Abstract
RNA binding proteins (RBPs) are involved in several post-transcriptional stages of gene expression and dictate the quality and quantity of the cellular proteome. When aberrantly expressed, they can lead to disease states as well as cancers. A basic requirement to understand their role in normal tissue development and cancer is the build of comprehensive gene expression maps. In this direction, we generated a list with 383 human RBPs based on the NCBI and EMSEMBL databases. SAGE and MPSS were then used to verify their levels of expression in normal tissues while SAGE and microarray datasets were used to perform comparisons between normal and tumor tissues. As main outcomes of our studies, we identified clusters of co-expressed or co-regulated genes that could act together in the development and maintenance of specific tissues; we also obtained a high confidence list of RBPs aberrantly expressed in several tumor types. This later list contains potential candidates to be explored as diagnostic and prognostic markers as well as putative targets for cancer therapy approaches.
Keywords: RNA binding proteins, post-transcriptional regulation, SAGE, MPSS, oncogenomics, cancer, gene expression analysis
Introduction
A decade of studies using high throughput genomics and proteomics data revealed that cancer cells differ from normal cells in both RNA and protein contents. Presumably, a substantial part of this difference originates from gene expression regulators. Mutations affecting their function as well as events that alter their expression levels can lead to a cascade of effects, compromising the expression of several direct and indirect target genes. Not surprisingly, a substantial number of tumor suppressor genes and oncogenes are in fact transcription factors (reviewed in ref. 1). Using the same rationale, post-transcriptional regulators are expected to be listed in the same categories. In fact, a good portion of aberrant protein expression in tumors has its origin at the post-transcriptional level. For instance, studies with lung adenocarcinomas have shown that there is only a 21% correlation between the transcriptome and the proteome in these cells.2
The two major types of regulators of post-transcriptional events are RNA binding proteins (RBPs) and non-coding RNAs, especially microRNAs (reviewed in ref. 3). With regard to RBPs, overexpression of several of them (YB1, hnRNPA1, PABP2, La, hnRNPE2, etc.) has been observed in different primary tumors and cancer cell lines.4–6 However, their role in tumor formation and progression is still poorly understood; careful analyses addressing the subject are rare. A good example is provided by a study carried out by Jeff Ross’ group on CRD-BP, an oncofetal protein that is known to be present in colon cancer (81%), breast cancer (58.5%) and sarcoma (73%).7 In this study, CRD-BP was expressed in mammary epithelial cells of adult transgenic mice. The incidence of mammary tumors was 95% and some of the tumors metastasized. The authors concluded that CRD-BP functions in fact as an oncogene.8 Four other recent studies demonstrated that, RBM3,9 PTB,10 Musashi111,12 and ASF/SF2,13 whose aberrant expression is seen in different tumor types, also have oncogenic properties. On the other hand, it has been shown that RMB5 or Luca-15 potentially functions as a tumor suppressor by increasing apoptotic signals and inhibiting cell proliferation.14,15
RBPs contribute to the quality and quantity of the proteome of a given cell by modulating cellular processes like splicing, translation, RNA stability, RNA transport and localization. Aside from the splicing process, RBPs function mainly by binding to cis-regulatory elements located on the untranslated regions (UTRs) of target mRNAs. In this regard, it is important to acknowledge the connection between UTR-mediated regulation and cancer. Approximately 10% of all mRNAs have atypically long 5′ UTRs, in most cases containing a variety of regulatory elements. 75% of them encode oncogenes and genes implicated in cell growth, death and proliferation (reviewed in refs. 16 and 17).
Mapping the expression of RBPs in normal and cancer tissues constitutes an important step towards a full understanding of their participation in normal tissue development and tumorigenesis. In this direction, we generated a comprehensive list of RBPs and performed a systematic analysis using SAGE (Serial Analysis of Gene Expression), MPSS (Massively Parallel Signature Sequencing) and microarray data. We provide here a detailed expression map of a representative group of RBPs in 33 different adult tissues. Moreover, an extensive comparison among different collections of normal and tumor tissues allowed us to generate a broad list of RBPs that are aberrantly expressed in several tumor types.
Results
Preparing a list of human RNA binding proteins
Although various databases provide lists of proteins/genes organized by functional domains, it is becoming evident that these lists are either incomplete or present redundant information. In order to circumvent these problems and create a comprehensive list of human RBPs, we searched both the NCBI and EBI databases for proteins containing the most characteristic domains that interact with RNA (RRM or RBD, dsRBD and KH) as well as for proteins whose description included the key word “RNA binding”. The files derived from each individual database were compared and compiled. Redundant proteins or alternative splicing variants were consolidated. Finally, a total of 932 protein IDs were grouped into 383 RBP genes (clusters) (Suppl. File 1). Even though, our collection is one of the most complete sets of RBPs available, it is far from being complete. As proteins become better characterized, more examples of proteins that do not have a classic RNA binding domain, but do interact with RNA are identified.
A Gene Ontology analysis using the cellular component parameter indicated that out of 383 proteins, 205 are supposed to have strict nuclear localization or nuclear/cytoplasmic localization (Suppl. File 2). Details about the list preparation are described in the methods section.
RBP expression in normal tissue
SAGE and MPSS are methodologies that have been used extensively to map gene expression in numerous tissues and to identify proteins implicated in tumorigenesis.18–22 The main advantage of these technologies is the possibility of performing multiple comparisons involving large sets of genes in a variety of tissue types. We combined these methods to map the expression of our list of RBPs in normal tissues.
In order to inquire the SAGE and MPSS libraries, each RBP transcript must be linked first to a reliable tag sequence (tag to gene assignment). Each tag short nucleotide sequence) corresponding to a transcript is used to determine the relative frequency of a given gene in a particular tissue or cell line. After listing all the tags for the RBPs present in our list, we employed an additional step to eliminate tags that present problematic sequence and/or ambiguous tag to gene assignment (see Methods for details). The analysis of some genes can be compromised due to problems with tag sequences23 or due to tag to gene assignment.24 There are cases for instance of genes that do not have a unique SAGE or MPSS tag. If a tag is shared by two different genes, the final tag counting will actually reflect the expression of these two genes. The best option in these cases is to eliminate the problematic tags/genes from the analysis. The screened tags cover a total of 363 RBPs; 305 RBPs have reliable tags for both SAGE and MPSS.
The reliable “tags” were used to determine the expression pattern of their corresponding RBPs in 31 normal adult tissues and in embryonic stem cells and placenta. We used MPSS libraries only in cases where a given tissue type was not covered by the SAGE libraries. The libraries and tags used in our analysis are listed in Supplementary files 3 and 4. Unfortunately, SAGE and MPSS data cannot be compared directly due to technical differences in the preparation of the libraries. This means that although vertical comparisons are possible (comparisons to assess differences in expression of two or more RBPs within the same tissue), horizontal comparisons (comparisons between different tissues to determine differences in expression of the same RBP) are only possible if they are done either between two or more SAGE libraries or between two or more MPSS libraries.
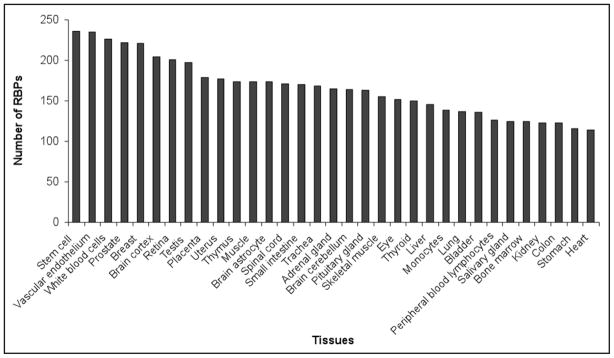
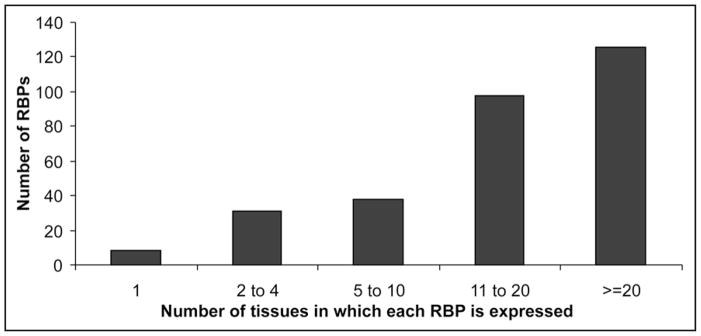
Figures 1 and 2 summarize the data we obtained for the 305 proteins that have both SAGE and MPSS tags. We did not take into consideration relative expression levels; proteins were listed as long as the numbers indicated their presence in a given tissue. Figure 1 shows the number of RBPs that were found to be expressed in each individual tissue. In Figure 2, the graph (number of RBPs vs. number of tissues) shows the distribution of the RBPs according to the number of tissues where they were found to be expressed. The raw data for Figures 1 and 2 are represented in Supplementary files 5 and 6 and the complete SAGE and MPSS analyses in Supplementary files 7 and 8. As can be observed, the great majority of the RBPs can be considered to be ubiquitously expressed; 227 RBPs are expressed in 10 tissues or more while proteins that have their expression restricted to only one tissue constitute ~2.5% of the RBPs analyzed. These “tissue specific” proteins are listed in Supplementary file 9.
Figure 1.
RBP expression in normal tissue. The graph represents the number of RBPs expressed in each of the 33 different normal tissues analyzed either by SAGE or MPSS. The levels of expression of each individual RBP in each individual tissue were not taken into consideration to prepare the graph. Proteins are listed as long as their expression was detected.
Figure 2.
RBP distribution in tissues. The graph represents the distribution of the RBPs present in our list according to the number of tissues in which they are expressed. The expression levels were not taken into consideration when preparing the graph. As can be observed, the great majority of the RBPs are ubiquitously expressed, while only a small subset can be considered tissue specific.
Finally, we performed a hierarchical clustering analysis to identify RBPs with similar pattern of expression in normal tissue—Supplementary file 10. These RBP clusters could be used to identify functional protein groups that co-regulate gene expression in a tissue specific fashion. The results corroborate this idea by showing that the clusters’ distribution follows the tissue embryonic origin. In this case, the RBP clusters could be categorized as ectodermic (cerebellum, astrocyte, cortex, stomach and colon), endodermic (thyroid, prostate, breast, lung and liver) and mesodermic (vascular endothelium and white blood cells).
Comparative analysis: RBP expression in normal tissues vs. RBP expression in tumors
As discussed in the introduction, since RBPs contribute substantially to the quality and quantity of the protein content of cells, it is reasonable to think that RBPs could be participating in tumor formation and progression. However, the number of RBPs described so far as either tumor suppressors or oncogenes is extremely small. The lack of studies specifically designed to understand the involvement of RBPs in cancer is responsible, at least in part, for this scenario. Although aberrant expression in tumor samples can not be considered as a definitive evidence of participation in tumorigenesis, it has been proved to function as a strong indicator. In fact, all the RBPs named in the introduction to be acting as either potential oncogenes or tumor suppressors were first identified in studies that showed that they were aberrantly expressed in a high number of tumor samples. Therefore, studies targeting the identification of RBPs that are either up or downregulated in tumor samples can direct cancer biologists to genes with high chances of being directly involved with malignancy. In this direction, we performed a study combining SAGE and microarray datasets. In order to increase the confidence on the results, we performed the analysis in two steps. First, we used SAGE libraries to perform comparisons between 11 normal and tumor tissues including: astrocyte (brain), cerebellum, cortex (brain), breast, colon, liver, lung, prostate, stomach, thyroid and white blood cells. A list was prepared with RBPs that were found to be up or downregulated (three-fold minimum cut off) in tumor samples. To increase our confidence, this list was rescreened using a different approach. We searched ONCOMINE (http://www.oncomine.org), a web-based resource that stores results of microarray studies. For most tumor types, SAGE and Oncomine use the same type of classification; in these cases, a direct comparison was made. Whenever there was an agreement between the SAGE and the Oncomine data, the RBP was passed to a final list. RBPs with contradictory microarray data or none listed data were eliminated. Due to differences in categorization, the RBPs that were identified by SAGE to be aberrantly expressed in tumors derived from neuronal tissues (astrocyte, medulla and cortex) had to be analyzed in a different way. We first compiled the RBPs that were determined to be aberrantly expressed in all three tumor types; they all correspond to upregulated proteins; we then retrieve data for these RBPs in microarray studies done with brain tumors. Finally, we passed to the final list only the RBPs determined to be upregulated in brain tumors in a minimum of three microarray studies. Table 1 contains the entire list of proteins for the combine SAGE-Oncomine analysis. Supplementary files 11 and 12 contain the detailed information regarding the SAGE analysis, while Supplementary file 13 contains the raw information for Oncomine analysis. Since SAGE and Oncomine only deal with transcriptomic data, we also collected protein expression data to corroborate some of our findings. We conducted a shotgun proteomics analysis of the highly tumorigenics lines: Daoy (medulloblastoma)25 and U251 (glioblastoma). We used APEX26 to estimate absolute protein concentrations. Of the 36 RBPs upregulated in brain tumors, 18 were present in Daoy and/or U251 data (Table 2). The 18 proteins were, on average, expressed at higher levels than other proteins detected in the respective datasets; in three datasets this difference was significant (t-test, p-value <0.05).
Table 1.
RBPs aberrantly expressed in tumors
| UP | DOWN | |
|---|---|---|
| breast | DRB1, ASCC1, ZNF638, RNASEL, COVA1, CUGBP1, HTATSF1, RBM27, FMR1, ELAVL4, MYEF2, NCBP1, SLC4A1AP, ZCRB1, RBM35B, DSRAD, JOSD2, DDX53, TIA1, POLDIP3, APOBEC1, ELAVL3, IGF2BP1, NCBP2, RBM35A, RBP56, PPARGC1B, PNPT1, RTCD1 | TNRC6C, RPS4Y1 |
| colon | SYNCRIP, TRBP2, SR140, CPSF5, FUBP2, HNRPDL, PAIP1, PPIE, PNPT1, HRB, NOL8, NCBP2, RPL22, PSMA1, FXR1, PTBP1, CSTF3, SRP72, FUS, KRR1, HNRPM, DHX9, PABP4, ILF3, SNRPB2, DKC1, NUFIP1, RBM9, HNRPUL1, FBL, EXOSC7, RBM8A, NOL3, FUBP1, HNRPG, HARS, HNRPU, EIF3S9, ElF3S4, RPL12, NKRF, NCL, LSM5, SAFB1, DSRAD, NHP2L1, NPM1, MARS, MRPL23, HNRPL, NONO, LA, TIAR, RPL34, MRPL3, RBM26, RPL13, RPL18, SLBP, RBPMS, RPLP2, RPL7, RPS5, SIAHBP1, RPL28, RPS17, RBM19, PCBP1, PABP2, RBM14, RBM10, RPS3, ELAV1 | DKFZp686F02235, RNASE1 |
| liver | RPL7, SIAHBP1, HNRPG, HNRPLL, SNRPA, NCBP2, PTBP1, SF3B14, SF3B4, RPS5, EIF3S9, PAPOLG, FUBP1, DKC1, MGC10433, IGF2BP2, NUFIP1, T1A1, PNPT1, RNMT, RCAN3, IGF2BP3, HNRPUL1, RBM9, CPSF6, RPL18, GARS, RBM8A, APOBEC1, RBM28, C20orf119, ANKRD17, NONO, MARS, TARDBP, SYNJ2, LRPPRC, RBM10 AKAP1, FXR1 | |
| prostate | ANKRD17, DICER, SNRPA, CSTF3, NOL8, C20orf119, STRBP, TUT1 RBM35A, CSTF2, SYNJ2 | TACC1, RBM38, IGF2BP3, IREB1, ACF |
| leukemia | RBM28 | U2AF2, ROD1, CPEB2, SAFB2, SRP14, SR140, SNRP70, SFRS12, TACC1, SLTM, RBMS1, MINT, LKAP, NOL8, MTHFSD, FMR1, ERAL1, LARP7, KRR1, RBM25, RBM23, RBM44, RBM26, ACIN1, DND1, RBM16, PSPC1 |
| nervous system* | ANKHD1, C20orf119, CNOT4, COVA1, CPEB2, CPSF6, DHX9, DKC1, EIF3S9, HNRPK, HNRPUL1, LA, LARP7, MSI1, MTHFSD, NOL8, NONO, NOVA, NPM1, NXF1, PAIP1, PPIL4, PRKRA, RBM15B, RBM25, RBM28, RBM39, RO60, RPL12, RPL13, RPL18A, RPL34, RPS17, RPS5, RPS9, SPF45 | |
| gastric | R06U, EIF3S9, NONO, FXR1, NKRF, CSTF2, FUBP1 | |
| thyroid | IREB1 | |
| lung | RPL18, RTCD1, PABP4, PSMA1, MRPL23, CNOT4, CSTF2, PTBP1, RBM14, SYNJ2, TRBP2, RBM34, SFR51, SART3, DKC1, TUT1, RBM16, MARS, RBM12, U2AF2, RENT1, RBM4, DAZAP1, MRPL12, SIAHBP1, AKAP1, CPSF6, LA, NCBP2, DSRAD, EIF3S4, GARS, FUBP1, FBL, FUBP2, HTF9C, FUS, GRSF1, STRBP, PPIE, ANKRD1 | PAPOLG, CDKN2AIP, CUGBP2, RNH1, RBM25, TACC1 |
We list all RBPs according to tissue type determined to be aberrantly expressed (over or under expression) in combined “normal vs. tumor” analyses performed with SAGE and microarray data obtained from the ONCOMINE resource.
Table 2.
RBPs expressed in the medulloblastoma Daoy and the glioblastoma U251 cell line
| Sample | Daoy cytosol | U251, replicate 1, cytosol | U251, replicate 2, cytosol | U251, replicate 1, pellet | U251, replicate 2, pellet |
|---|---|---|---|---|---|
| Total number of proteins | 1025 | 1160 | 813 | 1210 | 702 |
| Average expression level across all proteins | 9.79 | 11.02 | 11.46 | 11.28 | 11.47 |
| Standard deviation | 3.03 | 2.50 | 2.42 | 2.45 | 2.36 |
| Number of RBPs detected in dataset (out of 36) | 12 | 15 | 13 | 11 | 10 |
| Average expression level | 13.58 | 12.47 | 12.52 | 12.40 | 13.11 |
| Standard deviation | 2.53 | 2.32 | 2.66 | 2.77 | 2.00 |
| p-value (t-test) | 0.0002 | 0.0329 | 0.1843 | 0.2141 | 0.0241 |
| Name | Daoy cytosol | U251, replicate 1, cytosol | U251, replicate 2, cytosol | U251, replicate 1, pellet | U251, replicate 2, pellet |
| CPSF6 | 11.10 | 13.16 | 12.98 | 13.29 | 13.46 |
| DHX9 | 12.89 | 9.51 | 10.60 | 11.00 | 11.00 |
| HNRPK | 15.45 | 13.26 | 14.01 | 15.46 | 14.98 |
| HNRPUL1 | 11.29 | 11.61 | 10.24 | 11.65 | 11.26 |
| NONO | 12.89 | 12.81 | 13.48 | 14.25 | 14.32 |
| NPM1 | 18.39 | 16.29 | 16.66 | 17.09 | 17.11 |
| RPL12 | 14.33 | 15.79 | 15.24 | 10.56 | 13.75 |
| RPS17 | 15.24 | 15.00 | 13.83 | 13.99 | 12.51 |
| DKC1 | 8.81 | 10.82 | 12.23 | 11.49 | |
| RPS5 | 16.87 | 14.50 | 15.98 | ||
| RPS9 | 10.79 | 12.01 | 11.49 | ||
| EIF3S9 | 8.80 | 7.85 | |||
| PAIP1 | 11.49 | 9.56 | |||
| RBM39 | 9.15 | 11.21 | |||
| RPL18A | 10.50 | 12.32 | |||
| NXF1 | 7.72 | ||||
| RPL13 | 13.22 | ||||
| RPL34 | 11.71 |
We analyzed quantitative proteomics data from Daoy and U251 cell lines to test for over-expression of RBPs involved in brain tumor formation. The Daoy sample derives from a cytosolic fraction.25 The U251 sample derives from two biological replicates, fractionated into cytosol and pellet. Of the 36 RBPs, we identified 18 proteins across the five samples, and these proteins had higher expression levels than average in the dataset (p-value <0.05). The raw LC-MS/MS data and experimental methods are published at http://www.marcottelab.org/MSdata/. Expression levels were estimated using APEX,26 and are provided in (log base 2) molecules/cell.
It is expected that the chances of a given RBP to be involved in tumor formation increase if its aberrant expression is observed in several distinct tumor types. In order to identify this type of proteins, we compiled all the data obtained with the SAGE vs. Oncomine comparison. The results are illustrated in Table 3 and discussed below. We used then the literature based software Pathway Studio 6.0 (Ariadne Genomics, Inc.,) to verify any associations between the proteins listed in Table 3 and cancer related processes (apoptosis, cell cycle, cell proliferation, cell differentiation, cell survival and cell growth). The results are represented in Table 4 and several of the pertinent references are listed in Supplementary file 14. We should highlight the RBPs that have a high number of connections to distinct cancer related processes: PTBP1, CSTF2, SSB, NONO, PNPT1, ADAR, TACC1, ACO1, APOBEC1, NPM1 and RPS5.
Table 3.
Top aberrantly expressed RBPs in tumor
| Number of tissues with aberrant expression | RBPS |
|---|---|
| 4 | FUBP1, NCBP2, DKC1*, NONO*, EIF3S9* |
| 3 | CPSF6*, FXR1, PTBP1, STRBP, SYNJ2, SIAHBP1, RPL18, CSTF2, MRPL23, PNPT1, MARS, GARS, DSRAD, ANKRD17, LA*, C20orf119*, HNRPUL1*, NOL8*, NPM1*, PAIAP1*, RBM28*, RO60*, TACC1** |
| 2 | SNRPA, RBM8A, SLBP, RBM35A, RPS5*, RTCD1, HNRPG, RPL7, RBM14, FUS, RBM26, FUBP2, TRBP2, PPIE, CSTF3, RBM9, PSMA1, AKAP1, APOBEC1, NUFIP1, PABP4, RBM10, EIF3S4, TUT1, TIA1, RBM25**, IREB1**, NKRF |
The results obtained for each tumor type by the combined SAGE and microarray analyses were pulled together to determine RBPs that are aberrantly expressed in multiple tumor types. The genes labeled with *were determined to be upregulated in three different types of brain tumors and the genes labeled with **were determined to be downregulated in tumor samples.
Table 4.
Top aberrantly expressed RBPs in tumor and cancer related processes
| Biological process | Connectivity | Genes |
|---|---|---|
| Apoptosis | 19 | ACO1, ADAR, AKAP1, APOBEC1, NKRF, NOL8, NPM1, PABPC4, PNPT1, PTBP1, PUF60, RBM10, RBM25, RPL7, RPS5, SSB, TACC1, TIA1, TROVE2 |
| Cell cycle | 19 | ADAR, AKAP1, CSTF2, CSTF3, EIF3B, FUS, GARS, MARS, NONO, NPM1, PNPT1, PSMA1, PTBP1, RBM14, RPL7, RPS5, SLBP, SSB, TACC1 |
| Cell differentiation | 15 | ACO1, ADAR, APOBEC1, CSTF2, EIF3G, FUS, GARS, KHSRP, NONO, NPM1, PNPT1, PTBP1, RBM14, RPS5, SNRPA |
| Cell proliferation | 14 | ACO1, ADAR, APOBEC1, CPSF6, CSTF2, EIF3B, NKRF, NPM1, PTBP1, RBM14, RBMX, SLBP, SSB, TACC1 |
| Cell growth | 10 | ADAR, CSTF2, FXR1, NCBP2, NOL8, NPM1, PNPT1, RBM8A, RPL18, STRBP |
| Cell survival | 8 | ACO1, ADAR, AKAP1, NONO, NPM1, TACC1, TIA1, TROVE2 |
We used Pathway Studio 6 to identify connections between the selected RBPs and cancer related processes. Proteins are listed according to references that indicate participation in specific cellular functions.
One important issue in cancer is the identification of changes in the proteome that leads cells to metastasize. Focal adhesions (FA) play a critical role during cell invasion. They constitute specialized attachment and signaling centers that form at sites of cellular contact. Hoog et al.27 used a mass spectroscopic method named SILAC to identify and quantify proteins interacting in an attachment-dependent manner with focal adhesion proteins. This study revealed that a large portion of the proteins identified is constituted by RBPs, among them several are present also in the list of identified aberrantly expressed RBPs (Table 1): NONO, RPL7, RPL13, RPL22, RPL28, PTBP1 and U2AF.
Discussion
We have successfully mapped the expression of more than 300 RBPs in both normal and tumor tissue. We expect this dataset to become a powerful tool to help with the design of approaches intended to comprehend the function of RBPs in development, tissue differentiation and tumorigenesis.
The mapping of RBP expression in normal tissue revealed that the majority of the RBPs we analyzed are ubiquitously expressed. However, clear differences in expression levels among tissues could be noted for numerous proteins. This data reflects first, the well-known fact that several RBPs take part of basic molecular machineries like the spliceosome, responsible for controlling essential aspects of RNA processing in all cell types. Second, our data corroborates the idea that differences in the concentration of specific RBPs constitute the main mechanism to achieve variations in gene expression among tissues. Although, tissue specific RBP have important regulatory roles, our data suggests that they should have a minor contribution in the generation of tissue specific profiles.
Analysis of RBP expression in the developing brain showed that many RBPs have a similar pattern of neuronal expression; fact that indicates that multiple RBPs function concurrently to regulate the expression of specific RNA subsets.28 In agreement with this observation and corroborating the important role RBPs have in development and tissue differentiation, we determined that libraries from tissues of the same embryonic origin form clusters when organized in function of RBP expression (Suppl. File 10).
An important chapter of our analysis related to the identification of RBPs potentially involved in tumor formation. The fact that a protein is aberrantly expressed in several tumor types suggests a participation in cancer that deserves to be investigated. A combined analysis employing assays designed to test oncogenic or tumor suppressor properties as well as high throughput methods to determine the direct RNA targets of selected RBPs would be ideal to elucidate the impact of RBPs in tumor formation and progression (reviewed in ref. 3). The development of recent tools and methods like the RIP-Chip/Ribonomics assay, CLIP and alternative splicing microarrays29–32 will accelerate the understanding of RBPs participation in tumorigenesis.
On the list of RBPs aberrantly expressed in multiple tumor types, we observed that two protein families are particularly enriched. Ribosomal proteins constitute the first group. In agreement with the data we obtained, several recent reports have pointed to a possible participation of members of this family in tumor formation.33–36 It has been known for quite a while that ribosomes can vary in protein composition. Recent data from Pamela Silver’s laboratory showed that distinct ribosomal protein can affect the expression of specific gene subsets in yeast.37 Having said that, we suggest that an increase in expression of a specific group of ribosomal proteins could alter the cellular proteome in a qualitatively and quantitative manner; what could alter the expression of genes directly linked to proliferation, apoptosis and other cancer related processes. Another possibility is that ribosomal proteins could contribute to tumor formation by acting in other cellular processes outside translation. In fact, it was proposed several years ago that a group of ribosomal proteins may function as cell cycle checkpoints.38 The RBM family was the second group we identified as being enriched among proteins overexpressed in different tumor types. Most of its members are poorly characterized with the exception of RMB5, also known as Luca-15,15 and RBM3.9 While RMB5 is apparently involved in apoptosis and has been suggested to function as a tumor suppressor, RBM3 was suggested to function as a proto-oncogene. It remains to be investigated if other family members are also involved in tumor formation.
In respect to RBPs that were found to be upregulated in all three tumor types of the nervous system, NOL8 comes up as the first highlight. Among all identified proteins, it is the only one whose ratios (tumor vs. normal) exceed 10 fold in all three tissues in SAGE analysis. Despite the fact that the function of this protein is poorly understood, there is one report connecting NOL8 and cancer. Knockdown of this protein in three different gastric cancer cell lines affected cell growth and increased apoptosis.39 The presence of MSI1 was somehow expected since high levels of expression were previously observed in glioblastoma, medulloblastoma and astrocytoma.40–44 We have recently shown that MSI1 is potentially involved in medulloblastoma formation through its role as a regulator of “cancer stem like cells”.11 A few proteins were identified to be upregulated in other tissues as well as in brain tumors; these are the case of DKC1, EIF3S9, HNRPUL1, NONO, NPM1, N0L8, LA, PAIP1, RBM28, RO60, C20orf119, CPSF6 and RPS5. For most of them, we identified studies corroborating their upregulation in distinct tumor types; in the case of CPSF6, NPM1, NONO, NOL8 and RPS5, connections to cancer related processes have been also described. CPSF6 is a cleavage factor required for 3′ RNA cleavage and poly-adenylation processing. CPSF6 was recently described as being part of the “Poised Gene Cassette”, a set of cancer specific genes exhibiting precise transcriptional control in solid tumors whose expression could influence metastasis.45 NPM1 or Nucleophosmin is a relatively well studied gene in the context of cancer biology; being the most frequently mutated gene in acute myeloid leukemia (AML). NPM1 has been defined both as a putative proto-oncogene and tumor suppressor; it functions in several cellular processes that include ribosome biogenesis, regulation of chromosome duplication and cell proliferation (reviewed in ref. 46). In a study for bladder cancer marker identification, NONO was strongly correlated with vascular invasions and associated with a decreased probability of survival.47 RNAi knockdown of NOL8 inhibited cell growth of HeLa cells48 and induced apoptosis in three diffuse-type gastric cancer cells, St-4, MKN45 and TMK-1.39 Alteration in the pattern of expression of RPS5 was observed during differentiation and apoptosis in murine erythroleukemia cells.
In conclusion, the gene expression map that we produced in a total of 33 normal tissues will be important for the identification of RBPs and clusters of RBPs that are potentially required for tissue specific development and maintenance. In regard to cancer, RBPs that have altered expression in a large number of tumor tissues appear as future candidates to be explored as diagnostic and prognostic markers and to be tested as target candidates in cancer therapy approaches.
Material and Methods
Preparation of a list of human RNA binding proteins
The list of human RBPs was obtained from the EBI-InterPro (www.ebi.ac.uk/interpro/) and NCBI protein database (www.ncbi.nlm.nih.gov/sites/entrez?db=protein). In the EBI, we searched for proteins containing the most characteristic domains that interact with RNA (RRM or RBD, dsRBD and KH). In NCBI we also searched for proteins whose description includes the key word “RNA binding.” A manual inspection was performed to exclude non-RBP sequences.
Grouping RBP sequences
We performed a clustering of all sets of sequences present in our list of RBPs; each cluster corresponds to one RBP gene. First, we mapped all RBP sequences into the genome using BLAT.50 Second, we employed a cluster algorithm to analyze the genome mapping and to merge those sequences containing the same exon-intron structure (reviewed in ref. 51). Third, based on sequence annotation, we attributed a gene name to each RBP cluster. Finally, we made a semi-automatic analysis of all clusters, checking the sequence groups and verifying gene annotation.
Gene ontology analysis of RBPs to determine cellular localization
The cellular localization of RBPs was determined through the gene ontology information (GO).52 We made an association between the gene name and the GO cellular component (CC) term. All RBPs classified as “location in the nucleus” were selected.
Tag selection for SAGE and MPSS analysis
SAGE libraries were downloaded from SAGEGenie.40 MPSS libraries were obtained from http//mpss.licr.org. All tags were normalized as described in ref.21 Reliable tags to RBP genes were selected through of tag to gene information downloaded from ACTG.24 For most analyses presented in this paper, we selected only the 3′ most tag from mRNAs containing a poly(A) tail or poly(A) signal. Tags mapped to two or more genes were discarded.
Hierarchical clustering of the RBPs
Hierarchical clustering analysis is commonly used to identify patterns in a dataset. We used this method to analyze the expression profile of RBPs in the samples of normal tissues. The hierarchical clustering was performed by the heatplus package of R (http://www.r-project.org/) using Euclidean distance for dissimilarity between elements.
Method employed for comparison between normal and tumor tissue (SAGE)
Comparison of gene expression between tumor and normal tissue was performed based on SAGE tag frequency. The identification of genes differentially expressed in normal and tumor were done through three steps: (i) we used a local implementation of Monte Carlo simulation method described in53 to generate a list of genes differentially expressed (only differential expression supported by a p-value <0.05 were selected); (ii) from the list of genes differentially expressed, were classified as overexpressed in cancer those RBPs whose tags presented a cancer vs. normal ratio greater than three; (iii) from the list of genes differentially expressed, were classified as under expressed in cancer those RBPs whose tags that presented a normal vs. cancer ratio greater than three. Only tissues containing both cancer and normal SAGE libraries were analyzed. All SAGE libraries used in this analysis are listed in Supplementary Material.
Analysis of oncomine data and final list preparation
All RBPs that showed differential expression in the SAGE normal vs. tumor analysis were re-screened. These proteins were then checked against the Oncomine (http://www.oncomine.org) database of microarray studies using a p-value <=0.01. RBPs made the final list as long SAGE and Oncomine information matched. In cases of conflicting data (i.e., similar microarray studies showing both up and down-regulation) or lack of information, the RBP was discarded.
Proteomics analysis
The data for the cytosolic fraction of the Daoy cell lines was taken from ref.25 In the case of the U251 cell line, we analyzed two biological replicates, divided into cytosolic and pellet fraction. Experimental procedures are identical to those described in.54 All raw data is deposited at http://www.marcot-telab.org/MSdata/.
Supplementary Material
Acknowledgments
We would like to thank Carolina Livi, Yufei Huang, Julia P.C. da Cunha, Robson F. de Souza, Yufei Xiao, Suzanne Burns, Dat Vo and Tarea Burton for comments; Vishy Iyer and Patrick Killion for discussions on the experimental design and Lauren Johnston and Marilyn Asher for helping with the literature search. P.A.F.G. was supported by a PhD fellowship from FAPESP. P.A.F.G. was supported in part by grant 5D43TW007015-02 from the Fogarty International center, NIH. M.G. was supported by a fellowship of the Medical School (UTHSCSA). C.V. was supported by the International Human Frontier Science Program.
Footnotes
Note
Supplementary materials can be found at: www.landesbioscience.com/supplement/GalanteRNA6-4-Sup.pdf
References
- 1.Nebert DW. Transcription factors and cancer: an overview. Toxicology. 2002;181:131–41. doi: 10.1016/s0300-483x(02)00269-x. [DOI] [PubMed] [Google Scholar]
- 2.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–13. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Diaz P, Penalva LO. Post-transcription meets post-genomic: the saga of RNA binding proteins in a new era. RNA Biol. 2006;3:101–9. doi: 10.4161/rna.3.3.3373. [DOI] [PubMed] [Google Scholar]
- 4.Dua K, Williams TM, Beretta L. Translational control of the proteome: relevance to cancer. Proteomics. 2001;1:1191–9. doi: 10.1002/1615-9861(200110)1:10<1191::AID-PROT1191>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Meric F, Hunt KK. Translation initiation in cancer: a novel target for therapy. Mol Cancer Ther. 2002;1:971–9. [PubMed] [Google Scholar]
- 6.Perrotti D, Calabretta B. Translational regulation by the p210 BCR/ABL oncoprotein. Oncogene. 2004;23:3222–9. doi: 10.1038/sj.onc.1207543. [DOI] [PubMed] [Google Scholar]
- 7.Ioannidis P, Kottaridi C, Dimitriadis E, Courtis N, Mahaira L, Talieri M, et al. Expression of the RNA-binding protein CRD-BP in brain and non-small cell lung tumors. Cancer Lett. 2004;209:245–50. doi: 10.1016/j.canlet.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 8.Tessier CR, Doyle GA, Clark BA, Pitot HC, Ross J. Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res. 2004;64:209–14. doi: 10.1158/0008-5472.can-03-2927. [DOI] [PubMed] [Google Scholar]
- 9.Sureban SM, Ramalingam S, Natarajan G, May R, Subramaniam D, Bishnupuri KS, Morrison AR, Dieckgraefe BK, Brackett DJ, Postier RG, Houchen CW, Anant S. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008 Jul 31;27(33):4544–56. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, Pool M, Darcy KM, Lim SB, Auersperg N, Coon JS, et al. Knockdown of poly-pyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–8. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Diaz PC, Burton TL, Burns SC, Hung JY, Penalva LO. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC Cancer. 2008;8:280. doi: 10.1186/1471-2407-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, et al. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–58. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 13.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mourtada-Maarabouni M, Sutherland LC, Williams GT. Candidate tumour suppressor LUCA-15 can regulate multiple apoptotic pathways. Apoptosis. 2002;7:421–32. doi: 10.1023/a:1020083008017. [DOI] [PubMed] [Google Scholar]
- 15.Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J Cell Biochem. 2005;94:5–24. doi: 10.1002/jcb.20204. [DOI] [PubMed] [Google Scholar]
- 16.Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Lopez de Silanes I, Quesada MP, Esteller M. Aberrant regulation of messenger RNA 3′-untranslated region in human cancer. Cell Oncol. 2007;29:1–17. doi: 10.1155/2007/586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, et al. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–7. [PubMed] [Google Scholar]
- 19.Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–75. [PubMed] [Google Scholar]
- 20.Kirschbaum-Slager N, Lopes GM, Galante PA, Riggins GJ, de Souza SJ. Splicing factors are differentially expressed in tumors. Genet Mol Res. 2004;3:512–20. [PubMed] [Google Scholar]
- 21.Jongeneel CV, Delorenzi M, Iseli C, Zhou D, Haudenschild CD, Khrebtukova I, et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome research. 2005;15:1007–14. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, Hao P, Zhang YL, Deng FX, Deng Q, Hong Y, et al. Discovering multiple transcripts of human hepatocytes using massively parallel signature sequencing (MPSS) BMC Genomics. 2007;8:207. doi: 10.1186/1471-2164-8-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva AP, De Souza JE, Galante PA, Riggins GJ, De Souza SJ, Camargo AA. The impact of SNPs on the interpretation of SAGE and MPSS experimental data. Nucleic acids research. 2004;32:6104–10. doi: 10.1093/nar/gkh937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galante PA, Trimarchi J, Cepko CL, de Souza SJ, Ohno-Machado L, Kuo WP. Automatic correspondence of tags and genes (ACTG): a tool for the analysis of SAGE, MPSS and SBS data. Bioinformatics. 2007;23:903–5. doi: 10.1093/bioinformatics/btm023. [DOI] [PubMed] [Google Scholar]
- 25.Ramakrishnan SR, Vogel C, Prince JT, Li Z, Penalva LO, Myers M, Marcotte EM, Miranker DP. Integrating Shotgun Proteomics and mRNA expression data to Improve Protein Identification. Bioinformatics. 2009 Mar 24; doi: 10.1093/bioinformatics/btp168. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–24. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 27.de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–62. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 28.McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA. 2000;97:14085–90. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penalva LO, Tenenbaum SA, Keene JD. Gene expression analysis of messenger RNP complexes. Methods Mol Biol. 2004;257:125–34. doi: 10.1385/1-59259-750-5:125. [DOI] [PubMed] [Google Scholar]
- 31.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–86. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 32.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 33.Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi T, Sasaki Y, Oshima Y, Yamamoto H, Mita H, Suzuki H, et al. Activation of the ribosomal protein L13 gene in human gastrointestinal cancer. Int J Mol Med. 2006;18:161–70. [PubMed] [Google Scholar]
- 35.Bee A, Ke Y, Forootan S, Lin K, Beesley C, Forrest SE, et al. Ribosomal protein l19 is a prognostic marker for human prostate cancer. Clin Cancer Res. 2006;12:2061–5. doi: 10.1158/1078-0432.CCR-05-2445. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Zhao LN, Li KZ, Ling R, Li XJ, Wang L. Overexpression of ribosomal protein L15 is associated with cell proliferation in gastric cancer. BMC Cancer. 2006;6:91. doi: 10.1186/1471-2407-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–71. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen FW, Ioannou YA. Ribosomal proteins in cell proliferation and apoptosis. Int Rev Immunol. 1999;18:429–48. doi: 10.3109/08830189909088492. [DOI] [PubMed] [Google Scholar]
- 39.Jinawath N, Furukawa Y, Nakamura Y. Identification of NOL8, a nucleolar protein containing an RNA recognition motif (RRM), which was overexpressed in diffuse-type gastric cancer. Cancer science. 2004;95:430–5. doi: 10.1111/j.1349-7006.2004.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, et al. An anatomy of normal and malignant gene expression. Proc Natl Acad Sci USA. 2002;99:11287–92. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano A, Kanemura Y, Mori K, Kodama E, Yamamoto A, Sakamoto H, et al. Expression of the Neural RNA-binding protein Musashi1 in pediatric brain tumors. Pediatric neurosurgery. 2007;43:279–84. doi: 10.1159/000103307. [DOI] [PubMed] [Google Scholar]
- 42.Yokota N, Mainprize TG, Taylor MD, Kohata T, Loreto M, Ueda S, et al. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23:3444–53. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
- 43.Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, et al. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 44.Ma YH, Mentlein R, Knerlich F, Kruse ML, Mehdorn HM, Held-Feindt J. Expression of stem cell markers in human astrocytomas of different WHO grades. Journal of neuro-oncology. 2008;86:31–45. doi: 10.1007/s11060-007-9439-7. [DOI] [PubMed] [Google Scholar]
- 45.Yu K, Ganesan K, Tan LK, Laban M, Wu J, Zhao XD, et al. A precisely regulated gene expression cassette potently modulates metastasis and survival in multiple solid cancers. PLoS Genet. 2008;4:1000129. doi: 10.1371/journal.pgen.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 47.Barboro P, Rubagotti A, Orecchia P, Spina B, Truini M, Repaci E, et al. Differential proteomic analysis of nuclear matrix in muscle-invasive bladder cancer: potential to improve diagnosis and prognosis. Cell Oncol. 2008;30:13–26. doi: 10.1155/2008/686940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sekiguchi T, Todaka Y, Wang Y, Hirose E, Nakashima N, Nishimoto T. A novel human nucleolar protein, Nop132, binds to the G proteins, RRAG A/C/D. J Biol Chem. 2004;279:8343–50. doi: 10.1074/jbc.M305935200. [DOI] [PubMed] [Google Scholar]
- 49.Vizirianakis IS, Pappas IS, Gougoumas D, Tsiftsoglou AS. Expression of ribosomal protein S5 cloned gene during differentiation and apoptosis in murine erythroleukemia (MEL) cells. Oncol Res. 1999;11:409–19. [PubMed] [Google Scholar]
- 50.Kent WJ. BLAT—the BLAST-like alignment tool. Genome research. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galante PA, Vidal DO, de Souza JE, Camargo AA, de Souza SJ. Sense-antisense pairs in mammals: functional and evolutionary considerations. Genome biology. 2007;8:40. doi: 10.1186/gb-2007-8-3-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, et al. Gene expression profiles in normal and cancer cells. Science (New York, NY) 1997;276:1268–72. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 54.de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, Burns SC, Ko D, Burton TL, Vo DT, Chennasamudaram S, Le SY, Shapiro BA, Penalva LO. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J Biol Chem. 2009 May 1;284(18):12125–35. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.