one of AAA, the C-CRD pocket is less open than its counterpart due to the replacement of Phe 37 in N-CRD by the bulkier Trp 183. In addition, in the C-cdr4, Phe 220 makes an apolar contact with the fucose face B that in the N-CRD is provided by the unusual S-S bridge formed by consecutive cysteines (Cys 74 - Cys 75). The bulkier phenylalanine makes a close contact (distance 3.2 Å) with the fucose 2-OH group, shifting and tilting the monosaccharide out of the shallow binding pocket compared to the fucose on the N-CRD (Fig. 2c).
Figure 2. Comparison of the two binding sites between MsaFPB32 and AAA.
a) Side by side comparison of the fucose recognition by the (left) striped bass N-CRD (middle) AAA CRD and (right) striped bass C-CRD. b) Sequence alignment of binding site regions of striped bass lectin (MsaFPB32), Anguilla Anguilla Agglutinin (AAA) and other protein homologs: Streptococcus pneumoniae virulence factor spGH98, Horse-shoe crab Tachilectin-4 (tach4), Xenopus Laevis pentraxin (XLPXN-CDR5) and two Drosophila Melanogaster proteins: CG9095 and the furrower receptor.
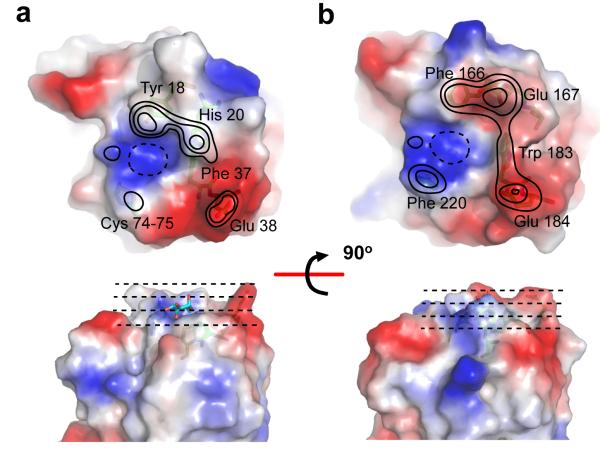
The solvent accessible surfaces (Figure 3) show the landscape of the MsaFPB32 recognition sites: a positively charged and shallow depression that recognizes an l-fucose (primary recognition site), is encircled by features, including ridges and valleys, which may contact epitopes of more complex carbohydrates (extended recognition site). For example, cdr1 residues form features that could contact a carbohydrate third moiety. An apolar patch between the bound fucose and the negatively-charged ridge of cdr2 defines a region that can accommodate an α-linked fourth moiety of the carbohydrate. Features of the cdr4 contact the first moiety of the carbohydrate and also large substituent groups of the second moiety. The landscapes of the N- and C-CRD recognition sites show remarkable differences: N-cdr1 residues, Tyr 18 and His 20, form a polar ridge and a valley different from the more apolar and broader ridge formed by C-cdr1 residues, Phe 166 and Glu 167. In the C-cdr2, the indole ring of Trp 183 make a protruding feature, not present in the N-CRD, which connects the C-cdr1 with the C-cdr2 ridge. In the N-cdr4, the disulfide-bridge forms a flat apolar surface; instead, in the C-cdr4, Phe 220 forms a protruding feature that narrows the recognition site and blocks substitution at the 2-OH of the fucose (Fig. 3b).
Figure 3. Landscape of the carboydrate recognition sites in MsaFPB32.
Solvent accessible surface of (a) N-CRD and (b) C-CRD recognition sites. The figure shows the top-view (top panel) and the side-view (bottom panel) recognition sites. The surfaces were colored using the electrostatic potential at the surface as criteria. Contour levels represent approximated elevations from the bottom of the binding site (drawn with dashed lines), where the axial 4-OH of the fucose binds. The second level corresponds to the elevation of the ring fucose. The third level corresponds to the elevation of the glycosidic bond oxygen and the two additional levels were drawn at equally spaced from the third. The external ring of the lowest level (dashed line) was removed on the top views for clarity.
Modeling the recognition of fucosylated-carbohydrates by MsaFBP32
To analyze the effect of the CRDs differences in their carbohydrate specificity, computer models of complexes of the lectin with well-characterized fucosylated carbohydrates were built based on the crystal structure reported here.
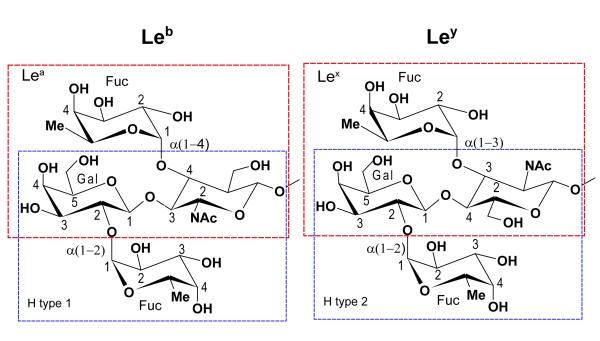
The number and density of interactions between the fucose and the binding site of MsaFPB32 observed in the structure of the complex, support a relatively strong primary recognition of the terminal fucose of a complex oligosaccharide by F-lectin compared to the secondary recognition of other carbohydrate moieties by the extended binding site. The binding site plus the adjacent area (Fig. 4) covers an approximated area slightly wider than the footprint of a fully fucosylated tetrasaccharide, allowing the interaction with the glycan backbone disaccharide and two terminal α-linked fucose units. The alpha-linked fucose units protrude from the glycan backbone and are easily recognized by lectins21. Interactions with other moieties extending the glycan backbone are expected to be weaker as the ones A-antigen in ref. 15. Lewis and H antigens contain fucose linked in α(1-2/3/4) to the Galβ(1-4/3)GlcNAc backbone. They are present in naturally occurring glycans from vertebrates and invertebrates, in eukaryotic parasites, and in pathogenic bacteria in which putative α(1-2) and α(1-3/4) fucosyl transferases have been identified (see ref. 20 for a review).
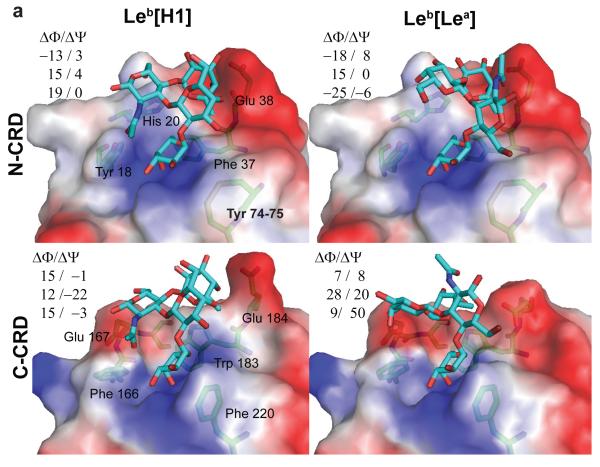
Figure 4. Models of the complexes of MsaFBP32 with tetrasaccharide antigens.
(Top) Leb and Ley bound to the N-CRD carbohydrate recognition site using each of the fucose terminals corresponding to the constituent trisaccharide (H and Lewis). (Bottom) Leb and Ley bound to the C-CRD carbohydrate recognition site. ΔΦ and ΔΨ are the differences of glycoside bond torsion angles between the free and bound conformation. Φ and Ψ are defined using the IUPAC-IUB convention, Φ = Θ(O[ring] – C1 – O[x] – C[x]) and Ψ = Θ (C1—O[x] – C[x] --- C[x-1]).
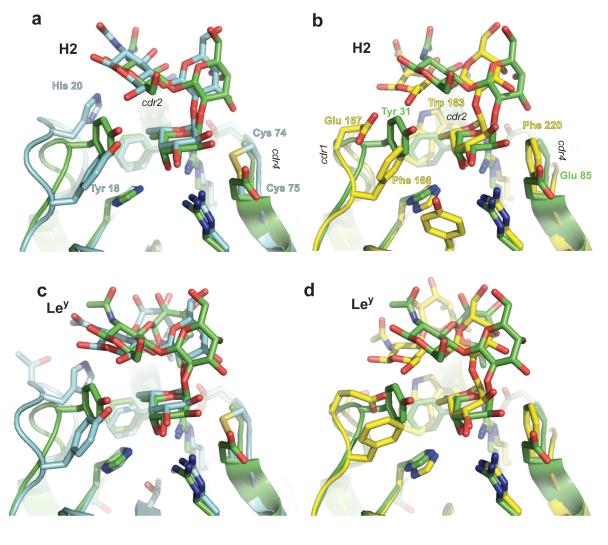
Figure 4 shows the models of complexes of both striped bass CRDs with two Lewis blood-group antigens, Lewis-b tetrasaccharide (Leb) and Lewis-y tetrasaccharide (Ley). These antigens can be divided in two different fucosylated-trisaccharides each (Fig. 5): Leb in H type 1 (H1) and Lewis-a (Lea) antigens, and Ley in H type 2 (H2) and Lewis-x (Lex) antigens. Each trisaccharide contains a terminal α-linked fucose that can bind to the primary recognition site. Consequently, in the case of the tetrasaccharides both modes of binding of the lectin, either with the fucose of the H-antigen –α2 linked-- or with the fucose of the Lewis-antigen – Lex α(1-3) linked or Lea α(1-4) linked--, were analyzed. Glycosidic angle differences in Figure 4 and the rms deviations in Table 2 quantify the changes in conformation between free and bound carbohydrates. Leb and Ley bind to the N-CRD with smaller perturbations of their conformation than when they bind to the C-CRD. In the case of the N-CRD/antigen complexes (Fig. 4), the bound carbohydrate conformations are close to those observed in other crystal structures such as the F-lectin-like spGH98 CBM (PDB accession codes: 2J1T, 2J1V and 2J1U15) and other lectins (PDB accession codes: 1W8H22; 1SL623) or close to those conformations predicted to occur in solution by theoretical calculations24. Figure 7 shows the overlap between the predicted H2 and Ley binding and the crystal structure of spGH98 complexes with these carbohydrates. In the N-CRD case (Figure 7a,c) the conformation of the carbohydrates is similar.
Figure 5. Blood group antigens.
Each tetra-saccharide can be divided in two distinct fucosylated tri-saccharides: Leb in a H type-1 tri-saccharide --Fucα2Galβ3GlcNAc— and in a Lewis-a (Lea) tri-saccharide --Galβ3[Fucα4GlcNAc]— and Ley in a H type-2 tri-saccharide --Fucα2Galβ4GlcNAc— or in a Lex tri-saccharide --Galβ4[Fucα3GlcNAc]—.
Table 2. Modeling MsaFPB32/Antigen interactions.
Summary of additional carbohydrate epitopes recognized by the two bass lectin CRDs and the rms deviations between the free and bound carbohydrate conformations. For the rms calculations, 36 and 46 heavy-atoms of the carbohydrates were used in the cases of trisaccharides or tetrasaccharides, respectively.
| CRD | cdr | Additional interactions: type / antigen-Moiety/ (Moiety-groups) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Resid. | H1 | H2 | Lea | Lex | Leb | Ley | ||||
| H1(¥) | Lea | H2 | Lex | |||||||
| N-CRD | cdr1 | Tyr 18 | GlcNAc (O7) |
None | Gal (6-OH) |
Gal (6-OH) |
GalNAc (O7) |
None | None | Gal (4-OH or 6-OH) |
| His 20 | GlcNAc (6-OH) |
None | Gal (4-OH) |
Gal (4-OH) |
GalNAc (6-OH) |
Apolar Fuc_2 (Face B) |
GlcNAc (2-NAc) |
Gal (3-OH or 4-OH) |
||
| cdr2 | Phe 37 | None | None | None | None | Apolar Fuc_2 (C6) |
Apolar Fuc_2 (C6) |
Apolar Fuc_2 (C6) |
Apolar Fuc_2 (C6) |
|
| Glu 38 | None | None | None | None | Fuc_2 (4-OH, 3-OH) |
None | Fuc_2 ( 4-OH) |
Fuc_2 (4-OH) |
||
| cdr4 | Cys 74 Cys 75 |
None | None | None | Apolar GlcNAc (2-NAc) |
None | None | None | Apolar GlcNAc (2-NAc) |
|
| rms (Å) | 0.63 | 0.4 | 0.5 | 0.3 | 0.73 | 0.7 | 0.43 | 0.5 | ||
| C-CRD | cdr1 | Phe166 |
Steric
GlcNAc (O7) |
None |
Steric
Gal (4-OH) |
Steric
Gal (4-OH, 5-COH) |
None |
Steric
Gal (4-OH) |
None |
Steric
Gal (4-OH) |
| Glu167 | GlcNAc (β1-OH) |
GlcNAc (6-OH) |
Gal (3-OH, 4-OH) |
Gal (3-OH, 4-OH) |
GlcNAc (β1-OH) |
Gal (3-OH) Fuc_2 (2-OH) |
None | Gal (3-OH) Fuc_2 (2-OH) |
||
| cdr2 | Glu184 | None | None | None | None | Polar Fuc_2 (4-OH) |
Polar Fuc_2 (4-OH) |
Fuc_2 (4-OH) |
None | |
| Trp183 | None | Apolar GlcNAc (face B) |
Gal (2-OH) |
None | None | Polar Fuc_2 (4-OH) |
Steric Fuc_2 GlnNAc |
Polar Fuc_2 (O5) |
||
| cdr4 | Phe220 | None | None | None | Steric GlnNAc (2-NAc) |
None | None | None | None | |
| rms (Å) | 0.54 | 0.7 | 0.7 | 1.0 | 0.78 | 0.71 | too large |
1.0 | ||
Trisaccharide whose Fuc is recognized by the CRD.
Figure 7. Comparison of the modeled complexes with the spGH98 complexes with H2 and Ley.
The models of fucosylated carbohydrates bound to the MsaFBP32 CRDs are compared to the same complexes observed for the CBM of spGH98. a) and b) corresponds to the complexes of the N- and C-CRD respectively with H2 (PDB accession 2J1V). c) and d) the same with Ley (2J1T). Carbon atoms, labels, and secondary structure are colored blue and yellow for MsaFBP32’s N- and C-CRD respectively and green for the spGH98 CBM.
The carbohydrates were docked readily in the binding sites of the lectin with their terminal fucose coinciding with that in the experimental MsaFPB32 structure. The analysis of the models identifies several residues of cdr1, 2 and 4 as participating in the secondary recognition of additional moieties of the carbohydrate: Tyr 18, His 20, Glu 38, Cys 74 and Cys 75 in the N-CRD, and Phe 166, Glu 167, Trp 183, Glu 184 and Phe 220 in the C-CRD. The modeled carbohydrates present their epitopes to binding site features in each CRD that have distinctive polar characteristics. The N-cdr1 has two H-bond forming groups, Tyr 18 and His 20, but, C-cdr1 has only one, Glu 167. At neutral pH, His 20 has its two nitrogen atoms free to interact alternatively with groups of the carbohydrate, one as proton acceptor and the other as a proton donor, allowing this residue to make H-bonds with a nitrogen atom or an OH group. Glu 167 of C-cdr1 is an H-bond acceptor and is expected to interact only with hydroxyls or other donor epitopes. Cdr4 interactions are limited to the primary fucose, potentially affecting all others, and to the 2-NAc group on the second moiety, if present. C-cdr4 Phe 220 tilts the carbohydrate against C-cdr1, enhancing the hindrance of cdr1 upon the carbohydrate (Fig. 4 bottom panels). Favorable direct polar interactions between Glu 38 (Glu 184 in the C-CRD) and hydroxyls groups of the second fucose are suggested by the model.
A preliminary rigid docking of Lewis tetrasaccharides on the MsaFPB32 binding sites suggests a high complementarity between these carbohydrates and the N-terminal site (results not shown). Models allowing flexible docking of ligands and an induced-fit of the binding site confirm that the Lewis antigens bind the N-CRD recognition site with only small disturbances in the protein or in the carbohydrate conformation (Fig. 4 top panels).
For example Lewis antigens with their Lewis-trisaccharides bound --using the α(1-3/4)) linked Fuc -- make at least two hydrogen-bonds with the N-cdr1 using the Gal groups 6-OH and 4-OH. The N-acetyl group of the GlcNAc moiety of Lex bound epitopes lies snugly on top of the N-cdr4. The α(q,2)–linked fucose of the tetrasaccharides lies in a pocket on top of the N-cdr2 Phe 37, which provides an apolar contact surface for its C6 group.
In contrast, the models show that binding of the Lewis tetrasaccharides to the C-CRD recognition site requires relatively large adjustments of both the carbohydrates and the binding site conformations to accommodate interactions between the third carbohydrate moiety and C-cdr1 and between the second fucose and Trp 183. The C-CRD recognition site cannot accommodate Lex epitopes, due to the hindrance of Phe220 upon GlcNAc’s 2-NAc group. In the C-cdr2 the bulky Trp 183 fills the C6-pocket hindering binding of the second fucose. However, in the case of Leb bound to the C-CRD using the α(1-4 ) linked fucose, Trp 183 might provide an additional H-bond to axial 4-OH of the other fucose (see Table 2). Together, these two features appear to select against binding of Lewis tetrasaccharides to the C-terminal domain of striped bass lectin.
H1 trisaccharides present a very polar face to the cdr1: the GlcNAc groups O5, β1-OH, 6-OH and O7. These groups may form H-bonds to Tyr 18 and His 20 of the N-cdr1 or to Glu 167 of the C-cdr1. In contrast, H2 trisaccharides can only interact with the cdr1 of the lectin through the GlcNAc group N6. The NAc group of H2 widens the carbohydrate and accentuates the hindrance of N-cdr1. This group only accepts interactions with N-cdr1 His 20 or C-cdr1 Glu 167. As consequence, binding to MsaFPB32 of H2, alone or as part of Ley, involves few or no H-bond interactions with the N-terminal extended-recognition site.
The modeling analysis suggests that binding of fucosylated antigens larger than disaccharides to the C-CRD is generally disadvantageous.
DISCUSSION
F-lectin sequences from organisms ranging from bacteria to amphibians and fish show F-type domains in arrangements of multiple contiguous CRDs, as well as forming part of mosaic proteins such as the Xenopus pentraxin-fusion protein, and the Drosophila “furrowed” protein and CG9095 putative receptor 7; 8; 10. The structure of the MsaFBP32 is the first one for an F-lectin with tandemly arrayed CRDs. The structure shows that the two CRDs are not arranged in a head-to-tail manner as found in multidomain proteins such as fibronectin and immunoglobulins. Instead, they are arranged tail-to-tail with binding sites pointing in opposite directions. The structure of DiscII also exhibits a similar trimeric subunit arrangement19, although in that case, each monomer of DiscII has two distinct CRDs, the N-terminal resembling an AAA trimer and the C-terminal resembling HPA (an H-type lectin25). The N- and the C-CRD of each MsaFBP32 monomer are only connected to each other via a flexible linker peptide. Given the flexibility of the hinge, it can be expected that only in the trimer does the relative orientation of the binding sites of the N- and C-CRD become fixed, pointing in opposite directions. The extended conformation observed in the crystal appears to be a consequence of the presence of the three linkers (one per chain) that connect the two halves of the trimer. Under conditions that do not favor trimer formation the monomer may be capable of undergoing ample hinge motions around residues 148 to 152 that can easily change the orientation of the binding sites in the two CRDs with respect to each other.
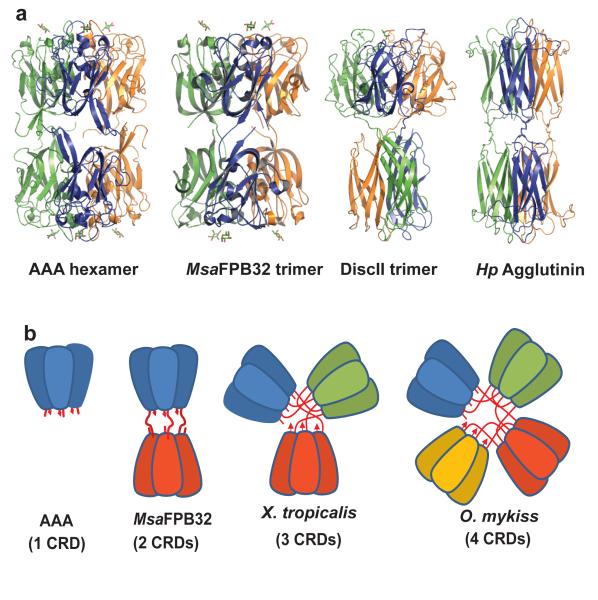
As mentioned above, the physiological AAA trimer and each of the two halves of the MsaFBP32 trimer are highly similar (rms differences: N-trimer to AAA 0.88 Å for 429 Cαs; C-trimer to AAA 1.54 Å for 414 Cαs). This, together with the similarity between the areas buried per MsaFPB32 trimer-half and that in the AAA physiological trimer (3772 Å2), suggest that trimerization is a conserved property of this family of lectins. Moreover, both halves of MsaFPB32 are related by approximate two-fold axes perpendicular to the three-fold axis, such that the trimer has pseudo 32-point group symmetry. Intriguingly, a similar arrangement of the CRDs was observed in the native oligomeric state of AAA and HPA. The AAA-crystal form belongs to space group R32, such that the hexameric CRD arrangement in the crystal unit cell has also 32-point group symmetry. The physiological unit of the HPA crystal is a disulfide cross-linked hexamer that also displays 32-point group symmetry25. The hexamers of AAA, DiscII, and HPA show two flat binding surfaces segregated to opposite ends of the trimer/hexamer, in an arrangement similar to that observed in the MsaFPB32-crystal asymmetric unit (Figure 6). HPA binding sites are 20 Å apart from each other in comparison with the 25 Å of the F-lectin. Alignment of the MsaFBP32 whole trimer with the six AAA molecules in the cell of the crystals show an rms difference of 2.1 Å for 820 Cα-atoms aligned. In light of the MsaFBP32 model, the diametric orientation of AAA trimers -thought to result from optimal packing during crystallization- may actually represent the physiological state of some isoforms of the Japanese eel lectin10 whose CRDs could be covalently linked through their C-terminal cysteines, like those in HPA25.
Figure 6. Quaternary structure of tandemly arrayed related CRDs.
a) side-by-side comparison between AAA hexamer, MsaFPB32 trimer, DiscII trimer and HPA hexamer. b) the quaternary arrangement of AAA and MsaFPB32, and, how similar arrangement may be extended to higher number of tandemly arrayed F-lectin CRDs for example african clawed frog (Xenopus tropicalis) with three CRDs and rainbow trout (Oncorhynchus mykis) with four CRDs.
AAA and MsaFPB32 trimers contain one metal (Ca2+) per domain and several Cl− ions placed on the three-fold axis. The weakly hydrated chloride anion readily forms ion pairs with cationic side chains. In this family of lectins, chloride induced oligomerization may provide a mechanism for increasing avidity when the protein is released to the plasma: the high concentration of chloride in the extracellular environment [~110 mM26] would favor trimerization while low concentration of chloride in the intracellular environment (~3 mM) would favor the monomeric form. It is noteworthy that chloride concentration has a key role in the activation of proteins of the coagulation cascade including Factor Va27. In the case of MsaFBP32, the fact that no trimers were detected in gel filtration and other solution experiments, even in the presence of Cl−, may indicate that binding of an endogenous small molecule is required for additional trimer stabilization in solution. Another possibility is that trimer formation in solution is concentration-dependent as has been observed in other lectins28. Thus, the crystal structures reported here and the native state of AAA, suggest an important role for trimeric association in the physiological functions of these lectins that might depend on their presence in particular microenvironments. This quaternary arrangement generates two distinct recognition surfaces 81Å apart at either end of the trimer, each of which recognizes simultaneously carbohydrates separated by 25 Å. The high similarity between the group of six CRDs in the MsaFBP32 structure with that observed in the AAA unit cell of the crystal suggests that this arrangement, consisting of two sets of three carbohydrate binding sites separated by 81 Å, may also be a functional feature of F-type of lectins with either one (AAA) or two (MsaFBP32) tandemly arranged CRDs. Interestingly, the proximity of the F-lectin fold terminals may allow for quaternary arrangements of F-lectin monomers with multiple (n) tandemly arrayed CRDs as n trimers of segregated CRDs, resulting in collectin-like “bouquet” CRD displays (Figure 6b).
MsaFPB32 CRDs fine specificity
The Cdr1 loop was proposed as a key determinant of oligosaccharide specificity in AAA7. This was based on the hypothesis that oligosaccharide preferences of AAA are a function of its protruding cdr1, which hinders binding of a third carbohydrate moiety. The sequences of the MsaFBP32 cdr1s are shorter than those in AAA, resulting in smaller cdr1s. This led to the suggestion that MsaFBP32 may have a broader specificity than AAA due to a decrease in the number of steric constraints for binding additional moieties of the carbohydrate29. Although this appears to be correct in the case of N-CRD, in the case of C-CRD, the bulkier Phe 220 may amplify the hindrance effect of the C-cdr1, because the phenylalanine tilts the carbohydrate against that loop.
Both MsaFBP32 CRDs conserve the apolar interaction with the fucose C6 provided by an aromatic residue in cdr2 (Phe 37 and Trp 183). However, because in the MsaFBP32 N-CRD the cdr1 is shorter than in AAA, the C6 pocket is more solvent-accessible. Because l-Gal has a 6-OH, this more solvent-exposed pocket may provide a rationale for the observed five-fold higher affinity of MsaFBP32 for l-Gal as compared to AAA8. O-antigens of pathogens containing α-linked l-Fuc, 2-acetoamido l-Fuc (FucNAc), and colitose (3- deoxy-l-fucose) as non-reducing terminal residues30 are probably the natural ligands of MsaFPB32, as proposed for the horseshoe crab F-lectins9. A bulky substitution at C2 as the NAc in FucNAc could establish favorable interactions with the cdr4 of N-CRD, but very unfavorable with the same loop in the C-CRD. l-Rha (6-deoxy-l-mannose), frequently observed in E. coli glycans, has a 2-OH axial group that may also be recognized by the MsaFPB32 primary site, although the 3-OH becomes buried. In the C-CRD, the hindrance to carbohydrate binding by c-cdr2 (Phe 220) is relaxed by the lack of that hydroxyl in l-Rha. Further, Phe 220 provides a hydrophobic contact with the 6-deoxy group.
Recognition of complex fucosylated-carbohydrates by MsaFBP32
Crystal structures and computational studies show consistently that the stable conformations of fucosylated oligosaccharides – such as Lewis-a (Lea) or Lewis-x (Lex) -- involve ring stacking using apolar areas of alternating moieties24; 31; 32. Due to the exo-anomeric effects, α-linkages such as those in H antigens and Lewis (Fucα2Gal and Fucα[3/4]GlcNAc linkages) have more restricted Φ than Ψ glycosidic-bond torsions (see Fig. 4 for torsion definitions). In β-linkages, however, Φ and Ψ torsions have similar flexibility, except when they belong to N-acetylated carbohydrates for which conformational space may be further restricted by the steric effect of the bulky 2-NAc group (i.e. in type-1 chains as Galβ3GlcNAc or type-2 chains as Galβ4GlcNAc). The structures of Ley and Leb have been determined in solution and were found to be essentially rigid molecules for the reasons outlined above33; 34. Lectin recognition sites are expected to be highly complementary to the stable carbohydrate conformations of their preferred carbohydrates because the energy cost of distortions in the carbohydrate or the protein upon ligand binding may overwhelm the small gain in interaction energy. In support of this hypothesis, the comparison of the crystal structures of complexes of spG98H and Fuc to complexes of the same lectin with blood antigens shows negligible changes in the protein binding site resulting from the interactions with the more complex carbohydrate. The N-CRD shows better complementarity to most of the terminal trisaccharides analyzed than the C-CRD (Figure 7), because it accommodates more readily polar interactions with groups that point towards the protein, such as the axial 4-OH and the equatorial 3-OH groups of the Gal moiety of 2′-, 3-fucosyllactoses, and of Lex or Lea. Thus, preference for this type or related carbohydrates can be expected for the N-CRD. However, apolar epitopes such as the C1-O5-C5 edge of the GlcNAc moiety in H-type II antigens have more favorable contact with the C-cdr1 than with the N-cdr1. An α reducing end can ameliorate or reverse this effect, but such a linkage is not expected in the most common oligoglycan backbones.
In the N-CRD, a methyl group of a second fucose may dock on top of Phe 37, but in the C-CRD, Trp 183 closes the pocket with its indole ring (Figure 7). This imposes steric constrains to the second fucose moiety of Lewis tetrasaccharides. Beta-linked carbohydrate moieties that branch from the backbones, like the A or B blood group antigens, extend to regions where no additional interaction with the protein can be expected.
It is possible that MsaFBP32 cross-links fucosylated glycans on bacteria or parasites35 to putative phagocyte receptors containing Lea, a blood group that may be also present in ectotherms36. Pre-exposure of E. coli to a binary tandem F-lectin from sea bass (Dicentrarchus labrax) significantly increases their phagocytosis by peritoneal macrophages relative to the unexposed bacteria, suggesting that F-lectins with multiple CRDs such as MsaFBP32 function as opsonins that mediate innate immune responses against microbial pathogens37.
Summary and conclusions
The back-to-back orientation of the binding surfaces of the trimeric MsaFBP32 supports the notion that the function of this lectin in circulation is to cross-link fucosylated glycoconjugates displayed on different cells with an epitope separation of 25 Å on the cell surface. In the intracellular environment, a low Cl− milieu, the monomer form of the protein will be favored. This form can undergo hinge-like motions around residues 148 to 152 such that the N- and C-CRDs may no longer point in opposite directions. Under these conditions the molecule may have a limited function in cross-linking carbohydrate determinants. It is only in the extracellular environment, at higher Cl− concentration, that the trimer is formed and can mediate cross-linking of carbohydrate moieties that are present on the surface of two different cells or soluble glycans.
Both CRDs of MsaFBP32 maintain the primary recognition of fucose by a triad of basic residues that is strongly conserved throughout the F-lectin family and by the apolar recognition of the fucose C6 group by a binding site pocket. However, the N- and C-CRD display differences in the characteristics of this apolar pocket that explain the differences in the monosaccharide specificity observed. In addition, variations in the extended recognition sites of MsaFBP32 suggest distinct ligand selectivity for fucosylated oligosaccharides between the two CRDs. In this regard, modeling suggests that: (a) the N-CRD can recognize oligosaccharides containing Lewis- and H- trisaccharides, and (b) the C-CRD prefers carbohydrates with less than two terminal Fuc moieties. These novel structural data, together with recent evidence from opsonization experiments suggest that, at least in teleosts, F-lectins cross-link fucosylated surface ligands and aid in “self”/“non-self” recognition in innate immune responses.
Methods
Protein Isolation, Purification and Crystallization
MsaFBP32 was purified from serum of the striped bass, Morone saxatilis as described before8. Crystals of the bass fucolectin were obtained by vapor diffusion at room temperature using the hanging drop method. Drops of equal amounts (1 μl) of protein (10 mg/ml in 10 mM Tris HCl, pH 7.0, 5 mM l-fucose) and reservoir (25 % (w/v) PEG 2K MME, 100 mM NaBicine, pH 9.0) solution were mixed and equilibrated against 1 mL of reservoir solution at 20 °C of temperature.
Crystal Structure Determination
Data collection on flash frozen MsaFB32/Fuc crystals was carried out at 100K using wavelength of 1.1 Å at the X25 beam line of the National Synchrotron Light Source of the Brookhaven National Laboratory. Processing and data reduction were performed with the program HKL2000 (HKL Research Inc.). The structure of the MsaFBP32/Fuc complex was determined by molecular replacement with the program MOLREP 38 using the AAA-CRD (PDB accession id 1K12) as the initial model of half of the MsaFBP32 molecule. The space group was determined to be P43212 by comparing the height of the translation peaks of the two possible enantiomorphs. The final model consisted of six such domains. Rebuilding of the complete molecular structure (residues 1- 293 for each of the three chains) was performed using the program O39 with sigmaA weighted maps. Clear density for a bound fucose was present in all six binding sites. The refinement was carried out with the program CNS v1.140. Area buried calculations were performed with the program NACCESS41.
Computational Modeling of complexes
Computer models of the MsaFBP32/antigens were built using the structure of the MsaFBP32/Fuc complex. The Leb and Ley blood antigens were built and/or modified using the program Quanta (Accelerys™) and docked in MsaFPB32’s CRD using the bound Fuc observed in the crystal structure as a guide; for this modeling it was assumed that they adopted one of the stable conformations calculated for these carbohydrates in solution24. After manual adjustments to improve interactions and avoid clashes, the coordinates of the resulting model were minimized locally (fixing residues not expected to be interacting with other moieties of the antigen) with CHARMm32 (Accelerys™) to optimize the carbohydrates interactions with the protein. Figures were drawn using the molecular graphics programs Molscript42; 43 and PyMol (© 2006 DeLano Scientific LLC).
Acknowledgements
We acknowledge the use of beamlines X25 at the Brookhaven National Laboratory. This work was supported by NIH grants: ARRA-1RO1NS061827 (LMA) and R01 GM070589 (GRV), and by National Science Foundation grants MCB 0077928 and IOS-0822257 (GRV).
Footnotes
Accession Code: Coordinates and structures factors of the MsaFPB32/Fuc complex were deposited in the Protein Data Bank (accession id 3CQO).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sharon N, Lis H. Lectins. Second edit Kuwer Academics; Dordrecht; Boston: 2003. [Google Scholar]
- 2.Drickamer K, Taylor ME. Biology of animal lectins. Annu Rev Cell Biol. 1993;9:237–64. doi: 10.1146/annurev.cb.09.110193.001321. [DOI] [PubMed] [Google Scholar]
- 3.Geijtenbeek TB, van Vliet SJ, Engering A, t Hart BA, van Kooyk Y. Self- and nonself-recognition by C-type lectins on dendritic cells. Annu Rev Immunol. 2004;22:33–54. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- 4.Fujita T, Matsushita M, Endo Y. The lectin-complement pathway--its role in innate immunity and evolution. Immunol Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 5.Rabinovich GA, Toscano MA, Jackson SS, Vasta GR. Functions of cell surface galectin-glycoprotein lattices. Curr Opin Struct Biol. 2007;17:513–20. doi: 10.1016/j.sbi.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabius HJ. Animal lectins. Eur J Biochem. 1997;243:543–76. doi: 10.1111/j.1432-1033.1997.t01-1-00543.x. [DOI] [PubMed] [Google Scholar]
- 7.Bianchet MA, Odom EW, Vasta GR, Amzel LM. A novel fucose recognition fold involved in innate immunity. Nat Struct Biol. 2002;9:628–34. doi: 10.1038/nsb817. [DOI] [PubMed] [Google Scholar]
- 8.Odom EW, Vasta GR. Characterization of a binary tandem domain F-type lectin from striped bass (Morone saxatilis) J Biol Chem. 2006;281:1698–713. doi: 10.1074/jbc.M507652200. [DOI] [PubMed] [Google Scholar]
- 9.Saito T, Hatada M, Iwanaga S, Kawabata S. A newly identified horseshoe crab lectin with binding specificity to O-antigen of bacterial lipopolysaccharides. J Biol Chem. 1997;272:30703–8. doi: 10.1074/jbc.272.49.30703. [DOI] [PubMed] [Google Scholar]
- 10.Honda S, Kashiwagi M, Miyamoto K, Takei Y, Hirose S. b2Multiplicity, structures, and endocrine and exocrine natures of eel fucose-binding lectins. J Biol Chem. 2000;275:33151–7. doi: 10.1074/jbc.M002337200. [DOI] [PubMed] [Google Scholar]
- 11.Cammarata M, Vazzana M, Chinnici C, Parrinello N. A serum fucolectin isolated and characterized from sea bass Dicentrarchus labrax. Biochim Biophys Acta. 2001;1528:196–202. doi: 10.1016/s0304-4165(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 12.Cammarata M, Benenati G, Odom EW, Salerno G, Vizzini A, Vasta GR, Parrinello N. Isolation and characterization of a fish F-type lectin from gilt head bream (Sparus aurata) serum. Biochim Biophys Acta. 2007;1770:150–5. doi: 10.1016/j.bbagen.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Ito N, Phillips SE, Stevens C, Ogel ZB, McPherson MJ, Keen JN, Yadav KD, Knowles PF. Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase. Nature. 1991;350:87–90. doi: 10.1038/350087a0. [DOI] [PubMed] [Google Scholar]
- 14.Gaskell A, Crennell S, Taylor G. The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure. 1995;3:1197–205. doi: 10.1016/s0969-2126(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 15.Boraston AB, Wang D, Burke RD. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J Biol Chem. 2006;281:35263–71. doi: 10.1074/jbc.M607620200. [DOI] [PubMed] [Google Scholar]
- 16.Weis WI, Drickamer K. Structural basis of lectin-carbohydrate recognition. Annu Rev Biochem. 1996;65:441–73. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 17.Dam TK, Brewer CF. Effects of clustered epitopes in multivalent ligand-receptor interactions. Biochemistry. 2008;47:8470–6. doi: 10.1021/bi801208b. [DOI] [PubMed] [Google Scholar]
- 18.Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–78. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 19.Aragao KS, Satre M, Imberty A, Varrot A. Structure determination of discoidin II from Dictyostelium discoideum and carbohydrate binding properties of the lectin domain. Proteins. 2008;73:43–52. doi: 10.1002/prot.22038. [DOI] [PubMed] [Google Scholar]
- 20.Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 21.Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291:2351–6. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 22.Perret S, Sabin C, Dumon C, Pokorna M, Gautier C, Galanina O, Ilia S, Bovin N, Nicaise M, Desmadril M, Gilboa-Garber N, Wimmerova M, Mitchell EP, Imberty A. Structural basis for the interaction between human milk oligosaccharides and the bacterial lectin PA-IIL of Pseudomonas aeruginosa. Biochem J. 2005;389:325–32. doi: 10.1042/BJ20050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Feinberg H, Conroy E, Mitchell DA, Alvarez R, Blixt O, Taylor ME, Weis WI, Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat Struct Mol Biol. 2004;11:591–8. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- 24.Imberty A, Mikros E, Koca J, Mollicone R, Oriol R, Perez S. Computer simulation of histo-blood group oligosaccharides: energy maps of all constituting disaccharides and potential energy surfaces of 14 ABH and Lewis carbohydrate antigens. Glycoconj J. 1995;12:331–49. doi: 10.1007/BF00731336. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez JF, Lescar J, Chazalet V, Audfray A, Gagnon J, Alvarez R, Breton C, Imberty A, Mitchell EP. Biochemical and structural analysis of Helix pomatia agglutinin. A hexameric lectin with a novel fold. J Biol Chem. 2006;281:20171–80. doi: 10.1074/jbc.M603452200. [DOI] [PubMed] [Google Scholar]
- 26.Nikinmaa M, Cech JJ, McEnroe M. Blood oxygen transport in stressed striped bass (Morone saxatilis): role of beta-adrenergic responses. Journal of Comparative Physiology B: Biochemical, Systemic, and Environmental Physiology. 1984;154:365–369. [Google Scholar]
- 27.Di Stasio E. Anionic regulation of biological systems: the special role of chloride in the coagulation cascade. Biophys Chem. 2004;112:245–52. doi: 10.1016/j.bpc.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 28.Vasta GR, Hunt JC, Marchalonis JJ, Fish WW. Galactosyl-binding lectins from the tunicate Didemnum candidum. Purification and physicochemical characterization. J Biol Chem. 1986;261:9174–81. [PubMed] [Google Scholar]
- 29.Wu AM, Wu JH, Singh T, Liu JH, Herp A. Lectinochemical studies on the affinity of Anguilla anguilla agglutinin for mammalian glycotopes. Life Sci. 2004;75:1085–103. doi: 10.1016/j.lfs.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 30.Stenutz R, Weintraub A, Widmalm G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol Rev. 2006;30:382–403. doi: 10.1111/j.1574-6976.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 31.Imberty A. Oligosaccharide structures: theory versus experiment. Curr Opin Struct Biol. 1997;7:617–23. doi: 10.1016/s0959-440x(97)80069-3. [DOI] [PubMed] [Google Scholar]
- 32.Azurmendi HF, Martin-Pastor M, Bush CA. Conformational studies of Lewis X and Lewis A trisaccharides using NMR residual dipolar couplings. Biopolymers. 2002;63:89–98. doi: 10.1002/bip.10015. [DOI] [PubMed] [Google Scholar]
- 33.Bekiroglu S, Kenne L, Sandstrom C. NMR study on the hydroxy protons of the Lewis X and Lewis Y oligosaccharides. Carbohydr Res. 2004;339:2465–8. doi: 10.1016/j.carres.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Bekiroglu S, Sandstrom C, Norberg T, Kenne L. Hydroxy protons in conformational study of a Lewis b tetrasaccharide derivative in aqueous solution by NMR spectroscopy. Carbohydr Res. 2000;328:409–18. doi: 10.1016/s0008-6215(00)00104-x. [DOI] [PubMed] [Google Scholar]
- 35.Cummings RD. Structure and function of the selectin ligand PSGL-1. Braz J Med Biol Res. 1999;32:519–28. doi: 10.1590/s0100-879x1999000500004. [DOI] [PubMed] [Google Scholar]
- 36.Guerardel Y, Petit D, Madigou T, Guillet B, Maes E, Maftah A, Boujard D, Strecker G, Kol O. Identification of the blood group Lewis(a) determinant in the oviducal mucins of Xenopus tropicalis. FEBS Lett. 2003;554:330–6. doi: 10.1016/s0014-5793(03)01183-9. [DOI] [PubMed] [Google Scholar]
- 37.Salerno G, Parisi MG, Parrinello D, Benenati G, Vizzini A, Vazzana M, Vasta GR, Cammarata M. F-type lectin from the sea bass (Dicentrarchus labrax): purification, cDNA cloning, tissue expression and localization, and opsonic activity. Fish Shellfish Immunol. 2009;27:143–53. doi: 10.1016/j.fsi.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 38.CCP4 The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallographica D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Brunger A, Adams P, Clore G, DeLano W, Gros P, Grosse -KR, Jiang J, Kuszewski J, Nilges M, Pannu N, Read R, Rice L, Simonson T, Warren G. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Hubbard SJ, Thornton JM. ‘NACCESS’, Computer program, Department of Biochemistry and Molecular Biology. University College; London: 1993. [Google Scholar]
- 42.Kraulis P. Molscript. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 43.Esnouf RM. Further additions to Molscript version 1.4, including reading and countouring of electron density maps. Acta Crystallographica D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]