Abstract
Diabetic retinopathy does not halt after hyperglycemia is terminated; the retina continues to experience increased oxidative stress, suggesting a memory phenomenon. Mitochondrial DNA (mtDNA) is highly sensitive to oxidative damage. The goal is to investigate the role of mtDNA damage in the development of diabetic retinopathy, and in the metabolic memory. mtDNA damage and its functional consequences on electron transport chain (ETC) were analyzed in the retina from streptozotocin-diabetic rats maintained in poor control (PC, glycated hemoglobin >11%) for 12 months or PC for 6 months followed by good control (GC, GHb < 6.5%) for 6 months. Diabetes damaged retinal mtDNA and elevated DNA repair enzymes (glycosylase). ETC proteins that were encoded by the mitochondrial genome and the glycosylases were compromised in the mitochondria. Re-institution of GC after 6 months of PC failed to protect mtDNA damage, and ETC proteins remained subnormal. Thus, mtDNA continues to be damaged even after PC is terminated. Although the retina tries to overcome mtDNA damage by inducing glycosylase, they remain deficient in the mitochondria with a compromised ETC system. The process is further exacerbated by subsequent increased mtDNA damage providing no relief to the retina from a continuous cycle of damage, and termination of hyperglycemia fails to arrest the progression of retinopathy. Antioxid. Redox Signal. 13, 797–805.
Introduction
Retinopathy is one of the most severe ocular complications of diabetes, and sustained hyperglycemic milieu exposes retina and its microvasculature to increased oxidative stress (25, 28, 38). Retinal mitochondria are dysfunctional in diabetes (27), superoxide levels are elevated, and the enzyme responsible for scavenging superoxide (MnSOD) is compromised (16, 21, 30). Mitochondria, in addition to serving as a site of oxidative stress induction, are also highly susceptible to reactive oxygen species (ROS). Diabetes decreases the activity of retinal complex III of the electron transport chain and overexpression of MnSOD prevents such inhibition (21). ROS are postulated to be the critical mediators in the activation of multiple biochemical pathways implicated in the pathogenesis of diabetic retinopathy (6). The exact mechanisms by which mitochondrial damage could contribute to the development of diabetic retinopathy, however, require further elucidation.
The Diabetes Control and Complications Trial (DCCT) has shown that intensive glycemic control prevents the development and progression of retinopathy in patients with diabetes (12). The follow-up Epidemiology of Diabetes Interventions and Complications (EDIC) study has demonstrated that, despite similar glycated hemoglobin levels during this follow-up period, the ongoing risk of all levels of retinopathy are significantly lower in the intensive control group compared with the conventional group, and the benefits of intensive glycemic control persist for some time even after its termination (13). This carry-over effect of prior glycemic control on the later manifestation of complications suggests a “metabolic memory” phenomenon. Recent follow-up EDIC studies have shown that this metabolic memory continues to persist for at least 10 years after termination of the desired glycemic control in diabetic patients (14). Using a rodent model of diabetic retinopathy, we have shown that the retina continues to experience increased oxidative damage, and histopathology characteristic of diabetic retinopathy does not halt after hyperglycemic insult is terminated in rats. The levels of nitrotyrosine remain elevated in their retinal microvasculature for at least 6 months (26, 31).
Unlike nuclear DNA (nDNA), which has introns and is packaged into nucleosomes, the mitochondrial DNA (mtDNA) is packed as nucleoid-like structures without any histones (8, 34). Due to its close proximity to the ROS-generating electron transport chain and lack of protective histones, mtDNA is particularly prone to oxidative damage (42). Oxidative damage to mtDNA initiates a vicious cycle leading to decreased transcription and protein synthesis, and this is further exacerbated due to subsequent decreased electron transport and increased ROS (43). For genomic maintenance, mitochondria utilize base excision repair (BER) as the primary DNA repair mechanism (15). Glycosylases, including 8-oxoguanine DNA glycosylase (OGG1), MutY homolog (MYH), and thymine DNA glycosylase (TDG), recognize and remove damaged or inappropriate bases (11, 18). Decreased DNA glycosylases are postulated to contribute to mtDNA damage in age-related macular degeneration (45). Studies using isolated retinal endothelial cells cultured in high glucose conditions have suggested that mtDNA damage could potentially contribute to mitochondrial dysfunction by decreasing mRNA abundance of electron transport subunits (46). Studies by Roy et al. have suggested that high glucose could be responsible for inducting self-perpetuating changes in the gene expression in the tissues that are the targets of diabetic complications (40). However, the contribution of mitochondrial genome in the metabolic memory phenomenon remains to be elucidated.
The objective of this study is to examine the effect of mtDNA damage and its encoded proteins in the resistance of retinopathy to arrest after re-establishment of good glycemic control. Using a rat model of diabetic retinopathy, we have investigated the effect of glycemic control on mitochondrial oxidative stress and its DNA damage and repair system. Also, the functional consequence of mtDNA is evaluated by investigating the proteins encoded by mtDNA.
Methods
Rats
Diabetes was induced in rats (male, Wistar, 200–220 g) with streptozotocin (55 mg/kg body weight). Animals were randomly assigned to be in one of three glycemic controls: poor glycemic control for 12 months (PC group), good glycemic control for 12 months (GC group), or poor control for 6 months followed by good control for 6 months (PC-GC). The choice of the durations of poor and good glycemic controls is based on previous reports showing that the activation of retinal caspase-3, an apoptosis execution enzyme, and capillary cell apoptosis are observed in rats around 6–8 months of diabetes, and the histopathology characteristic of diabetic retinopathy around 10–12 months of diabetes (24, 33). Rats maintained in poor glycemic control received 1–2 U insulin, 4 to 5 times per week to allow for slow weight gain while maintaining hyperglycemia and preventing ketosis. Those maintained in good glycemic control received insulin twice daily (7-8 units total). Each group had 10 or more rats, and age-matched normal rats served as controls. All animals were housed in metabolism cages and received powdered diet (PURINA 5001), their body weights were measured twice per week, blood glucose once a week (Glucometer Elite, Bayer Corporation, IN), and glycated hemoglobin (GHb) every 2–3 months using a kit from Helena Laboratories (Beaumont, TX). All experiments were performed in accordance to the Association of Research in Vision and Ophthalmology on Treatment of Animals in Research and the Wayne State University's Animal Care and Use Committee guidelines. At the end of the desired duration, animals were sacrificed by pentobarbital overdose, and the retina was harvested. Retina were either processed for mitochondrial isolation or stored at −80°C for further analyses.
Mitochondria isolation
To quantify oxidative stress, the mitochondria-enriched fraction was isolated by differential centrifugation as reported earlier (4, 6). Briefly, retina were suspended in mitochondria isolation buffer (25 mM Tris-HCl, pH 7.4, 250 mM sucrose, 2 mM EDTA, and protease inhibitor cocktail) and gently homogenized with a Dounce homogenizer. The homogenate was centrifuged at 750 g for 5 min to remove cell debris, and the supernatant was then centrifuged 10,000 g for 15 min at 4°C to pellet mitochondria. The pellet was washed by centrifugation, and then suspended in a mitochondria isolation buffer. This fraction was enriched in mitochondria with some nuclear contamination.
To determine the protein expression of DNA glycosylases, mitochondria were isolated using a Mitochondria Isolation kit from Pierce (Rockford, IL). Briefly, freshly isolated retina was washed with ice-cold phosphate buffered saline, cut into small pieces and homogenized using 10–12 strokes in BSA-reagent A in a cold Dounce homogenizer. An equal volume of reagent C was added to the homogenate, and after mixing, centrifuged at 700 g for 10 min at 4°C. The supernatant was centrifuged at 12,000 g for 15 min, and the pellet (mitochondria fraction) was washed with phosphate buffered saline. Mitochondrial pellet was suspended in 2% CHAPS in TBS (25 mM Tris, 0.15 M NaCl; pH 7.2) and vortex for 1 min. Protein concentration was quantified by using Bicinchoninic Acid Solution (Sigma Aldrich, St Louis). Mitochondria prepared by this method were largely devoid of nuclear contamination as determined by quantifying the expression of histone H2B.
Oxidative stress in the mitochondria
Superoxide levels were quantified using 2–5 μg of mitochondria-enriched fraction in 100 μl assay volume containing 10 mM MOPS (pH 7.6), 0.5 mM EDTA, and 20 μM lucigenin (bis-N-methylacridinium nitrate). The reaction was started by the addition of 70 μM of NADH, and luminescence was monitored for 5 min with the readings recorded every 20 sec. Values obtained from the appropriate blank containing all of the assay components except the protein were subtracted from each sample (21).
Reduced glutathione (GSH) was measured in retinal mitochondria-enriched fraction using an enzymatic recycling method (Cayman Chemical, Ann Arbor, MI). Protein (3–5 μg) was deproteinized with phosphoric acid, and GSH concentration was quantified in the deproteinized samples using DTNB (5, 5’-dithiobis-2-nitrobenzoic acid) (21).
8-hydroxy-2′-deoxyguanosine (8-OHdG) levels were measured in the DNA isolated from retinal mitochondria-enriched fraction using an ELISA kit (Oxis International, Beverly Hills, CA), as routinely employed in our laboratory (30). Mitochondrial DNA (2–10 μg) or 8-OHdG standard (0.5–40 ng/ml) was incubated with monoclonal antibody against 8-OHdG. The final color was developed by the addition of 3,3’,5,5’-tetramethylbenzidine, and absorbance was measured at 450 nm.
Mitochondrial DNA damage and repair
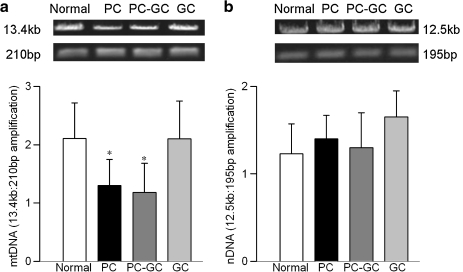
DNA was isolated from retina with the DNeasy kit (Qiagen, Valenci, CA), according to the manufacturer's protocol, and quantified using the Quant-iT dsDNA assay (Invitrogen, Carlsbad, CA). Genome-specific (mitochondrial and nuclear) quantitative extended length PCR was performed with the GeneAmp XL PCR kit (Applied Biosystems, Foster City, CA), following the method described by others (1, 45). Briefly, 15 ng DNA was amplified in a reaction mixture containing 1x XL PCR buffer II, 200 μM dNTP, 1.1 mM Mg(OAc)2, 1 unit rTth DNA polymerase, and 0.1 μM genome specific primers: mtDNA long, forward-AAA ATC CCC GCA AAC AAT GAC CAC CCC and reverse-GGC AAT TAA GAG TGG GAT GGA GCC AA; mtDNA short, forward-CCT CCC ATT CAT TAT CGC CGC CCT TGC and reverse-GTC TGG GTC TCC TAG TAG GTC TGG GAA. After 1 min at 75°C, polymerase was added followed by 1 min at 94°C, cycles of 94°C for 15 sec and 65°C for 12 min, and a final extension at 72°C for 12 min. Mitochondrial DNA and nDNA were amplified with 24 and 28 cycles, respectively, and PCR products were resolved on agarose gel containing 0.5 μg/ml ethidium bromide. To determine the correct number of cycles for amplification, we initiated with the combination previously used by others (1, 44). This was followed by testing several other combinations until we had amplification at a quantifiable level but not at saturation level. The gels were imaged with a UV transilluminator and bands were digitized and quantified using Un-Scan-It software (Silk Scientific; Orem, UT). Relative amplification was quantified by normalizing the intensity of the long product to the short product (mtDNA = 13.4 kb/210 bp and nDNA = 12.5 kb/195 bp).
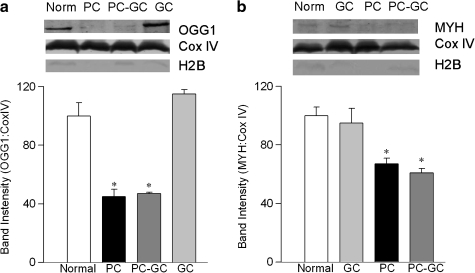
Protein expressions of OGG1 and MYH were determined in retinal mitochondria by SDS-PAGE using polyclonal antibodies against OGG1 and MYH (Santa Cruz Biotechnology, Santa Cruz, CA). Mitochondria-specific cytochrome c oxidase subunit IV (Cox IV, Invitrogen) was used as a loading control and histone H2B (Abcam Inc, Cambridge, MA) to screen for nuclear contamination.
Mitochondrial DNA encoded proteins
Relative transcript abundance of subunits 1, 4, and 6 of NADH dehydrogenase of complex I (ND1, ND4, and ND6, respectively) and cytochrome b of complex III were quantified in the retinal mitochondria by semiquantitative PCR using the gene-specific primers: ND1 forward-TTC TAT GAT CGG GAT GAG CC and reverse-GTT GGG AAT GGA GCG TAG AA; ND4 forward-TAC TAA TAA TCG CCC ACG GC and reverse-AAT TCT CGT GTG TGG GAA GG; ND6 forward-CCC AGC CAC CAC TAT CAT TC and reverse-CAT CGT ACT CCT GCT TGC TG; and cytochrome b-forward-CCC ACA GGA TTA AAC TCC GA and reverse-GTT GGG AAT GGA GCG TAG AA. The RT-PCR was performed under the following thermal profile: initial denaturation at 95°C for 2 min, followed by 28 cycles of 92°C for 1 min, 50°C for 2 min, 72°C for 1 min, and a final extension for 5 min at 72°C. PCR products were visualized by agarose gel electrophoresis and imaged with a UV transilluminator. Relative amplification was quantified by normalizing the gene-specific band intensity to that of β-actin.
Respiratory chain complex activity
Activity of complex I was quantified in the mitochondria-enriched fraction by measuring the consumption of NADH at 340 nm and that of complex III by measuring the reduction of cytochrome c at 550 nm, as previously described by us (21).
Statistical analysis
Data are expressed as mean + standard deviation. Statistical analysis was performed using the nonparametric Kruskal–Wallis test, followed by Mann–Whitney U test, and also by parametric test-analysis of variance with post hoc Bonferroni comparisons. Similar results were obtained by these methods. P < 0.05 was considered as significant.
Results
Severity of hyperglycemia in rats
Average body weights of the rats in PC group were significantly lower compared to their age-matched normal rats (PC = 318 ± 38 g, and Normal = 505 ± 82 g). In contrast, the rats that were maintained in good glycemic control for the entire duration of the experiment (12 months) had similar average body weights (527 ± 47 g) as their normal control counterparts. The average body weight of rats in the PC-GC group before initiation of GC was 337 ± 25 g, and was increased to 491 ± 26 g at the end of 6 months of GC. Routine blood glucose ranged from 75 to 130 mg/dl for rats that were normal or in GC, and 300–450 mg/dl for rats in PC. Average GHb value for rats in PC group was 11.7 ± 0.7% compared to 6.3 ± 1.5% for normal control rats and 5.2 ± 0.4 in GC rats. GHb value before initiation of GC in the rats that were maintained in PC for 6 months, followed by 6 additional months of GC (PC-GC) was 12.0 ± 1.1% GHb, but was reduced to about 4.8 ± 0.5% at 2 months after initiation of good metabolic control, and remained ∼5% for the entire duration of additional good metabolic control.
Effect of poor glycemic control on retinal mitochondrial genome
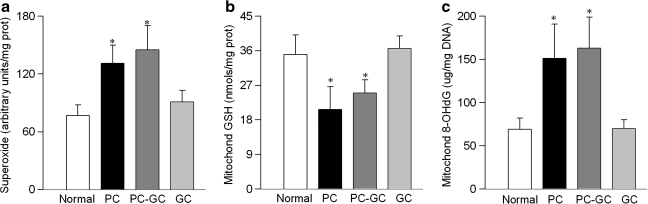
Diabetes increased levels of superoxide in retinal mitochondria by over 70% (Fig. 1A). The levels of intracellular antioxidant, GSH, were decreased by about 40% (Fig. 1B), and those of oxidatively modified DNA (8-OHdG) were increased by over 2-fold (Fig. 1C) compared to the values obtained from the age-matched normal rats. Since the mitochondria-enriched fraction was used to measure oxidative stress, we needed to acknowledge that this could have resulted in underestimating the absolute values for the parameters of oxidative stress, however, without affecting the outcome of the results. DNA damage due to oxidative modification prevents the progression of polymerase along the DNA template, and this phenomenon was exploited using extended length PCR with genome specific primers (44, 46). As shown in Figure 2A, mtDNA amplification was reduced by over 40% in the retina obtained from diabetic rats. However, in the same animals there was no significant change in nDNA damage (Fig. 2B).
FIG. 1.
Retinal mitochondrial oxidative stress is increased in diabetes. Freshly isolated retinal mitochondria were used to measure (a) superoxide levels by lucigenin method, (b) GSH levels by a colorimetric method, and (c) 8-OHdG by an ELISA. Results are mean ± SD of the measurements made in duplicate in 5–8 rats in each group. Normal, age-matched normal control; PC, poor glycemic control; PC-GC, 6 months poor glycemic control followed by 6 months good glycemic control; GC, good glycemic control for the entire 12 months. *p< 0.05 compared to normal.
FIG. 2.
Termination of hyperglycemia fails to reverse diabetes-induced retinal mtDNA damage. (a) MtDNA damage was assessed using 15 ng total retinal DNA and mitochondrial-specific primers for long and short PCR product: long, forward-AAA ATC CCC GCA AAC AAT GAC CAC CCC and reverse-GGC AAT TAA GAG TGG GAT GGA GCC AA; and short, forward-CCT CCC ATT CAT TAT CGC CGC CCT TGC and reverse-GTC TGG GTC TCC TAG TAG GTC TGG GAA, respectively. The relative amplification was calculated by normalizing the intensity of the 13.4 kb product to the 210 bp product. (b) For nDNA damage the primers used were: long, forward-AGA CGG GTG AGA CAG CTG CAC CTT TTC and reverse-CGA GAG CAT CAA GTG CAG GCA TTA GAG, and short, forward-GGT GTA CTT GAG CAG AGC GCT ATA AAT and reverse-CAC TTA CCC ACG GCA GCT CTC TAC, and the relative amplification of 12.5 kb and 195 bp products was calculated. Results represent values obtained from 5 or more rats in each group. Normal, age-matched normal control; PC, poor glycemic control; PC-GC, 6 months poor glycemic control followed by 6 months good glycemic control; GC, good glycemic control for the entire duration of diabetes. *p < 0.05 compared to normal.
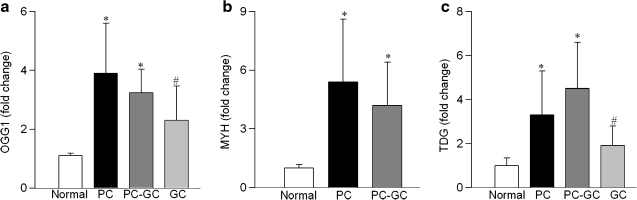
Transcripts of OGG1, MYH, and TDG, members of the DNA glycosylase family that recognize and remove modified bases, were significantly elevated in the retina in diabetes (Figs. 3A–3C). In contrast, despite similar expressions of COX IV in the retinal mitochondria obtained from normal rats and diabetic rats, the protein abundance of OGG1 and MYH was decreased by 35%–50% in the retinal mitochondria obtained from diabetic rats (Figs. 4A and 4B). Thus, although transcript abundance of retinal DNA glycosylases is increased in diabetes, their protein expressions remain subnormal in the mitochondria. We were unable to get a clear band for TDG; the reason for this could be the poor quality of the anti-TDG antibody or undetectable protein expression of this glycolyase in the mitochondria.
FIG. 3.
Diabetes increases the gene expressions of DNA repair enzymes in the retina. RNA isolated from the retina was assessed by real-time RT-PCR for (a) OGG1, (b) MYH, and (c) TDG, and was normalized to B2M in each sample. Fold-change relative to normal age-matched controls was calculated using the ddCt method. Results are from the measurements made in 5 or more rats in each group. *p < 0.05 compared to normal and #p < 0.05 compared to PC.
FIG. 4.
Decreased protein abundance of DNA glycosylases in retinal mitochondria is not reversed by good glycemic control. Retinal mitochondrial protein (20 μg) was separated on 10% polyacrylamide gels, transferred to nitrocellulose, and analyzed by Western blot using polyclonal antibodies for OGG1 (a) and MYH (b) with Cox IV as a loading control. To evaluate nuclear contamination, the membranes were screened for Histone H2B expression. Histograms represent band intensities of OGG1 and MYH, respectively, normalized to that of Cox IV. The values are presented as the mean + SD of 4 rats in each group. *p < 0.05 compared to normal.
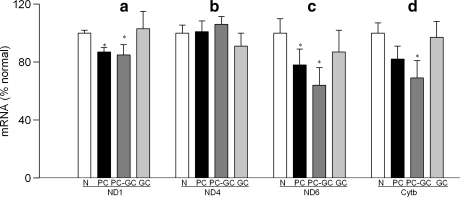
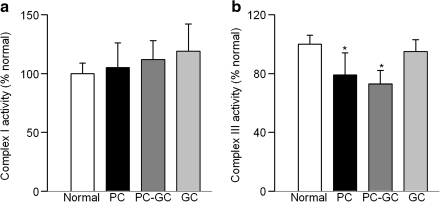
Mitochondrial DNA encodes only 13 proteins, and these are important in the formation of electron transport chain complexes. Gene expressions of ND1 and ND6 of complex I (Figs. 5A and 5C) and cytochrome b of complex III (Fig. 5D) were significantly decreased in diabetes as compared to the values obtained from age-matched normal rats (P < 0.05), but the transcripts of ND4 were not altered (Fig. 5B). In the same animals, diabetes had no significant effect on the activity of mitochondrial complex I (Fig. 6A),but the activity of complex III was decreased by 20% (Fig. 6B).
FIG. 5.
Mitochondrial-encoded genes continue to be altered in the retina after hyperglycemia is terminated. Transcript abundance was assessed in DNase-treated RNA isolated from retina using conventional RT-PCR for (a) ND1, (b) ND4, (c) ND6 of complex I, and (d) cytochrome b of complex III. Relative mRNA abundance was quantified using Un-Scan-It Gel digitizing software, and the values in the figures are presented as mean band intensity of the target gene normalized by the intensity of β-actin. The values obtained from normal rats are considered 100%. Results are from the measurements made in 4–6 rats in each group. N, age-matched normal control; PC, poor glycemic control; PC-GC, 6 months poor glycemic control followed by 6 months good glycemic control; *p < 0.05 compared to normal.
FIG. 6.
Reversal of hyperglycemia does not normalize the reduction in retinal complex III activity. (a) Complex I activity was assayed in retinal mitochondria by measuring the consumption of NADH at 340 nm. (b) Complex III activity was assessed by measuring the reduction of cytochrome c at 550 nm. Values from normal rat retina mitochondria are considered 100%. *p < 0.05 compared to normal.
Effect of reversal of hyperglycemia on mtDNA damage and repair mechanisms
Re-institution of good glycemic control after 6 months of poor glycemic control in rats failed to reverse increased mitochondrial oxidative stress; the levels of superoxide remain elevated and those of GSH remained subnormal in the PC-GC group (Figs. 1A and 1B), and this was further confirmed by increased levels of 8-OHdG (Fig. 1C)
In the same group of rats, termination of hyperglycemia also failed to help ameliorate mtDNA damage; retinal mtDNA damage in rats from the PC and PC-GC groups was not different from each other, but the values were significantly higher than from the age-matched normal rats (Fig. 2A). The transcripts of DNA glycosylases, OGG1, MYH, and TDG remained elevated in the retina for at least 6 months after hyperglycemia was terminated (Fig. 3). In contrast, their protein abundance in the mitochondria remained compromised, and the expressions were similar to those obtained from rats in PC group (Fig. 4).
The transcripts of ND1, ND6, and cytochrome b, which were decreased in the retinal mitochondria of PC rats, continued to be subnormal 6 months after reversal of hyperglycemia (Fig. 5), and the activity of complex III remained attenuated (Fig. 6), suggesting that the re-institution of good glycemic control for 6 months did not provide any relief to the mitochondrial electron transport system. In contrast, reduction in ND6 gene expression was ameliorated.
However, when the rats were allowed to maintain good glycemic control for the entire 12 months duration of the experiment (GC group), superoxide, GSH and 8-OHdG levels were similar to those obtained from age-matched normal rats (Figs. 1A–1C), and mtDNA damage was significantly lower compared to the rats in PC group. Further, re-institution of good glycemic control soon after induction of diabetes also protected the DNA repair system and electron transport complexes, and the transcripts of cytochrome b and the activity of complex III were not different from those obtained from age-matched normal rats (Figs. 4–6). These data imply that the increased insulin administered to maintain good glycemic control did not influence these parameters.
Discussion
Diabetic retinopathy is a slow progressing and duration-dependent disease; it takes over decades in humans and about one year in rodents to develop (14, 24, 31, 37). While histopathology characteristic of diabetic retinopathy is not observed till 10–12 months of diabetes in rats, retinal capillary cell apoptosis can be seen around 6–8 months after induction of diabetes (24, 37). Our previous work has demonstrated that the retinal capillary cell apoptosis (as demonstrated by TUNEL staining and caspase-3 activity) and the histopathology of diabetic retinopathy continue even after 6 months of good glycemic control in rats that has followed 6 months of poor glycemic control (29, 31, 33). The results presented here are the first to illustrate the role of mtDNA in the development of diabetic retinopathy, and show that the retinal mitochondria remains compromised for some time after hyperglycemia is terminated in diabetic rats; superoxide and 8-OHdG levels continue to be elevated and GSH levels remain subnormal for at least 6 months after initiation of normal glycemic control in diabetic rats. Their mtDNA remains damaged with the repair system compromised, and the transcripts of the mtDNA encoded proteins attenuated. The activity of complex III also fails to benefit from re-institution of good glycemic control. However, if good glycemic control is initiated soon after induction of diabetes, retinal mitochondria are protected from increased oxidative damage with stable mtDNA. These results suggest that the retinal mtDNA damage, which is present at duration of diabetes in rats when apoptosis of capillary cells and histopathology can be detected (24, 37), and is experienced in high glucose conditions by the retinal endothelial cells (36), does not correct by reversal of hyperglycemia. Thus, mtDNA could be an important factor in the metabolic memory phenomenon associated with the continued progression of diabetic retinopathy. In support, our previous studies have shown that the oxidative stress remains elevated in the retina, glyceraldehyde-3-phosphate dehydrogenase inactivated and nitrotyrosine accumulated in the retinal microvasculature for at least 6 months after hyperglycemia is terminated in diabetic rats (23, 26, 31).
Superoxide radicals are considered to act as causal links between elevated glucose and the major abnormalities associated with the vascular complications of diabetes (6). We show that re-institution of good glycemic control after 6 months of poor control does not provide any benefit to mitochondrial superoxide levels, and this sustained oxidative insult to mitochondria is further exacerbated by decreased GSH. These data imply that the retinal mitochondria, in addition to producing increased superoxide, continues to experience increased oxidative stress due to its inefficient redox (GSH) and scavenging (MnSOD) mechanisms (8, 30).
The close proximity to the electron transport chain and less tightly packaging by DNA transaction factors and chaperones (8) makes mtDNA more vulnerable to damage from insults generated by the electron transport chain than nDNA (4, 31, 34). Although mtDNA damage after a short insult is ultimately repaired, longer insults lead to persistent damage that could subsequently result in apoptosis (47), and the damage is postulated to contribute to another chronic ocular disease, age-related macular degeneration, (36, 46, 47). Our study is the first to demonstrate the overall mtDNA damage in the retina in animal models of diabetic retinopathy, and shows that this damage continues even after hyperglycemia is reversed, suggesting a strong role for mtDNA damage in the development, and also in the metabolic memory associated with the progression of diabetic retinopathy. However, our study did not identify the specific transition mutation, and the possibility that the damage to mtDNA could include contributions from multiple DNA lesions including strand breaks and base modifications (3) cannot be ruled out. Mitochondrial genome has much higher turnover than nuclear genome (10); our results show that the levels of superoxide do not normalize after reversal of hyperglycemia, resulting in a continuous damage of mtDNA. This could result in passing forward the damaged DNA which does not get a chance to be repaired after hyperglycemia is terminated.
The base excision repair (BER) pathway, because of its ability to recognize oxidized and inappropriate bases and single-strand breaks, is one of the most often employed pathway for mtDNA damage repair (18). DNA glycosylases perform the critical first step of base recognition and removal, and each distinct DNA glycosylase recognizes an individual alteration in damaged DNA (18, 41). A rapid increase in OGG1 levels is seen in the retina after a short period of bright light damage (9), but chronic insult (aging) downregulates retinal OGG1 and MYH (45), suggesting that the DNA repair is rapidly initiated by acute insult but is impaired by chronic insult. Our results provide an interesting disparity; in diabetes, a chronic disease, retinal DNA glycosylase transcripts are significantly elevated, but their protein expression in the mitochondria is compromised, and this process is not benefited by the termination of hyperglycemia. This suggests that, although the mitochondria attempt to initiate repair mechanisms by inducing gene expressions of glycosylases, their protein abundance in the mitochondria remains deficient in maintaining the stability, and this process does not reverse after hyperglycemia is terminated. The plausible mechanisms for such disparity could be the impaired trafficking of the enzymes to the mitochondria. In support, mitochondrial membrane potential is considered to serve as one of the major driving forces for the trafficking of these nuclear encoded glycosylases to the mitochondria (19), and we have shown that retinal mitochondrial membrane potential is impaired in diabetes (21).
The major functional consequences of mtDNA damage are subnormal complex I and III and impairment in membrane potential (46). Complex I has 7 ND subunits encoded by mtDNA, but it also has at least 38 subunits encoded by nDNA. Our data show that the transcript abundance of mtDNA encoded subunits ND1 and ND6 is decreased in the retina in diabetes, but ND4 remains unchanged. The possible reason for ND4 escaping damage could be the random nature of the oxidative DNA damage. However, though we observed decreased mRNA of mitochondrial encoded ND1 and ND6, complex I activity is not compromised in diabetes. The reason for such disparity could be that in diabetes the amount of functional complex I remain sufficient to sustain normal activity. In addition, others have shown that both ND1 and ND 6 are important in the assembly of complex I, but do not have much effect on NADH-dehydrogenase activity (2, 5, 7).
Cytochrome b is essential for the formation and activity of complex III (3, 39), and the functional consequence of the reduction in cytochrome b transcript is its impaired activity. Here we show that the complex III activity is decreased in the retina in diabetes, and this continues even after hyperglycemia is terminated. This modest decrease could be sufficient to increase ROS because ROS produced by complex III are directed away from the antioxidant defenses of the matrix. Our previous studies have shown that complex III is one of the major sources of increased retinal superoxide in diabetes (21). Thus, this continued mtDNA damage could result in a perpetual loop: hyperglycemia increases superoxide, this damages mtDNA, and damaged mtDNA compromises the electron transport system. The decline in flux through the electron transport chain increases diversion of electrons to oxygen and this result in additional superoxide, and the cycle continues.
DCCT studies have demonstrated the importance of early and sustained intensive glycemic control in diabetic patients (12–14). Here we show that retinal mtDNA escapes damage and complex III activity remains normal if good glycemic control is instituted soon after the induction of diabetes (GC group). The results also demonstrate that the damage, once initiated, is not easily reversible, and further strengthens the importance of early and sustained good glycemic control for diabetic patients.
Due to technical limitations in preparing isolated retinal microvessels (22), the mitochondria fraction used in the present study was isolated from the whole retina. This did not allow the identification of the specific cell types undergoing DNA damage. Our recent study has shown that re-institution of good glycemic control in the retinal microvasculature of the same rat model, fails to normalize increased accumulation of peroxynitrite, and also the development of retinopathy (23, 31). Here we show that retinal mtDNA damage is also not reversed after reversal of hyperglycemia. This strongly supports that retinal microvasculature is one of the targets experiencing mtDNA damage in diabetes, and strengthens its role in the development and progression of retinopathy.
In summary, our study is the first to show that the failure of mtDNA damage to reverse has an important role in the continued progression of retinopathy after hyperglycemia is terminated; damaged mtDNA and impaired DNA repair system continue to compromise the mitochondrial electron transport system during normal glycemic control. This normal glycemic control that has followed a period of poor glycemic control provides no relief to the mitochondria from continuous damage from superoxide, and the activation of the major pathways implicated in the development of diabetic retinopathy initiated during hyperglycemia does not halt. As we try to understand this process, repair and maintenance of mtDNA may serve as a potential therapeutic target to combat the development and progression of diabetic retinopathy.
Abbreviations Used
- 8-OHdG
8-hydroxy-2′-deoxyguanosine
- BER
base excision repair
- Cox IV
cytochrome c oxidase subunit IV
- DCCT
Diabetes Control and Complications Trial
- DTNB
5, 5′-dithiobis-2-nitrobenzoic acid
- EDIC
Epidemiology of Diabetes Interventions and Complications
- ETC
electron transport chain
- GC
good glycemic control
- GHb
glycated hemoglobin
- GSH
reduced glutathione
- MnSOD
manganese superoxide dismutase
- MOPS
4-morpholinepropanesulfonic acid
- mtDNA
mitochondrial DNA
- MYH
MutY homolog
- nDNA
nuclear DNA
- OGG1
8-oxoguanine DNA glycosylase
- PC
poor glycemic control
- PC-GC
poor control for 6 months followed by good control for 6 additional months
- ROS
reactive oxygen species
- TDG
thymine DNA glycosylase
Acknowledgments
The authors sincerely appreciate the technical assistance of Dr. Srirupa Das, Mr. Divyesh Sarman, and Yakov Shamailov. This study was supported in part by grants from the National Institutes of Health, Juvenile Diabetes Research Foundation, the Thomas Foundation, and Research to Prevent Blindness.
Authors Disclosure Statement
No competing financial interests exist for Sally A. Madsen–Bouterse, Ghulam Mohammad, Mamta Kanwar, and Renu A. Kowluru.
References
- 1.Ayala–Torres S. Chen Y. Svoboda T. Rosenblatt J. Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22:135–147. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- 2.Bai Y. Attardi G. The mtDNA-encoded ND6 subunit of mitochondrial NADH dehydrogenase is essential for the assembly of the membrane arm and the respiratory function of the enzyme. EMBO J. 1998;17:4848–4858. doi: 10.1093/emboj/17.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballinger SW. Bouder TG. Davis GS. Judice SA. Nicklas JA. Albertini RJ. Mitochondrial genome damage associated with cigarette smoking. Cancer Res. 1996;56:5692–5697. [PubMed] [Google Scholar]
- 4.Ballinger SW. Van Houtens B. Jin GF. Conklin CA. Godley BF. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp Eye Res. 1999;68:765–772. doi: 10.1006/exer.1998.0661. [DOI] [PubMed] [Google Scholar]
- 5.Bourges I. Ramus C. Mousson de Camaret B. Beugnot R. Remacle C. Cardol P. Hofhaus G. Issartel JP. Structural organization of mitochondrial human complex. I: Role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochem J. 2004;383:491–799. doi: 10.1042/BJ20040256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownlee M. The pathobiology of diabetic complications: A unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 7.Cardol P. Matagne RF. Remacle C. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas. Implication for the structural organization of the enzyme. J Mol Biol. 2002;19:1211–1221. doi: 10.1016/S0022-2836(02)00407-2. [DOI] [PubMed] [Google Scholar]
- 8.Chen XJ. Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2008;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 9.Cortina MS. Gordon WC. Lukiw WJ. Bazan NG. Oxidative stress-induced retinal damage up-regulates DNA polymerase gamma and 8-oxoguanine-DNA-glycosylase in photoreceptor synaptic mitochondria. Exp Eye Res. 2005;81:742–750. doi: 10.1016/j.exer.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Cortopassi G. Wang E. Modeling the effects of age-related mtDNA mutation accumulation; complex I deficiency, superoxide and cell death. Biochim Biophys Acta. 1995;1271:171–176. doi: 10.1016/0925-4439(95)00025-y. [DOI] [PubMed] [Google Scholar]
- 11.de Souza–Pinto NC. Wilson DM., 3rd Stevnsner TV. Bohr VA. Mitochondrial DNA, base excision repair and neurodegeneration. DNA Repair (Amst) 2008;7:91098–1109. doi: 10.1016/j.dnarep.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diabetes Control. Complications Trial Research Group. The effect of intensive treatment of diabetes on the development of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med. 1993;329:977–986. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- 13.Diabetes Control Complications Trial/Epidemiology of Diabetes Interventions Complications Research Group. Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. N Engl J Med. 2000;342:381–389. doi: 10.1056/NEJM200002103420603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diabetes Control Complications Trial/Epidemiology of Diabetes Interventions Complications Research Group. Prolonged effect of intensive therapy on the risk of retinopathy complications in patients with type 1 diabetes mellitus: 10 years after the Diabetes Control and Complications Trial. Arch Ophthalmol. 2008;126:1707–1715. doi: 10.1001/archopht.126.12.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Druzhyna NM. Wilson GL. LeDoux SP. Mitochondrial DNA repair in aging and disease. Mech Ageing Dev. 2008;129:383–390. doi: 10.1016/j.mad.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Y. Miller CM. Kern TS. Hyperglycemia increases mitochondrial superoxide in retina and retinal cells. Free Radic Biol Med. 2003;35:1491–1499. doi: 10.1016/j.freeradbiomed.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Godley BF. Shamsi FA. Liang FQ. Jarrett SG. Davies S. Boulton M. Blue light induces mitochondrial DNA damage and free radical production in epithelial cells. J Biol Chem. 2005;280:21061–21066. doi: 10.1074/jbc.M502194200. [DOI] [PubMed] [Google Scholar]
- 18.Hegde ML. Hazra TK. Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrmann JM. Neupert W. What fuels polypeptide translocation? An energetical view on mitochondrial protein sorting. Biochem Biophys Acta. 2000;1459:331–338. doi: 10.1016/s0005-2728(00)00169-9. [DOI] [PubMed] [Google Scholar]
- 20.Jarrett SG. Albon J. Boulton M. The contribution of DNA repair and antioxidants in determining cell type-specific resistance to oxidative stress. Free Radic Res. 2006;40:155–1165. doi: 10.1080/10715760600876613. [DOI] [PubMed] [Google Scholar]
- 21.Kanwar M. Chan PS. Kern TS. Kowluru RA. Oxidative damage in the retinal mitochondria of diabetic mice: Possible protection by superoxide dismutase. Invest Ophthalmol Vis Sci. 2007;48:3805–3811. doi: 10.1167/iovs.06-1280. [DOI] [PubMed] [Google Scholar]
- 22.Kanwar M. Kowluru RA. Diabetes regulates small molecular weight G-protein, H-Ras, in the microvasculature of the retina: Implication in the development of retinopathy. Microv Res. 2008;76:189–193. doi: 10.1016/j.mvr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanwar M. Kowluru RA. Role of glyceraldehyde 3-phosphate dehydrogenase in the development and progression of diabetic retinopathy. Diabetes. 2009;58:227–234. doi: 10.2337/db08-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kern TS. Tang J. Mizutani M. Kowluru RA. Nagaraj RH. Romeo G. Podesta F. Lorenzi M. Response of capillary cell death to aminoguanidine predicts the development of retinopathy: Comparison of diabetes and galactosemia. Invest Ophthalmol Vis Sci. 2000;41:3972–3978. [PubMed] [Google Scholar]
- 25.Kowluru RA. Tang J. Kern TS. Abnormalities of retinal metabolism in diabetes and experimental galactosemia. VII. Effect of long-term administration of antioxidants on the development of retinopathy. Diabetes. 2001;50:1938–1942. doi: 10.2337/diabetes.50.8.1938. [DOI] [PubMed] [Google Scholar]
- 26.Kowluru RA. Effect of re-institution of good glycemic control on retinal oxidative stress and nitrative stress in diabetic rats. Diabetes. 2003;52:818–823. doi: 10.2337/diabetes.52.3.818. [DOI] [PubMed] [Google Scholar]
- 27.Kowluru RA. Abbas SN. Diabetes-induced mitochondrial dysfunction in the retina. Invest Ophthalmol Vis Sci. 2003;44:5327–5334. doi: 10.1167/iovs.03-0353. [DOI] [PubMed] [Google Scholar]
- 28.Kowluru RA. Koppolu P. Chakrabarti S. Chen S. Diabetes-induced activation of nuclear transcriptional factor in the retina, and its inhibition by antioxidants. Free Radic Res. 2003;37:1169–1180. doi: 10.1080/10715760310001604189. [DOI] [PubMed] [Google Scholar]
- 29.Kowluru RA. Chakrabarti S. Chen S. Re-institution of good metabolic control in diabetic rats and activation of caspase-3 and nuclear transcriptional factor in the retina. Acta Diabetologica. 2004;44:194–199. doi: 10.1007/s00592-004-0165-8. [DOI] [PubMed] [Google Scholar]
- 30.Kowluru RA. Atasi L. Ho YS. Role of mitochondrial superoxide dismutase in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2006;47:1594–1599. doi: 10.1167/iovs.05-1276. [DOI] [PubMed] [Google Scholar]
- 31.Kowluru RA. Kanwar M. Kennedy A. Metabolic memory phenomenon and accumulation of peroxynitrite in retinal capillaries. Exp Diab Res. 2007;2007:21976. doi: 10.1155/2007/21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowluru RA. Kanwar M. Chan PS. Zhang JP. Inhibition of retinopathy and retinal metabolic abnormalities in diabetic rats with AREDS-based micronutrients. Arch Ophthalmol. 2008;126:1266–1272. doi: 10.1001/archopht.126.9.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowluru RA. Chan PS. Metabolic memory in diabetes–from in vitro oddity to in vivo problem: Role of Apoptosis. Brain Res Bull. 2010;87:2297–302. doi: 10.1016/j.brainresbull.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kucej M. Kucejova B. Subramanian R. Chen XJ. Butow RA. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J Cell Sci. 2008;121:1861–1868. doi: 10.1242/jcs.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang FQ. Godley BF. Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: A possible mechanism for RPE aging and age-related macular degeneration. Exp Eye Res. 2003;76:397–403. doi: 10.1016/s0014-4835(03)00023-x. [DOI] [PubMed] [Google Scholar]
- 36.Madsen–Bouterse SA. Zhong Q. Mohammad G. Ho YS. Kowluru RA. Diabetic retinopathy: Protection of oxidative damage of mitochondrial DNA by manganese superoxide dismutase. Free Radic Res. 2010;44:313–321. doi: 10.3109/10715760903494168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizutani M. Kern TS. Lorenzi M. Accelerated death of retinal microvascular cells in human and experimental diabetic retinopathy. J Clin Invest. 1996;97:2883–2890. doi: 10.1172/JCI118746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obrosova IG. Minchenko AG. Marinescu V. Fathallah L. Kennedy A. Stockert CM. Frank RN. Stevens MJ. Antioxidants attenuate early up regulation of retinal vascular endothelial growth factor in streptozotocin-diabetic rats. Diabetologia. 2001;44:1102–1110. doi: 10.1007/s001250100631. [DOI] [PubMed] [Google Scholar]
- 39.Rana M. de Coo I. Diaz F. Smeets H. Moraes CT. An out-of-frame cytochrome b gene deletion from a patient with parkinsonism is associated with impaired complex III assembly and an increase in free radical production. Ann Neurol. 2000;48:774–781. [PubMed] [Google Scholar]
- 40.Roy S. Sala R. Cagliero E. Lorenzi M. Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci USA. 1990;87:404–408. doi: 10.1073/pnas.87.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schärer OD. Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 42.Stuart JA. Brown MF. Mitochondrial DNA maintenance and bioenergetics. Biochim Biophys Acta. 2006;1757:79–89. doi: 10.1016/j.bbabio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Van Houten B. Woshner V. Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 2005;5:145–152. doi: 10.1016/j.dnarep.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Wang AL. Lukas TJ. Yuan M. Neufeld AH. Increased mitochondrial DNA damage and down-regulation of DNA repair enzymes in aged rodent retinal pigment epithelium and choroid. Mol Vis. 2008;14:644–651. [PMC free article] [PubMed] [Google Scholar]
- 45.Wang AL. Lukas TJ. Yuan M. Neufeld AH. Age-related increase in mitochondrial DNA damage and loss of DNA repair capacity in the neural retina. Neurobiol Aging. 2008. Dec 10. [Epub ahead of print] [DOI] [PubMed]
- 46.Xie L. Zhu X. Hu Y. Li T. Gao Y. Shi Y. Tang S. Mitochondrial DNA oxidative damage triggering mitochondrial dysfunction and apoptosis in high glucose-induced HRECs. Invest Ophthalmol Vis Sci. 2008;49:4203–4209. doi: 10.1167/iovs.07-1364. [DOI] [PubMed] [Google Scholar]
- 47.Yakes FM. Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]