Abstract
Background
Human-like H3N2 influenza viruses have repeatedly been transmitted to domestic pigs in different regions of the world, but it is still uncertain whether any of these variants could become established in pig populations. The fact that different subtypes of influenza viruses have been detected in pigs makes them an ideal candidate for the genesis of a possible reassortant virus with both human and avian origins. However, the determination of whether pigs can act as a “mixing vessel” for a possible future pandemic virus is still pending an answer. This prompted us to gather the epidemiological information and investigate the genetic evolution of swine influenza viruses in Jilin, China.
Methods
Nasopharyngeal swabs were collected from pigs with respiratory illness in Jilin province, China from July 2007 to October 2008. All samples were screened for influenza A viruses. Three H3N2 swine influenza virus isolates were analyzed genetically and phylogenetically.
Results
Influenza surveillance of pigs in Jilin province, China revealed that H3N2 influenza viruses were regularly detected from domestic pigs during 2007 to 2008. Phylogenetic analysis revealed that two distinguishable groups of H3N2 influenza viruses were present in pigs: the wholly contemporary human-like H3N2 viruses (represented by the Moscow/10/99-like sublineage) and double-reassortant viruses containing genes from contemporary human H3N2 viruses and avian H5 viruses, both co-circulating in pig populations.
Conclusions
The present study reports for the first time the coexistence of wholly human-like H3N2 viruses and double-reassortant viruses that have emerged in pigs in Jilin, China. It provides updated information on the role of pigs in interspecies transmission and genetic reassortment of influenza viruses.
Introduction
The pig is considered to be an important host of influenza A viruses as it might be associated with the generation of human pandemic influenza strains [1]. Historically, two human pandemic viruses, H1N1 in 1918 and H3N2 in 1968, were almost simultaneously detected both in humans and pigs [2], [3]. Although the pandemic H3N2 virus was initially detected in humans and the full genome of the 1918 H1N1 pandemic virus was recently decoded [4], it is still unknown whether these viruses were first introduced into humans or into pigs before they became human pandemic strains.
Currently, H1N1, H1N2 and H3N2 influenza subtype viruses co-circulate in pigs widely throughout the world. All of these viruses were the result of either interspecies transmission or reassortment events [5]–[7]. The Sydney-like H3N2 variants from pigs in the U.S.A. in 1998 were double and triple reassortants containing viral genes of human, swine and avian origin [8]. This highlights the complex and dynamic influenza ecology in pig populations.
Here we present the results of the genetic and phylogenetic characterization of swine H3N2 influenza viruses isolated from 2007 to 2008 in Jilin province of China. Genetic analysis showed that wholly contemporary human-like H3N2 viruses and double-reassortant viruses containing genes from contemporary human (PB2, PB1, PA, HA, NP, and NA) and avian H5 (M and NS) viruses were co-circulating in pig populations. This is the first description of an instance of reassortment between mammal H3N2 and avian H5 influenza viruses. The coexistence of entirely contemporary human-like viruses and double-reassortant viruses provides further evidence that pigs serve as intermediate hosts or mixing vessels. This emphasizes the importance of reinforcing swine influenza virus surveillance.
Results
HA1 amino acid analysis
To investigate all of the detailed genetic characteristics, we compared the deduced amino acid sequences of the hemagglutinin 1 (HA1) gene from the three swine H3N2 isolates against the representatives of five lineages (avian, European swine, earliest human, early human and contemporary human) available in GenBank. For the five lineages, the three H3N2 isolates showed a close relationship to HA1 genes from the contemporary human lineage with a sequence similarity of 94.8∼97.6% to Moscow/10/99, while there was 80.9∼87.2% similarity compared with the representatives of four other lineages (Dk/Hong Kong/7/75 (ABB88256.1), Sw/Italy/1461/96 (CAC40048.1), Hong Kong/1/68 (ACU79871.1), Port Chalmers/1/73 (AAC78096.1), and Victoria/3/75 (CAA24270.1)).
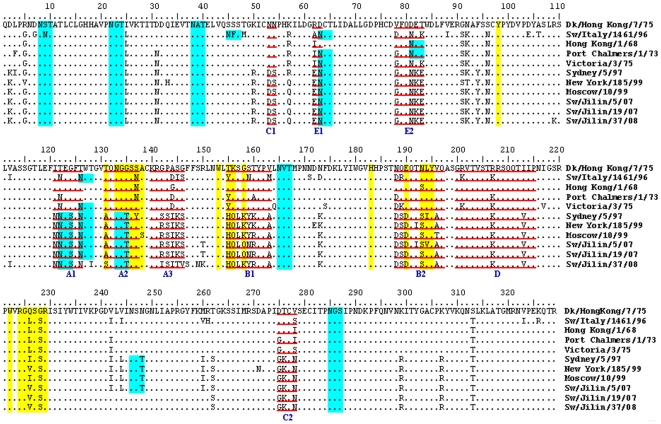
Analysis of amino acid variations of the proposed antigenic sites [9]–[11], receptor-binding sites [12], and potential glycosylation sites was conducted and variations are shown in Figure 1. The HA1 domain of HA, the major antigenic protein of influenza A viruses, contains all the antigenic sites of HA and is under continual immune-driven selection. For H3N2 viruses, the antigenic sites A∼E have been described [9]–[11]. All variations were accumulated at the antigenic sites A and B, while C, D and E were relatively conserved for the contemporary human-like H3N2 lineage. Concerning the three swine H3N2 isolates, two or four amino acid substitutions were observed at the major antigenic sites (A and B) of the HA1 molecule compared with Moscow/10/99, the representative for contemporary human lineage (Figure 1 and 2).
Figure 1. Alignment of the HA1 amino acid sequences of three swine H3N2 influenza virus isolates and the representatives of five H3N2 lineages.
The underlined residues represent the antigenic sites (lowercase letters indicate discrete antigenic sites), residues in green represent the potential glycosylation sites, and residues in yellow shade denote the receptor-binding sites.
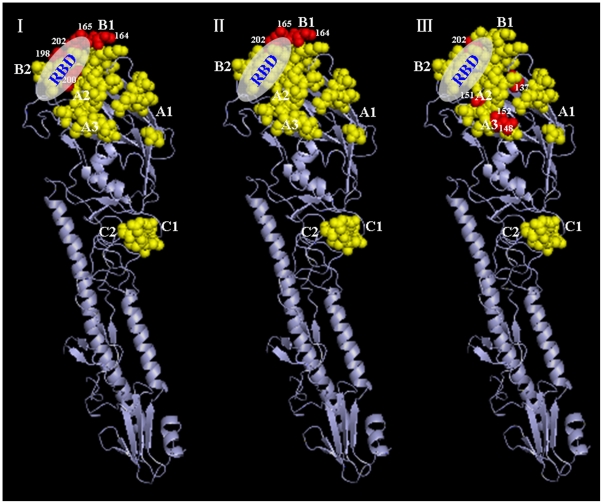
Figure 2. Cartoon diagram representing the amino acid changes at the HA1 molecule of H3 subtype influenza viruses.
RBD: receptor-binding domain. A, B and C: major antigenic sites at the HA molecules. Yellow color: conserved amino acids and red color: changed amino acids. (|) Sw/Jilin/5/07 vs. Moscow/10/99, (||) Sw/Jilin/19/07 vs. Moscow/10/99, (|||) Sw/Jilin/37/08 vs. Moscow/10/99.
Amino acids at the receptor-binding sites of the HA1 protein are associated with the differences in the receptor-binding specificity [13]. All of the H3N2 viruses had relatively conserved receptor-binding sites at Y98, G134, S136, W153, H183, Y195, and R224. Eight obvious mutations (A/S131T, T135G, H155T/Y, Q156K, K/Q158G, D190E, S193N, I/V194L) occurred between the contemporary human lineage and other lineages; these mutations were unique to the contemporary human lineage. Residues responsible for the sialic acid-α2,6-galactose (SAα2,6Gal) of H3N2 are L226 and S228 [14]. The viruses of avian lineage had Q at position 226, while the viruses of European swine, earliest human, and early human lineages had L, and the viruses of contemporary human lineage had I/V. The viruses of avian lineage had G at position 228, with other four lineages being S. In this study, all of the swine H3N2 isolates contained V instead of L at position 226.
It has been considered that carbohydrate side chains might affect receptor-binding capacity and antigenicity [11], [15], [16]. Analysis of potential glycosylation sites in the HAs of the three H3N2 isolates revealed eight common sites (N8, 22, 38, 63, 122, 133, 165 and 285, respectively) with the NXT/S motif (in which X may be any amino acid except aspartic acid and proline). Furthermore, as shown in Figure 1, both Sw/Jilin/5/07 and Sw/Jilin/19/07 possessed an additional glycosylation site at position 126, and Sw/Jilin/5/07 acquired another one at position 246.
Phylogenetic analysis
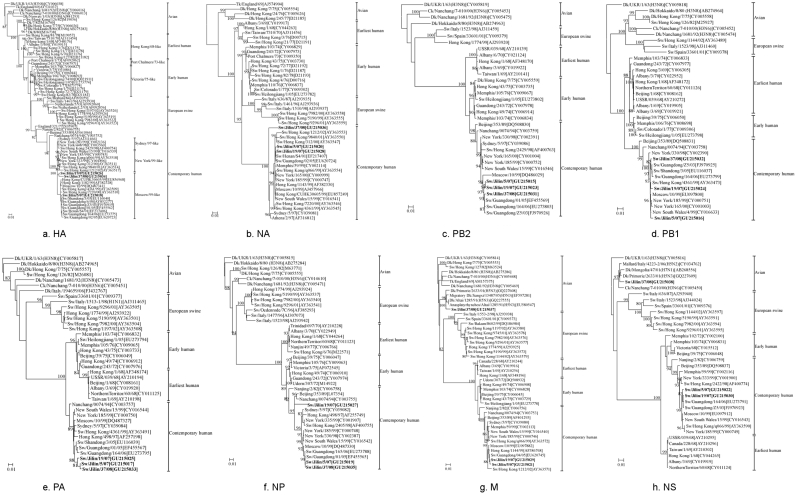
Phylogenetic analysis and identification of antigenically different strains are necessary to monitor the evolution of influenza virus. The phylogenetic relationships among swine H3N2 viruses prevalent in Jilin province compared to the selected reference strains available in the GenBank were estimated from the nucleotide sequences of each viral gene. The phylograms for all genes are shown in Figures 3a∼3h.
Figure 3. Phylogenetic trees of the eight gene segments of the influenza viruses.
The tree was created by the neighbor-joining method and bootstrapped with 1,000 replicates. The bootstrap numbers are given for each node. Only bootstrap values above 80 are shown. Viruses isolated in the present study are in bold. Abbreviations used in virus designations are as follows: Ck, chicken; Dk, duck; Sw, swine; Tk, turkey.
Phylogenetic analysis of H3 HAs showed that H3N2 viruses could be segregated into five distinct lineages, including avian strains, European swine strains, earliest human strains, early human strains, and contemporary human strains (Figure 3a). The swine viruses of earliest human lineage seemed to be derived from the human strain A/Hong Kong/1/68. The early-human-derived swine viruses were closely related to A/Victoria/75-like viruses. The swine viruses of European swine lineage were probably derived from European H3N2 swine influenza viruses, for which the HA genes originated from A/Victoria/3/75. The contemporary human lineage included three sublineages (represented by Sydney/97-like, New York/99-like and Moscow/99-like viruses). The HAs of the three swine H3N2 virus isolates tested in this study clustered into the Moscow/99-like virus sublineage.
The phylogeny of the NA and internal genes of H3N2 viruses paralleled to that of the HA genes, in which five different lineages were defined. The NA, PB2, PB1, PA, and NP genes of the three isolates in this study belonged to the contemporary human lineage. Additionally, in the NA tree, Sw/Jilin/37/08 was equally closely related to the early and contemporary H3 strains as well as an intermediate swine virus between the early lineage and the contemporary human lineage. In the M and NS trees, Sw/Jilin/5/07 and Sw/Jilin/19/07 still clustered in the contemporary human lineage, whereas Sw/Jilin/37/08 was incorporated into the avian lineage (Figures 3b∼3h).
The results of the published data in GenBank revealed that in recent years H3N2 influenza viruses were cocirculating with H5 influenza viruses, raising the possibility of genetic exchange between these viruses. The evolutionary trees revealed that the M gene of Sw/Jilin/37/08 formed a sublineage with H5N3 viruses (A/Dk/Altai/1285/1991, A/Dk/Primorie/2633/2001, A/Anasplatyrhynchos/Altai/1285/1991, and A/Migratory Dk/Jiangxi/13487/2005 (Figure 3g)). The NS gene of Sw/Jilin/37/08 had a close relationship with A/Mallard/Italy/4223-2/2006 (H5N2) and A/Dk/Primorie/2633/2001 (H5N3) (Figure 3h).
Discussion
The rapid evolution of influenza viruses occurs both clonally and non-clonally through a variety of genetic mechanisms and selection pressure. Three mechanisms on the non-clonal evolution of influenza viruses have been proposed: (i) relatively frequent reassortment among gene segments of multiple host types, (ii) possibly but rarely non-homologous recombination, in which short regions of sequence are transferred among different segments, and (iii) controversial and extremely rare homologous recombination within segments [17]. Reassortment, therefore, plays an important role in the non-clonal evolution of the influenza viruses.
Infection of pigs with H1, H3, H5 and H9 subtype influenza viruses has occurred frequently on multiple occasions [5], [18]–[23]. As pigs can serve as intermediate hosts or mixing vessels for the reassortment of human and avian influenza viruses, swine influenza infections have been the focus of increasing attention. It often appeared that multiple reassortments occurred among the same subtype or different subtypes of human, swine and avian viruses in pig populations. The best examples are the swine H1N2 isolates and the pandemic novel H1N1 influenza virus that emerged in the human population. Peiris et al [24] were the first to confirm that cocirculation of contemporary human H3N2 viruses and avian H9N2 in pigs had occurred in southeastern China. Xu et al [25] reported that the avian H9N2 viruses caused pig disease and death in clinics, and deduced that it probably originated from a reassortant of avian H5 influenza virus and H9N2 viruses. All of these situations indicated that the cocirculation of H1, H3, H5 and H9 viruses in pigs would provide an opportunity for genetic reassortment, leading to the emergence of viruses with pandemic potential. Avian influenza viruses, especially when avian H5 subtype viruses reassorted with suitable surface glycoproteins, can infect humans [26]. Yu et al [27] described the coexistence of wholly human-like H3N2 viruses, double-reassortant and triple-reassortant H3N2 viruses in pigs in China from 1970 to 2006. However, reassortment was not found between mammal H3N2 viruses and avian H5 viruses. From July 2007 to October 2008, we collected 279 nasopharyngeal swabs from pigs diagnosed with respiratory tract illness on different pig farms in Jilin province. The H3N2 influenza virus infections in the pigs were diagnosed by virus isolation. The results showed that one Moscow/10/99-like reassortant isolate, A/swine/Jilin/37/2008, contained avian H5-like M and NS genes, suggesting that after introduction to pigs, avian H5 viruses further reassorted with contemporary human-like H3N2 viruses. To our knowledge, such reassortment between mammal H3N2 viruses and avian H5 viruses has not been described in existing publications.
A comparison of the amino acid sequences of the HA1 region of our isolates with the representatives of avian lineage (Dk/Hong Kong/7/75), European swine lineage (Sw/Italy/1461/96), earliest human lineage (Hong Kong/1/68), early human lineage (Victoria/3/75 and Port Chalmers/1/73), and contemporary human lineage (Sydney/5/97, New York/185/99, and Moscow/10/99) showed that the three H3N2 swine isolates were more closely related to Moscow/10/99 (94.8∼97.6% amino acid similarity), indicating that the HAs of them seemed to be derived from those of the contemporary human lineage. Phylogenetic analysis of the HAs also suggested that the three H3N2 isolates had been introduced into pigs at several points in time from the human side.
The comparison of antigenic sites of HA1 regions revealed evolution by antigenic drift of their HA genes. Codons under positive selection were associated with antigenic site A and B [28]. Sw/Jilin/19/07 had two variations at the antigenic sites A and B, while Sw/Jilin/5/07 and Sw/Jilin/37/08 accumulated four variations at the antigenic sites A and B. The gradual mutations of HA might generate new antigenic strains. Wilson and Cox [29] proposed that a drift variant with ≥4 amino acid changes at ≥2 out of 5 antigenic sites would be of epidemiologic importance. It has been also observed that new antigenic variations are created either when ≥2 variations occur in antigenic sites or when one variation occurs in an antigenic site and one in a sialic acid receptor-binding site [30].
Although the molecular basis of host-range restrictions is not completely defined, the compatibility between the HA protein of the virus and its corresponding receptor, sialic acid, on the host cell is thought to contribute in part to the infection of the virus in a specific host [31], [32]. Pigs, unlike humans, seem to be readily infected by most, if not all, mammalian and avian influenza viruses. The susceptibility of pigs to both mammalian and avian viruses is due to the presence of receptors for both lineages of virus in the pig trachea [33]. For H3N2 viruses, residues 226 and 228 on the receptor-binding domain of the HA1 molecule were shown to play a critical role in determining receptor specificity [14], [34], [35]. The three H3N2 isolates possess V226 and S228, which are the same as those of both turkey and swine triple reassortants. While L/I226 and S228 are usually expressed in human viruses [36], Q226 and G228 are usually found in avian viruses [14]. V, L and I are neutral non-polar amino acids, and substitutions between them most likely maintain the hydrophobic interactions and the proper 3D conformation at the binding domain [37].
Carbohydrate side chains are important for the structure and stability of glycoproteins [38], [39]. Changes in carbohydrate side chains are an important mechanism in the structural variation underlying antigenic drift. In this study, the three swine H3N2 isolates had eight to ten glycosylation sites, although all of them may not be used. Of these sites, positions 122, 133, and 246 were unique to the contemporary human lineage. In addition, the lineages of European swine, earliest human, early human, and contemporary human had more glycosylation sites than the avian lineage. The addition of new carbohydrate side chains to HA may have provided the viruses with the ability to evade antibody pressure by changing the antigenicity and by having an increased ability to prevail. For instance, human virus variants which cocirculated in the epidemic area with a higher number of glycosylation sites appeared to prevail at the end of the outbreak [40]. Conclusively, molecular analysis of the hemagglutinin gene showed that the 2007–2008 H3N2 influenza viruses circulating in swine populations in Jilin province accumulate variations at the antigenic sites A and B, receptor-binding sites, as well as glycosylation sites.
Phylogenies of the whole genome of the swine H3N2 influenza viruses in Jilin province, China during 2007 to 2008 provided evidence of the persistence of both the contemporary human lineage and the avian lineage in pig populations. These results also revealed multiple interspecies transmissions of influenza viruses from human and avian to pig and subsequent reassortment events, particularly together with the participation of internal genes from the avian H5 lineage in this region. This is the first report on reassortment between mammal H3N2 and avian H5 viruses.
Experimental infection suggested that introduction of certain H5 viral segments into circulating human H3N2 viruses may increase their virulence for mice and perhaps other mammalian species [41]. Further research is needed to determine whether Sw/Jilin/37/08 has higher virulence than Sw/Jilin/5/07 and Sw/Jilin/19/07. The principal evolutionary mechanism of influenza virus is by antigenic drift, creating small progressive antigenic changes in the hemagglutinin and neuraminidase surface antigens [42]. However, genetic reassortment readily occurs between influenza viruses and may also contribute to the evolution of new strains. Therefore Sw/Jilin/37/08 may represent a potential threat in the emergence of new human viruses.
There have been numerous descriptions of human infection with swine influenza viruses [5]. The swine populations have become a reservoir of a much more diverse array of influenza viruses. The replicative gene constellation of H3N2 viruses has the capacity to reassort among avian, swine and human viruses. This means that activity in swine virus reservoirs is of concern for human health. The above study provides additional evidence for continuing interspecies transmission and reassortment events occurring in pigs, which naturally increased the possibility of pigs as an important host for the emergence of novel reassortants with genes adapted for replication in pigs or even humans. The coexistence of reassortant viruses, especially reassortants of H3 and H5 viruses, emphasizes that genetic reassortment is an important factor in the evolution of H3N2 viruses and a formal surveillance system is needed for swine and avian influenza. It is only through such a system that cross-interspecies transmission and novel reassortment events will be identified in a timely fashion.
Materials and Methods
Viruses
A total of 279 nasopharyngeal swabs were collected in Jilin province, China from July 2007 to October 2008. The initial isolation of the viruses was performed in Madin-Darby canine kidney (MDCK) cells. The viruses were grown in Eagle minimal essential medium (GIBCO/BRL) supplemented with 5% fetal bovine serum (GIBCO/BRL), penicillin-streptomycin (GIBCO/BRL), amphotericin B (Fungizone; GIBCO/BRL) and tolylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin (1 µg/ml; Worthington Biochemical Corporation, Lakewood, N.J.). Subtype identification of these viruses were determined by standard hemagglutination inhibition tests and neuraminidase inhibition tests with a panel of reference antisera recommended by the World Health Organization (http://www.who.Int/csr/resources/publications/en/#influenza). Viral fluids were harvested for MDCK-passaged viruses and used as stock for sequence analysis. The three H3N2 virus isolates obtained in this study were named as follows: A/swine/Jilin/5/2007 (Sw/Jilin/5/07), A/swine/Jilin/19/2007 (Sw/Jilin/19/07), and A/swine/Jilin/37/2008 (Sw/Jilin/37/08).
Gene sequencing and phylogenetic analysis
Viral RNA was extracted using Trizol reagents (GIBCO/BRL) and reverse transcription was performed using oligonucleotide influenza universal primer Uni12: 5′-AGC AAA AGC AGG-3′ [43]. After reverse transcription, PCR was done as described by Shu [44] using primers (Table 1) specific for each of the eight RNA segments. PCR products were purified with the QIA quick PCR purification kit (Qiagen). The purified PCR products were then partially sequenced using an Amersham ET Dye terminator kit and analyzed with an ABI 3730 DNA sequencer (Perkin-Elmer Appllied Biosystems, Foster City, CA, USA).
Table 1. The primer sequences used for PCR amplification of H3N2 influenza virus genes.
| Gene | Forward primer 5′→3′ | Reverse primer 5′→3′ | Expected size (bp) |
| PB2 | GCTGATAGTGAGTGGAAGAGACGAACA | AGTAGAAAAAGGTCGTTTTTAAACTATTC | 1166 |
| PB1 | TGCGAGCTGACTGATTCAATCTGGATA | AGTAGAAACAAGGCATTTTTTCATGAA | 2321 |
| PA | TGCGAGCTGACTGATTCAATCTGGATA | AGTAGAAACAAGGTACTTTTTTGGACA | 967 |
| HA | ATGAAGACTATCATTGCTTTGAGCTAC | TCAAATGCAAATGTTGCACCTAATG | 1701 |
| NP | AGCAAAAGCAGGGTAGATAATCACTCA | AGTAGAAACAAGGGTATTTTTCTTTAA | 1565 |
| NA | AGCAAAAGCAGGAGTAAAGATGAAT | AAGCTTATATAGGCATGAGATTGAGG | 1433 |
| M | ATATTGAAAGATGAGCCTTCTAACCG | ACTCCAACTCTATGCTGACAAAATGAC | 990 |
| NS | AGCAAAAGCAGGGTGACAAAGACATAA | AGTAGAAACAAGGGTGTTTTTTATTAT | 890 |
Assembly of sequences, translation of nucleotide sequences into protein sequences, and initial multiple sequence alignments were performed with the Clustal V method using MegAlign software version 1.03 (SNAStar Inc., Madison, WI).
The reference strains selected for phylogenetic analysis are based on the following criteria: 1. Using a blast search (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the most genetically closest segment sequence is selected. 2. The selected strains are well-characterized phylogenetically, so that they can represent their lineage and host origin, such as avian, pig or human. 3. The general topology of the phylogenetic tree constructed using the selected reference strains is consistent with previously well-recognized evolutionary analysis. Bootstrap support for tree topologies was accomplished using the Neighbor-joining (NJ) methods implemented in MEGA 4.0 with 1,000 iterations [45]. Genetic distances based on NJ phylogenetic trees were calculated applying Kimura's two-parameter method. In this study, the nucleotide sequences used for the phylogenetic analysis are as follows: PB2 1185-2306, PB1 64-2241, PA 1266-1579, HA 78-1064, NP 46-1504, NA 89-1399, M 35-990, and NS 27-822.
Molecular graphic visualization
Amino acid changes at the five antigenic sites (A∼E) [9]–[11] of the HA1 molecule were determined by amino acid sequence alignment using the MegAlign program. Changes at the major antigenic sites (A, B and C) of the HA monomer were located using PyMOL software (v0.99) (DeLano Scientific LLC, South San Francisco, California, U.S.A.) based on the HA structure of the H5 subtype influenza virus, A/duck/Singapore/3/97 [46], (1JSM) downloaded from the Protein Data Bank website (http://www.rcsb.org/pdb/home/home.do). The H5 structure was used because it is available as a monomer structure and it would be clearer to visualize the amino acid changes on it than the H3 structure, which is only available as a trimer structure.
Nucleotide sequence accession numbers
The nucleotide sequences for all H3N2 influenza virus isolates analyzed in this study are available from GenBank under accession numbers GU215015 to GU215038 (Table 2).
Table 2. The accession numbers of protein and nucleotide acid of swine H3N2 influenza viruses.
| A/swine/Jilin/5/2007 | A/swine/Jilin/19/2007 | A/swine/Jilin/37/2008 | ||||||||||
| Protein | Nucleotide acid | Protein | Nucleotide acid | Protein | Nucleotide acid | |||||||
| Accession | Length (aa) | Accession | Length(bp) | Accession | Length(aa) | Accession | Length(bp) | Accession | Length(aa) | Accession | Length(bp) | |
| PB2 | ACZ53952 | 373 | GU215015 | 1121 | ACZ53963 | 373 | GU215023 | 1121 | ACZ53974 | 373 | GU215031 | 1121 |
| PB1 | ACZ53953 | 757 | GU215016 | 2317 | ACZ53964 | 757 | GU215024 | 2317 | ACZ53975 | 757 | GU215032 | 2317 |
| PA | ACZ53955 | 302 | GU215017 | 930 | ACZ53966 | 302 | GU215025 | 930 | ACZ53977 | 302 | GU215033 | 930 |
| HA | ACZ53956 | 566 | GU215018 | 1701 | ACZ53967 | 566 | GU215026 | 1701 | ACZ53978 | 566 | GU215034 | 1701 |
| NP | ACZ53957 | 498 | GU215019 | 1541 | ACZ53968 | 498 | GU215027 | 1541 | ACZ53979 | 498 | GU215035 | 1541 |
| NA | ACZ53958 | 469 | GU215020 | 1414 | ACZ53969 | 469 | GU215028 | 1414 | ACZ53980 | 469 | GU215036 | 1414 |
| M1 | ACZ53959 | 249 | GU215021 | 957 | ACZ53970 | 249 | GU215029 | 957 | ACZ53981 | 249 | GU215037 | 957 |
| NS1 | ACZ53961 | 230 | GU215022 | 890 | ACZ53972 | 230 | GU215030 | 890 | ACZ53983 | 230 | GU215038 | 890 |
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by the Jilin University Scientific Research Startup Fund (4305050102B6), the Jilin University Basic Science Research Fund (200903334) and the National Natural Scientific Fund (30972192). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ludwig S, Stitz L, Planz O, Van H, Fitch WM, et al. European swine virus as a possible source for the next influenza pandemic? Virology. 1995;212:555–561. doi: 10.1006/viro.1995.1513. [DOI] [PubMed] [Google Scholar]
- 2.Kundin WD. Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature. 1970;228:857. doi: 10.1038/228857a0. [DOI] [PubMed] [Google Scholar]
- 3.Shortridge KF, Webster RG, Butterfield WK, Campbell CH. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977;196:1454–1455. doi: 10.1126/science.867041. [DOI] [PubMed] [Google Scholar]
- 4.Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci U S A. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol. 2000;74:29–46. doi: 10.1016/s0378-1135(00)00164-4. [DOI] [PubMed] [Google Scholar]
- 6.Brown IH, Harris PA, McCauley JW, Alexander DJ. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J Gen Virol. 1998;79 (Pt 12):2947–2955. doi: 10.1099/0022-1317-79-12-2947. [DOI] [PubMed] [Google Scholar]
- 7.Campitelli L, Donatelli I, Foni E, Castrucci MR, Fabiani C, et al. Continued evolution of H1N1 and H3N2 influenza viruses in pigs in Italy. Virology. 1997;232:310–318. doi: 10.1006/viro.1997.8514. [DOI] [PubMed] [Google Scholar]
- 8.Karasin AI, Schutten MM, Cooper LA, Smith CB, Subbarao K, et al. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 2000;68:71–85. doi: 10.1016/s0168-1702(00)00154-4. [DOI] [PubMed] [Google Scholar]
- 9.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima K, Nobusawa E, Tonegawa K, Nakajima S. Restriction of amino acid change in influenza A virus H3HA: comparison of amino acid changes observed in nature and in vitro. J Virol. 2003;77:10088–10098. doi: 10.1128/JVI.77.18.10088-10098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 12.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 13.Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, et al. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 14.Vines A, Wells K, Matrosovich M, Castrucci MR, Ito T, et al. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. 1998;72:7626–7631. doi: 10.1128/jvi.72.9.7626-7631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohuchi M, Ohuchi R, Feldmann A, Klenk HD. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J Virol. 1997;71:8377–8384. doi: 10.1128/jvi.71.11.8377-8384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–8856. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boni MF, de Jong MD, van Doorn HR, Holmes EC. Guidelines for Identifying Homologous Recombination Events in Influenza A Virus. Plos ONE. 2010;5:e10434. doi: 10.1371/journal.pone.0010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown IH. The pig as an intermediate host for influenza A viruses between birds and humans. Int Congr Ser. 2001;1219:173–178. [Google Scholar]
- 19.Cong YL, Pu J, Liu QF, Wang S, Zhang GZ, et al. Antigenic and genetic characterization of H9N2 swine influenza viruses in China. J Gen Virol. 2007;88:2035–2041. doi: 10.1099/vir.0.82783-0. [DOI] [PubMed] [Google Scholar]
- 20.Marozin S, Gregory V, Cameron K, Bennett M, Valette M, et al. Antigenic and genetic diversity among swine influenza A H1N1 and H1N2 viruses in Europe. J Gen Virol. 2002;83:735–745. doi: 10.1099/0022-1317-83-4-735. [DOI] [PubMed] [Google Scholar]
- 21.Reeth KV, Brown I, Essen S, Pensaert M. Genetic relationships, serological cross-reaction and cross-protection between H1N2 and other influenza A virus subtypes endemic in European pigs. Virus Res. 2004;103:115–124. doi: 10.1016/j.virusres.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Yu H, Hua RH, Wei TC, Zhou YJ, Tian ZJ, et al. Isolation and genetic characterization of avian origin H9N2 influenza viruses from pigs in China. Vet Microbiol. 2008;131:82–92. doi: 10.1016/j.vetmic.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 23.Bi YH, Fu GH, Chen J, Peng JS, Sun YP, et al. Novel Swine Influenza Virus Reassortants in Pigs, China. Emerg Infect Dis. 2010;16:1162–1164. doi: 10.3201/eid1607.091881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, et al. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol. 2001;75:9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu C, Fan W, Wei R, Zhao H. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003(H9N2) virus. Microbes Infect. 2004;6:919–925. doi: 10.1016/j.micinf.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Claas EC, Kawaoka Y, de Jong JC, Masurel N, Webster RG. Infection of children with avian-human reassortant influenza virus from pigs in Europe. Virology. 1994;204:453–457. doi: 10.1006/viro.1994.1553. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Hua RH, Zhang Q, Liu TQ, Liu HL, et al. Genetic evolution of swine influenza A (H3N2) viruses in China from 1970 to 2006. J Clin Microbiol. 2008;46:1067–1075. doi: 10.1128/JCM.01257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science. 1999;286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 29.Wilson IA, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol. 1990;8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 30.Shih AC, Hsiao TC, Ho MS, Li WH. Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc Natl Acad Sci U S A. 2007;104:6283–6288. doi: 10.1073/pnas.0701396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito T. Interspecies transmission and receptor recognition of influenza A viruses. Microbiol Immunol. 2000;44:423–430. doi: 10.1111/j.1348-0421.2000.tb02516.x. [DOI] [PubMed] [Google Scholar]
- 32.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res. 2004;103:67–73. doi: 10.1016/j.virusres.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 35.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, et al. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 36.Lindstrom S, Sugita S, Endo A, Ishida M, Huang P, et al. Evolutionary characterization of recent human H3N2 influenza A isolates from Japan and China: novel changes in the receptor binding domain. Arch Virol. 1996;141:1349–1355. doi: 10.1007/BF01718836. [DOI] [PubMed] [Google Scholar]
- 37.Yassine HM, Lee CW, Suarez DL, Saif YM. Genetic and antigenic relatedness of H3 subtype influenza A viruses isolated from avian and mammalian species. Vaccine. 2008;26:966–977. doi: 10.1016/j.vaccine.2007.11.094. [DOI] [PubMed] [Google Scholar]
- 38.Gallagher PJ, Henneberry JM, Sambrook JF, Gething MJ. Glycosylation requirements for intracellular transport and function of the hemagglutinin of influenza virus. J Virol. 1992;66:7136–7145. doi: 10.1128/jvi.66.12.7136-7145.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olden K, Parent JB, White SL. Carbohydrate moieties of glycoproteins. A re-evaluation of their function. Biochim Biophys Acta. 1982;650:209–232. doi: 10.1016/0304-4157(82)90017-x. [DOI] [PubMed] [Google Scholar]
- 40.Seidel W, Kunkel F, Geisler B, Garten W, Herrmann B, et al. Intraepidemic variants of influenza virus H3 hemagglutinin differing in the number of carbohydrate side chains. Arch Virol. 1991;120:289–296. doi: 10.1007/BF01310484. [DOI] [PubMed] [Google Scholar]
- 41.Li MC, Todd Davis C, Zhou H, Nancy JCox, Donis RO. Genetic Compatibility and Virulence of Reassortants Derived from Contemporary Avian H5N1 and Human H3N2 Influenza A Viruses. Plos Phathogens. 2008;4:1–10. doi: 10.1371/journal.ppat.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daly JM, Wood JM, Robertson JS. Nicholson KG, Webster RG, Hay AJ, editors. Co-circulation and divergence ofhumaninfluenza viruses. Textbook of influenzaOxford: Backwell Science. 1998. pp. 168–177.
- 43.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 44.Shu LL, Lin YP, Wright SM, Shortridge KF, Webster RG. Evidence for interspecies transmission and reassortment of influenza A viruses in pigs in southern China. Virology. 1994;202:825–833. doi: 10.1006/viro.1994.1404. [DOI] [PubMed] [Google Scholar]
- 45.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 46.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. H5 avian and H9 swine influenza virus haemagglutinin structures: possible origin of influenza subtypes. Embo J. 2002;21:865–875. doi: 10.1093/emboj/21.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]