Abstract
Summary: Genome-wide association studies (GWAS) have revealed hundreds of loci associated with common human genetic diseases and traits. We have developed a web-based plotting tool that provides fast visual display of GWAS results in a publication-ready format. LocusZoom visually displays regional information such as the strength and extent of the association signal relative to genomic position, local linkage disequilibrium (LD) and recombination patterns and the positions of genes in the region.
Availability: LocusZoom can be accessed from a web interface at http://csg.sph.umich.edu/locuszoom. Users may generate a single plot using a web form, or many plots using batch mode. The software utilizes LD information from HapMap Phase II (CEU, YRI and JPT+CHB) or 1000 Genomes (CEU) and gene information from the UCSC browser, and will accept SNP identifiers in dbSNP or 1000 Genomes format. Single plots are generated in ∼20 s. Source code and associated databases are available for download and local installation, and full documentation is available online.
Contact: cristen@umich.edu
1 INTRODUCTION
Genome-wide association studies (GWAS) have identified hundreds of loci associated with complex human diseases and traits (Manolio et al., 2009). GWAS test for association with dichotomous or quantitative traits at millions of SNPs across the genome and can identify variants many hundreds of kilobases away from any known gene. The next challenge in human genetics will be to identify the causal variants and genes responsible for disease association at the many disease-associated loci identified from GWAS. An associated region may contain only a single strongly associated SNP, or more commonly, a set of SNPs with varying degrees of association due to local linkage disequilibrium (LD) patterns. When examining results from a GWAS, it is important to visually inspect regions showing association to determine the extent of the association signal and the position relative to nearby genes. Genes several hundred kb from an associated SNP may be functionally relevant (Loos et al., 2008). We have developed a web-based tool that provides graphical display of locus-specific association results and gives an overview of the extent of LD and the position relative to nearby genes and local recombination hotspots.
2 IMPLEMENTATION
2.1 Features and functionality
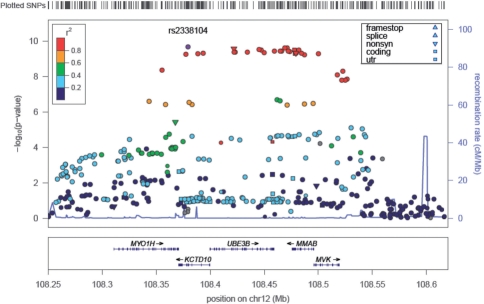
The main panel of a LocusZoom plot shows association P-values on the −log10 scale on the vertical axis, and the chromosomal position along the horizontal axis (Fig. 1). The user can specify the region to display in one of three ways: (i) an index SNP and a window size, (ii) the chromosome together with start and stop positions or (iii) gene name and size of flanking region. We allow for the display of a ‘rug’ above the main panel which gives a tick for any SNP in the results file, or for all SNPs from HapMap Phase II. The plots were designed to display ∼1 Mb windows of the genome, although for regions with several association signals or long-range LD patterns, plots extending further can be drawn.
Fig. 1.
An example LocusZoom plot showing the HDL cholesterol-associated region near the MMAB gene (Kathiresan et al., 2009).
To identify SNPs that may be potentially causative, LocusZoom plots show not only the magnitude of association for each SNP, but also the pairwise LD pattern with the most strongly associated SNP or another user-specified SNP. Quick inspection can reveal the extent of the associated region and the location and number of SNPs in strong LD with the index SNP. In addition, a locus may show strongly associated variants that are weakly correlated, suggesting the presence of multiple independent association signals.
Users may choose to display LD (r2 or D′) estimates from HapMap Phase II (CEU, YRI or JPT + CHB) or from the 1000 Genomes Project. LocusZoom is compatible with 1000 Genomes SNP naming format (chr:position) and will plot association results for novel SNPs identified by sequencing studies.
We provide an option for the data point symbol to reflect genomic annotation (nonsense, non-synonymous, coding, UTR, splice variants, transcription factor binding sites and multi-species conservation), which is available for all SNPs in dbSNP or the 1000 Genomes Project (August 2009 release). The size of the data points can optionally reflect the square root of the sample size.
The bottom panel of a LocusZoom plot shows the name and location of genes in the UCSC Genome Browser (Kent et al., 2002). Positions of exons are displayed, and the transcribed strand is indicated with an arrow. This allows the visual comparison of association results relative to coding regions. Gene names are automatically spaced relative to one another to avoid overlap.
Currently used plotting tools include regional association plotter SNAP (Johnson et al., 2008) and LD-based viewers such as LD-Plus (Bush et al., 2010), CandiSNPer (Schmitt et al., 2010) and VALID (Jorgenson et al., 2009). LocusZoom provides additional features not currently available in any other single tool, such as: (i) the display of 1000 Genomes or novel SNPs from sequencing studies, (ii) functional annotation of SNPs, (iii) exon/intron distinction and automated gene spacing, (iv) ability to plot regions larger than 500 kb, (v) no pre-selection of input files and (vi) web-based batch mode and availability of source code and databases for download and local installation of LocusZoom.
2.2 Usage
LocusZoom was written in R using the grid and lattice graphics packages and runs within a Python wrapper. SQLite tables with relevant data for recombination rate, SNP position, annotation and gene information can be accessed using Python's built-in SQLite tools. A simple plot can be generated from the web form by uploading a file with SNP names and P-values, and specifying the region to be plotted and optional features using drop-down buttons. Typical run time for a single plot returned to the browser window is ∼20 s, not including time required to upload a meta-analysis file, which varies according to the user's internet upload speed and file size. To reduce upload time, users may choose to restrict data files to the region being plotted. To generate a series of locus plots from the web form, users can submit a specification file where custom specifications for each plot can be listed. When a specification file is used to draw many plots, a single pdf containing all generated plots is returned to the user by e-mail. Finally, users can download our scripts, which require R and Python, and associated databases in SQLite format to enable plot generation on their local unix machine. Full documentation of all features is available on the LocusZoom website.
The LocusZoom webpage comes pre-loaded with genome-wide association results for HDL cholesterol, LDL cholesterol and triglycerides in ∼20 000 individuals of European ancestry (Kathiresan et al., 2009).
3 CONCLUSIONS
We have created a user-friendly tool to generate regional plots of association results in their genomic context. LocusZoom allows for quick visual inspection of the strength of association evidence, the extent of the association signal and LD, and the position of the associated SNPs relative to genes in the region. LocusZoom plots provide an option to size the data points relative to sample size and can display functional annotation. LocusZoom can be accessed from a simple web-based form with drop-down menus or by uploading a specification file to generate many plots at once. LocusZoom Python application, source code in R, and associated databases are available for download and we provide instruction for users to create custom database tables. It is anticipated that, in the future, additional publicly available result sets will be available for convenient viewing.
ACKNOWLEDGEMENTS
The authors thank Anne Jackson, Karen Mohlke and Laura Scott for ideas to improve LocusZoom. The UCSC Browser can be found at http://genome.ucsc.edu/. LocusZoom can be found at http://csg.sph.umich.edu/locuszoom.
Funding: R.J.P. is supported by a Research Fellowship from Calvin College. M.B. and T.M.T. are supported by grants from the National Institue of Diabetes and Digestive and Kidney Diseases (DK062370, PI M.B.). M.B., T.N.T. and G.R.A. are supported by the National Human Genome Research Institute (HG000376, PI M.B.; for T.M.T. HG000040, PI M.B.; HG002651, PI G.R.A. and HG005214, PI G.R.A.). G.R.A. is additionally funded by the National Institute of Mental Health (MH084698). C.J.W. is funded by a Pathway to Independence Award from the National Heart, Lung and Blood Institute (K99HL094535, PI C.J.W.).
Conflict of Interest: none declared.
REFERENCES
- Bush WS, et al. Visualizing SNP statistics in the context of linkage disequilibrium using LD-Plus. Bioinformatics. 2010;26:578–579. doi: 10.1093/bioinformatics/btp678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson E, et al. VALID: visualization of association study results and linkage disequilibrium. Genet. Epidemiol. 2009;33:599–603. doi: 10.1002/gepi.20411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AO, et al. CandiSNPer: a web tool for the identification of candidate SNPs for causal variants. Bioinformatics. 2010;26:969–970. doi: 10.1093/bioinformatics/btq068. [DOI] [PubMed] [Google Scholar]