Abstract
Motivation: Earlier studies of protein structure revealed closed loops with a characteristic size 25–30 residues and ring-like shape as a basic universal structural element of globular proteins. Elementary functional loops (EFLs) have specific signatures and provide functional residues important for binding/activation and principal chemical transformation steps of the enzymatic reaction. The goal of this work is to show how these functional loops evolved from pre-domain peptides and to find a set of prototypes from which the EFLs of contemporary proteins originated.
Results: This article describes a computational method for deriving prototypes of EFLs based on the sequences of complete genomes. The procedure comprises the iterative derivation of sequence profiles followed by their hierarchical clustering. The scoring function takes into account information content on profile positions, thus preserving the signature. The statistical significance of scores is evaluated from the empirical distribution of scores of the background model. A set of prototypes of EFLs from archaeal proteomes is derived. This set delineates evolutionary connections between major functions and illuminates how folds and functions emerged in pre-domain evolution as a combination of prototypes.
Contact: Igor.Berezovsky@uni.no
1 INTRODUCTION
Enzymes are involved in all processes in living organisms. Well before the first protein sequence and structure were determined (Sanger, 1952), the function of enzymes became one of the central questions in biochemical studies. Despite the wealth of experimental data available nowadays, the functions of the majority of proteins are still uncharacterized (Levitt, 2009). Since the presence of certain biochemical activities is typically sought for, while all other possible activities (e.g. promiscuous functions) are ignored (Furnham et al., 2009), experimental determination of enzymatic function is in most cases confirmative. Besides, biochemical assays are expensive, are subject to in vitro experimental conditions, and they can not be run on a genomic scale. All the above makes prediction of enzymatic function with computational methods an important alternative approach. There are several general assumptions on which such methods are based: (i) homologous proteins have similar functions; (ii) most of the functional variants emerged as a result of divergence from a common ancestor; (iii) structural homologs, so-called fold superfamilies, persist down to 25% of sequence identity; (iv) divergence below 25% of sequence identity leads to the emergence of families with different organism, substrate, and/or tissue specificities (Lo Conte et al., 2000). Though enzymatic function can be inferred by sequence and structure similarity, the relations between sequence, structure and function are far from being completely understood. Many folds display vast functional diversity. For example, structurally similar β/α barrels provide scaffolds to a number of biochemical functions (Nagano et al., 2002), while particular biochemical functions can be performed by different protein folds, e.g. hydrolase (Lo Conte et al., 2000).
The contemporary evolution of protein structure and function takes place through mutations (Aharoni et al., 2005), recombination and domain swapping and/or interactions (Chothia et al., 2003). However, it is rather obvious that this process was preceded by the emergence of a first set of protein domain structures/folds with a limited repertoire of biochemical functions. These structures emerged from peptides with rudimentary non-specific catalytic activities in the pre-domain stage of evolution (Lupas et al., 2001). Understanding of this process is important for characterizing the most ancient functions and their connections to the modern proteins. The difficulties in understanding and, more importantly, in predicting protein function, are well reflected in the diversity of their descriptions. Enzymatic reactions are classified in enzyme nomenclature (EC) by the biochemical transformation and the substrate (Bairoch, 2000). According to MACiE database, there could be different mechanisms employed for the same transformation (Holliday et al., 2007, 2009). Different biochemical reactions can have the same core mechanism, as it is exemplified by mechanistically diverse superfamilies (Glasner et al., 2006). In order to reconcile different approaches and to develop a generic description of enzymatic functions, one has to start from considering their elementary units which provide binding/activation and principal chemical transformation steps of the whole reaction. Then it should be found out how combinations of these units result in a variety of enzymatic reactions, and how protein folds restrict the possibility of performing a particular biochemical transformation or binding a certain substrate.
The first question that arises in this context is what elements of protein folds serve as elementary units of function. What were the structures of these units in pre-domain evolution, and how did they affect the structures of modern proteins? Earlier studies have shown that soluble proteins contain a basic universal element, stemming from the polymer nature of polypeptide chains, namely closed loops or returns of the polypeptide chain backbone with a typical size of 25–30 amino acid residues (Berezovsky and Trifonov, 2001; Berezovsky et al., 2000). Any protein fold can be decomposed into sets of consecutively connected closed loops (Berezovsky, 2003), indicating their independence in the evolutionary past (Trifonov and Berezovsky, 2003). Can we reconstruct the pre-biotic peptides that gave rise to the elementary functional units of modern proteins? Our hypothesis is that a functional signature revealing the type of binding/activation or principal chemical transformation of the loop can be obtained from the contemporary proteins. This signature complements the description of the closed loop, hence it is an elementary functional loop (EFL). Therefore, the first goal of this study is to investigate how these functional loops evolved from pre-domain peptides, and to find a set of prototypes from which the EFLs of contemporary proteins originate. In order to draw a picture of folds and functions emerging as combinations of EFLs, prototypes of the EFLs will be derived and the corresponding EFLs will be detected in proteins with known biochemical functions. The presence of EFLs in distinct folds and functions unravel evolutionary relations between them and can hint on recipes for protein function (re)design.
The specific nature of prototypes calls for developing a new computational procedure for their derivation and characterization. Indeed, we seek for entities which do not exist in modern proteins, but are represented by their descendants, EFLs. The EFLs themselves presumably have low sequence identity to each other, and, therefore, evolutionary connections between them are not obvious. In this work, we propose a computational procedure to derive prototypes of EFLs from the sequences of complete proteomes. We expect these prototypes to be of closed-loop size (25–30 residues), ring-like shape and to have distinct functional signatures, where several conserved positions in the profile describe chemically active amino acids which are involved in binding/activation steps and/or take part in principal chemical transformations of the substrate.
We illustrate our approach by reconstructing prototypes from complete archaeal proteomes and analyzing connections between functions and folds found by the reconstructed prototypes. In particular, we show examples for three characteristic cases: (i) nucleotide–triphosphate binding and hydrolyzing loop, called p-loop (Rossmann et al., 1974); (ii) a loop found in functionally diverse proteins having β/α barrel fold; (iii) prototypes of two EFLs involved into binding of ADP and glucose which form an enzymatic domain in glycosyltransferases (glycogen synthase).
2 MATERIALS AND METHODS
We describe a computational method for deriving prototypes of EFLs based on the sequences of complete genomes. The procedure comprises the iterative derivation of sequence profiles followed by the hierarchical clustering of profiles. We propose a scoring function that weights profile positions proportional to the information content on position, allowing to discrimine between matches that carry a specific signature from non-specific ones. The statistical significance of the scores is calculated from the empirical distribution of the reshuffled profile scores used as a control. We generalize the profiles and remove the remaining redundancies by clustering the profiles. The distance measure used in clustering also takes into account the information content on profile positions.
2.1 Derivation of prototypes from complete proteomes
Complete sets of protein coding sequences of 68 archaeal organisms (listed in Supplementary Material) are obtained from Genbank (Benson et al., 2009). We use one proteome representing each phylum of the archaeal superkingdom to produce a set of origins for the prototype derivation procedure. Proteomic sequences contain many sources of biases and redundancies: (i) homologous proteins; (ii) domain swapping, recombination and multiplication. These redundancies have to be removed, as they reflect recent events in the evolution of proteins (Chothia et al., 2003). The average domain size is 80–150 residues (Gerstein, 1998; Jones et al., 1998; Svedberg 1929; Wheelan et al., 2000); therefore, in order to remove redundancy originated from domain swapping, we compare 80-residue long sequence segments for identity. Sequences are clustered with CD-HIT (Li and Godzik, 2006) several times to gradually remove redundancy between domains down to 40% identity. Low-complexity regions in sequences which contain repeats or have highly biased amino acid composition are masked with SEG (Wootton and Federhen, 1996). Non-redundant domains are cut with 10-residue steps into overlapping 50-residue segments. These segments contain two 10-residue flanks, which can be adjusted in order to obtain a final 30-residue prototype. Based on the observation that gaps are not distributed uniformly, and multiple sequence alignments of remote homologs (below 25% sequence identity) contain well-aligned blocks without gaps (Kann et al., 2007) we consider that the cost of insertion or deletion in a functional signature is higher compared to at an arbitrary position in the whole protein sequence, therefore we do not allow gaps in profiles of EFLs.
The procedure starts from the search for sequences in the complete archaeal proteomes that are most closely related to the initial sequence segments (origins) in order to construct the seed alignment with a frequency matrix constituting a profile. The obtained profiles are then matched to the complete proteomes again, in order to find additional sequence matches and to update the profile. This profile-sequence search is repeated until the profile no longer changes, and, therefore, considered converged (Supplementary Figure S1). The profiles represent families of EFLs with specific signatures. The iterative procedure allows a profile to gradually expand to more distantly related, but statistically significant matches. Although the procedure resembles PSI-BLAST (Altschul et al., 1997), it has some notable differences originating from the specific requirements of the prototype derivation task. These differences are discussed in more detail in Supplementary Material.
2.2 Weighting of profile positions by information
We calculate position specific scoring matrices (PSSM) to score the profiles (for details of PSSM calculation see Supplementary Material). The profile-scoring function has to rank the matches according to the similarity of the sequence segment to the signature of the profile. An uneven contribution of positions in the profile should be taken into account. Degeneration of the profile towards the random compositional background or rare amino acids because of overestimation of pseudocounts (Altschul et al., 2009) should be prevented. Decrease of the profile sensitivity because of underestimation of pseudocounts should also be avoided. Therefore, in order to discriminate between matches that carry a specific signature from non-specific ones, we weight positions proportional to Kullback–Leibler divergence (DKL) (Kullback and Leibler, 1951), which reflects the information content on position i relative to the random background:
where f is observed amino acid frequencies on position i and c is proteomic amino acid composition. The score of a sequence segment q to profile P(n) will become:
where [mi,j]i=1,…,n;j=1,…,20 is the corresponding PSSM with n positions. Profile positions with low-information content (DKL < 1 bit) are not contributing to the overall score and are omitted.
2.3 Empirical calculation of the background
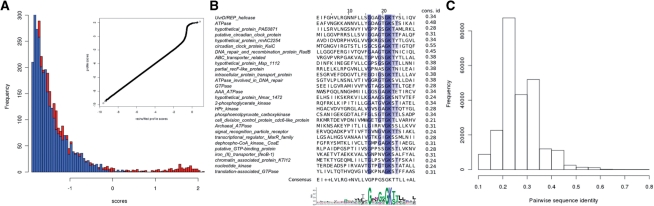
The significance of a score is characterized by an E-value, which is the number of false positive or unrelated matches above this particular score. The E-value is evaluated by comparing distributions of scores of the profile (s) with scores of the reshuffled profile (sR):E(s) = Np = N(1 − ecdf(sR)), where p is the P-value, N is the size of the combined proteome and ecdf(sR) is an empirical cumulative distribution function of the reshuffled profile scores. If, instead of the reshuffled profile, a randomized proteome is used as control, then all the biases except amino acid composition are lost, resulting in overestimation of significance of sequence matches containing non-specific signals. Complete positional permutations destroying relative distances between profile positions, similar to the combinatorial problem of pattern-avoiding permutations (Atkinson, 1999), give a robust estimation of the E-values (data not shown). Figure 1A shows an example of the score distributions for a derived profile and the reshuffled one. The difference exists only in the right-most tail of the distributions, showing that there are specific signatures in the profile, and that they are destroyed by reshuffling. All collected profile matches with the E-value below a certain significance threshold (e.g. E ≤ 1) are used to construct the updated profile. A sample of significant matches of the p-loop prototype and the corresponding sequence logo are shown in Figure 1B. Although the matches are found in proteins with different biochemical function, they all possess nucleotide-binding activity, which is presumably an elementary function of this prototype. It is important to note that some of the profile positions have much higher information content than others, and these positions constitute the signature. Comparison of all significant sequence matches to the consensus shows that the sequence identity is low on average and has a large variance (Fig. 1B and C). Therefore, for proper E-value estimation, i.e. for proper separation of related matches from the unrelated ones, discrimination between informative and non-informative positions is necessary.
Fig. 1.
Scoring and comparison of sequence-profile matches. (A) Histograms show tails of distributions of proteomic scores for the p-loop prototype (red) and for the same prototype, but with the reshuffled positions (blue) where the functional signature is destroyed, while the amino acid composition of the profile is preserved. Inset shows quantile–quantile plot of complete distributions. (B) Selected matches (28 of 689) of the p-loop prototype (shown as logo). Numbers indicate identity to the consensus sequence (shown below). Sequence label is Genbank description of the protein from which these matches were taken. (C) Histogram shows pair-wise sequence identity of all the significant matches of the p-loop prototype.
2.4 Hierarchical clustering of converged profiles
The iterative procedure described above results in a set of converged profiles that should be analyzed further. First, there is a redundancy between these profiles, caused by the way the origins are obtained: they overlap with a step of 10 residues. Redundancy also stems from the fact that different origins can actually converge to the same or very similar profiles. It means that these origins correspond to evolutionary connected EFLs, but their similarity can only be detected with the help of the profile. We introduce a distance measure that takes into account all possible profile–profile alignments in order to hierarchically cluster the profiles. Profile–profile comparison is more sensitive (Panchenko, 2003) than profile-sequence comparison, thus more distant relations could be detected during profile clustering. This procedure results in the removal of redundancy and further generalization of the profiles. The profiles with the most generic signatures represent the functional characteristics of presumably original prototypes. All possible profile–profile alignments without gaps are performed by sliding one 50-residue-long profile [A]50 against the other profile [A]50 and calculating pair-wise positional distances between all possible 30-residue windows [a]30 and [b]30, respectively. Distances between the pairs of corresponding positions are weighted proportionally to the information at each position (DKL):
The distance between two 50-residue profiles [A]50 and [B]50 is equal to the minimal distance between all possible sliding windows of size 30:
Hierarchical clustering of profiles is an iterative procedure where the most similar profiles (min[D(A,B)]) are consecutively merged together, resulting in a new, more generic profile (Supplementary Figure S3).
2.5 Characterization of prototypes
We characterize prototypes by looking for sequence matches in crystallized enzymes from ASTRAL/SCOP database (Brenner et al., 2000; Lo Conte et al., 2000). These matches describe descendant EFLs that diverged from the prototype. We assign elementary functions for derived prototypes and determine characteristic positions in their signatures based on the known enzymatic mechanisms in crystallized proteins.
Protein function is typically annotated by homology, although neither high-sequence identity (<50%), nor low BLAST E-values (below 10−50) guarantee the conservation of biochemical function (Rost, 1999, 2002). Here we annotate functional units of sub-domain size. Conventional homology detection methods, which operate on the level of whole proteins or domains, consider connections between SCOP superfamilies as false positives (Gough et al., 2001). It becomes obvious, that analysis of evolutionary relationships on the level of functional closed loops requires a special approach (Andreeva et al., 2007; Fong and Marchler-Bauer, 2009; Xie and Bourne, 2008). Although most of the derived prototypes (>70%, data not shown) have matches in Pfam, Prosite and CDD, functional annotation can not be directly transferred from the databases defining the function on whole-protein or domain level (Bateman et al., 2004; Lo Conte et al., 2000; Marchler-Bauer, et al., 2009; Sigrist et al., 2010), resulting in ambiguous annotations, and requiring additional manual curation. SCOP superfamilies can be used as a reference of the function for crystallized protein domains. CDD and Swissprot features can be used as more precise indicators of the elementary function of EFLs. Other databases describe enzymatic function on the residue level. For example, CSA (Gutteridge and Thornton, 2005), and MACiE (Holliday et al., 2007) databases describe experimentally determined roles of functional residues in the biochemical reactions and its mechanisms. Thus, via sequences of the crystallized structures this annotation can be transferred to the prototype's signature.
2.6 Statistics
Non-redundant archaeal proteome has 20 × 106 sequence segments of length 50. Starting from 175 458 origins extracted from four archaeal organisms, we end up with 8327 converged profiles having > 100 matches in the archaeal proteome and containing at least one position with four bits of information in their signature. These profiles were clustered for 120 iterations, which resulted in 138 profiles, from which the strongest 43 were selected for further consideration. The resulting 43 profiles are considered to be the most abundant ones and were used in the analysis. The ASTRAL sequence database based on SCOP release 1.75 contains 16 712 non-redundant domains at 95% sequence identity.
3 RESULTS AND DISCUSSION
We developed a computational procedure for deriving prototypes of EFLs, obtained prototypes from the set of archaeal proteomes, considered several prototypes in detail, delineated connections between domains superfamilies using the most abundant prototypes, and exemplified how combinations of EFLs result in specific enzymatic function.
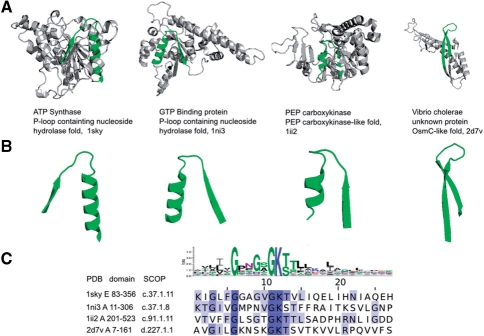
Figure 2 shows several representatives of the p-loop prototype and exemplifies detection of EFLs in different folds and biochemical functions. The signature of the prototype reads G-X-X-G-X-G-K-[TS] and is known to be the signature of nucleotide–triphosphate binding (Rossmann et al., 1974). We show EFLs corresponding to the prototype in four different proteins representing three different folds (p-loop containing nucleoside hydrolase, PEP carboxykinase-like fold, OsmC-like fold). Though sequence alignment of EFLs reveals high conservation in key sequence positions, the rest of the loop can diverge significantly. The degree of divergence of EFLs from the prototype is also indicated in the difference between corresponding structural segments: in the hydrolase and PEP-carboxykinase-like folds the structure resembles β-turn-α, while in OsmC-like fold it resembles β-hairpin. It is important to note, however, that despite the different structures of the EFL, naturally affected by the rest of the fold (Minor and Kim, 1996), the functional signature is always located in the elbow between the two elements of secondary structure and is highly conserved. EFLs representing this prototype universally provide the elementary function of nucleotide–triphosphate binding via interaction with the phosphate groups and with a Mg2+ ion, and also take part in phosphate hydrolysis. The combination of a specific sequence signature with its structural location emphasizes the conservation of the closed-loop structure, regardless of the exact secondary structural content of the loop and interaction of this loop with its structural environment. The diversity of folds containing this loop suggests that in the pre-domain stage of protein evolution the prototype of the p-loop was included into structurally and biochemically different folds, acquiring different elements of secondary structure and mutations in sequences, but preserving the active residues and their relative locations in sequence and space. The fourth structure in Figure 2 is a protein from V. cholerae with unknown function. With the help of the prototype it is now possible to hypothesize the function of this protein to a certain extent. It could be predicted, for example, that this V. cholerae protein has a nucleotide–triphosphate binding, and, perhaps, hydrolyzing activity. The combination with other EFLs detected in this protein can complete description of its possible biochemical function.
Fig. 2.
Matches of the nucleotide–triphosphate-binding (p-loop) prototype in crystal structures. Four matches of nucleotide–triphosphate-binding prototype are shown. The fold is displayed in cartoon and the structural loop corresponding to p-loop prototype is highlighted in green. The structures of the EFLs are also displayed. The logo of the prototype and the alignment of sequences of the corresponding EFLs highlight the functionally important residues involved in nucleotide–triphosphate binding and hydrolysis. PDB ID, SCOP ID and the coordinates of the sequence segments corresponding to the domains which contain the EFLs on display, are shown in the bottom.
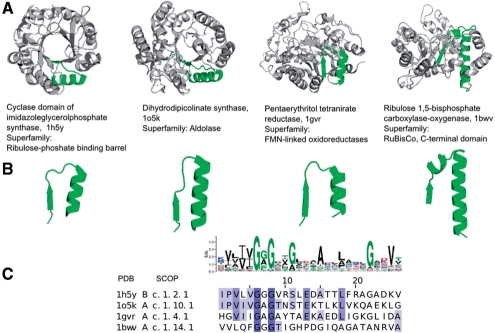
Figure 3 shows proteins that share the same β/α-barrel fold, which, in turn, has > 30 superfamilies in SCOP. This fold also serves as a scaffold for a variety of functions (Nagano et al., 2002), therefore the fold is an important target in protein (re)design experiments (Bershtein and Tawfik, 2008; Tokuriki and Tawfik, 2009). Based on the derived prototypes, β/α-barrels can be decomposed into a set of β-turn-α subunits, some of these subunits are directly involved in catalysis and, therefore, carry the functional signatures. The Glycine-rich prototype (Fig. 3) is an illustration of functional connections in an abundant β/α-barrel fold. The structure of the loops is β-turn-α, and the functionally important residues are located in the turn. The elementary function of the Glycine-rich pre-domain prototype is related to redox reactions revealing evolutionary connection between β/α barrels with different enzymatic functions. The biochemical functions of the enzymes where the loop is found are typically various oxidoreductases, dehydrogenases and synthases.
Fig. 3.
Matches of the Glycine-rich prototype in crystal structures of β/α-barrels. Four matches of the prototype involved presumably in redox reaction are shown. All structures have β/α-barrel fold, but perform different biochemical functions and belong, therefore, to different SCOP superfamilies.
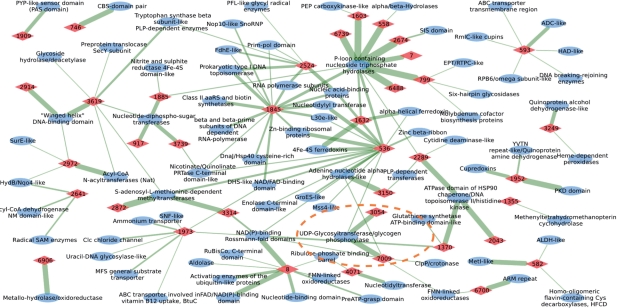
A set of abundant prototypes derived on the archaeal proteomes is used to delineate evolutionary connections between functional superfamilies of structural domains. Figure 4 illustrates connections revealed by a subset of the strongest prototypes (43 prototypes) in form of a graph. Nodes are prototypes (red diamonds) and protein domain superfamilies (blue ovals) according to SCOP classification. The edges represent matches between the superfamilies and the prototypes, where thickness of an edge is proportional to the logarithm of the number of matches found in the non-redundant set of protein domains derived from ASTRAL/SCOP database (Brenner et al., 2000; Lo Conte et al., 2000). Since a non-redundant set of protein domains was used in analysis, thickness is a rough indicator of the diversity of proteins in the superfamily. Each prototype is referred to by its number (numbers in red diamonds) and has a functional signature represented in form of a sequence profile. The logos of the corresponding profiles are listed in Supplementary Material.
Fig. 4.
Protein functions connected by prototypes of EFLs. Red diamonds represent prototypes and blue ovals represent protein domain superfamilies with the corresponding SCOP superfamily names. Edges are matches detected in the non-redundant sequences of protein domains. Thickness of edges characterizes the number of matches found.
The functional connections exemplified by the p-loop prototype and Glycine-rich prototype can be seen here in a larger context of archaeal (and homologous to archaeal) domains. The Glycine-rich prototype (Fig. 3) with the number 8 in the graph has five connections to folds other than β/α-barrel fold: NAD(P)-binding Rossmann fold, Activating enzymes of the Ubiquitin-like proteins, PreATP-grasp domain, Nucleotide-binding domain, FAD/NAD(P)-binding domain. Since all these folds have nucleotide–phosphate binding in common, these connections suggest that the elementary function of prototype 8 is related to nucleotide–phosphate binding. As a result, functional connections inside the β/α-barrel fold as well as connections between the β/α-barrel and other folds are found, unraveling nucleotide–phosphate binding as one of basic elementary functions crucial in the emergence of folds. Another interesting case is a p-loop containing nucleoside triphosphate hydrolase considered earlier (Fig. 2) which is an example of a fold with different biochemical functions. The particular biochemical function, in turn, is determined by the unique combination of EFLs, which is reflected as a group of prototypes gathered around the superfamily and connected by thick edges. One of the prototypes around the superfamily is prototype 1603, considered earlier (Fig. 2). The connection between p-loop containing nucleoside triphosphate hydrolase and PEP carboxykinase-like folds via p-loop prototype (Fig. 2) indicates that p-loop as EFL is an essential functional element of enzymes belonging to different superfamilies with different folds. It also suggests an important role of prototype 1603 in pre-domain evolution of folds and superfamilies. Some prototypes are present in a variety of superfamilies. For example, the Cysteine-rich metal binding loop (number 1845) which corresponds to EFLs forming a nest with cysteines co-ordinating a metal ion (typically Zn2+) facilitating nucleic acid binding. These EFLs are present in various superfamilies mainly related to nucleic acid binding, which is reflected in the graph by edges connecting prototype 1845 to thirteen superfamilies.
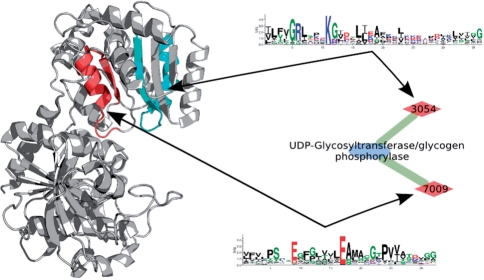
By representing proteins folds and functions in form of a graph connected by the prototypes one can proceed to explore the emergence of protein functions as combinations of prototypes. We illustrate how a functional domain emerges as combination of prototypes by the example of Glycosyltransferase superfamily (orange oval in Fig. 4). Figure 5 shows glycogen synthase (PDB 1rzu), which catalyzes the elongation of alpha-1,4-glucose backbone. The enzyme binds ADP-glucose and [Glucose]n−1 as substrates and transfers glycosyl group to form [Glucose]n as product and ADP. One of the enzyme's domains contains the EFLs corresponding to sequence prototypes 3054 (cyan) and 7009 (red). The elementary chemical functions of the prototypes are assigned according to the description of interactions with co-crystallized ligands analogous to the products and the substrates of the enzyme (Buschiazzo et al., 2004; Sheng et al., 2009). The elementary function of prototype 3054 is ADP binding: Arg-299 and Lys-304 interact with the phosphate, Ile-297 (second in position of 3054's PSSM) with the base and Ser-298 (third in 3054's PSSM) with the sugar in ADP. Prototype 7009 is also involved in ADP binding, its characteristic elementary function is glucose binding: Glu-376 interacts with phosphate and Thr-381 (third in 7009's PSSM) with the base of ADP. Besides, residue Glu-376 also plays an important catalytic role in glycosyltransferase activity. Finally, these two prototypes also interact with each other, forming a stabilizing salt bridge between Lys-304 and Glu-376. This example shows how the emergence of enzymatic function can be explored based on signatures of the prototypes and their elementary chemical functions. The two-domain nature of glycosyltransferase also points out that analysis of individual folds and their enzymatic functions should be followed by the exploration of recombination events in case of multi-domain proteins.
Fig. 5.
Two EFLs combine to form the active site in glycosyltransferase. The structure of glycogen synthase (PDB 1rzu) is shown in cartoon representation. The prototypes are shown as sequence logos, and the corresponding EFLs are highlighted in the structure.
4 CONCLUSIONS
The existence of EFLs in different folds and functions makes it possible to survey subtle evolutionary relations, originating from the pre-domain evolution of protein structure. It suggests that contemporary enzymatic functions are constructs of different sets and combinations of elementary chemical functions. It also shows that most of the enzymatic functions are performed by abundant prototypes (e.g. p-loop and Cysteine-containing prototype), which provide common reaction steps of different functions existing in different folds. An exhaustive description of a protein fold and its enzymatic function as a combination of EFLs illuminates how this fold emerged in pre-domain evolution by fusion of prototype genes. Therefore, obtaining the full collection of prototypes with elementary functions will make it possible to (i) predict enzymatic functions based on the sequences via determining EFLs corresponding to prototypes and their relative positions revealing structure of the fold; (ii) (re)design folds with desired functions by building constructs from necessary EFLs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Simon Mitternacht for helpful comments and suggestions.
Funding: FUGE-II Norwegian functional genomics platform.
Conflict of Interest: none declared.
REFERENCES
- Aharoni A, et al. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. PSI-BLAST pseudocounts and the minimum description length principle. Nucleic Acids Res. 2009;37:815–824. doi: 10.1093/nar/gkn981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, et al. SISYPHUS–structural alignments for proteins with non-trivial relationships. Nucleic Acids Res. 2007;35:D253–D259. doi: 10.1093/nar/gkl746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson MD. Restricted permutations. Discrete Math. 1999;195:27–38. [Google Scholar]
- Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, et al. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovsky IN. Discrete structure of van der Waals domains in globular proteins. Protein Eng. 2003;16:161–167. doi: 10.1093/proeng/gzg026. [DOI] [PubMed] [Google Scholar]
- Berezovsky IN, et al. Closed loops of nearly standard size: common basic element of protein structure. FEBS Lett. 2000;466:283–286. doi: 10.1016/s0014-5793(00)01091-7. [DOI] [PubMed] [Google Scholar]
- Berezovsky IN, Trifonov EN. Van der Waals locks: loop-n-lock structure of globular proteins. J. Mol. Biol. 2001;307:1419–1426. doi: 10.1006/jmbi.2001.4554. [DOI] [PubMed] [Google Scholar]
- Bershtein S, Tawfik DS. Advances in laboratory evolution of enzymes. Curr. Opin. Chem. Biol. 2008;12:151–158. doi: 10.1016/j.cbpa.2008.01.027. [DOI] [PubMed] [Google Scholar]
- Brenner SE, et al. The ASTRAL compendium for protein structure and sequence analysis. Nucleic Acids Res. 2000;28:254–256. doi: 10.1093/nar/28.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschiazzo A, et al. Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation. EMBO J. 2004;23:3196–3205. doi: 10.1038/sj.emboj.7600324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, et al. Evolution of the Protein Repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- Fong JH, Marchler-Bauer A. CORAL: aligning conserved core regions across domain families. Bioinformatics. 2009;25:1862–1868. doi: 10.1093/bioinformatics/btp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnham N, et al. Missing in action: enzyme functional annotations in biological databases. Nat. Chem. Biol. 2009;5:521–525. doi: 10.1038/nchembio0809-521. [DOI] [PubMed] [Google Scholar]
- Gerstein M. How representative are the known structures of the proteins in a complete genome? A comprehensive structural census. Fold Des. 1998;3:497–512. doi: 10.1016/S1359-0278(98)00066-2. [DOI] [PubMed] [Google Scholar]
- Glasner ME, et al. Evolution of enzyme superfamilies. Curr. Opin. Chem. Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Gough J, et al. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Gutteridge A, Thornton JM. Understanding nature's catalytic toolkit. Trends Biochem. Sci. 2005;30:622–629. doi: 10.1016/j.tibs.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Holliday GL, et al. The chemistry of protein catalysis. J. Mol. Biol. 2007;372:1261–1277. doi: 10.1016/j.jmb.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Holliday GL, et al. Understanding the functional roles of amino acid residues in enzyme catalysis. J. Mol. Biol. 2009;390:560–577. doi: 10.1016/j.jmb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Jones S, et al. Domain assignment for protein structures using a consensus approach: characterization and analysis. Protein Sci. 1998;7:233–242. doi: 10.1002/pro.5560070202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann MG, et al. The identification of complete domains within protein sequences using accurate E-values for semi-global alignment. Nucleic Acids Res. 2007;35:4678–4685. doi: 10.1093/nar/gkm414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullback S, Leibler RA. On Information and Sufficiency. Ann. Math Stat. 1951;22:142–143. [Google Scholar]
- Levitt M. Nature of the protein universe. Proc. Natl Acad. Sci. USA. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lo Conte L, et al. SCOP: a structural classification of proteins database. Nucleic Acids Res. 2000;28:257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, et al. On the evolution of protein folds: are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J. Struct. Biol. 2001;134:191–203. doi: 10.1006/jsbi.2001.4393. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor DL, Jr, Kim PS. Context-dependent secondary structure formation of a designed protein sequence. Nature. 1996;380:730–734. doi: 10.1038/380730a0. [DOI] [PubMed] [Google Scholar]
- Nagano N, et al. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- Panchenko AR. Finding weak similarities between proteins by sequence profile comparison. Nucleic Acids Res. 2003;31:683–689. doi: 10.1093/nar/gkg154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, et al. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Rost B. Enzyme function less conserved than anticipated. J. Mol. Biol. 2002;318:595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- Sanger F. The arrangement of amino acids in proteins. Adv. Protein Chem. 1952;7:1–67. doi: 10.1016/s0065-3233(08)60017-0. [DOI] [PubMed] [Google Scholar]
- Sheng F, et al. The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase. J. Biol. Chem. 2009;284:17796–17807. doi: 10.1074/jbc.M809804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedberg T. Mass and Size of Protein Molecules. Nature. 1929;123:871. [Google Scholar]
- Tokuriki N, Tawfik DS. Protein Dynamism and Evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- Trifonov EN, Berezovsky IN. Evolutionary aspects of protein structure and folding. Curr. Opin. Struct. Biol. 2003;13:110–114. doi: 10.1016/s0959-440x(03)00005-8. [DOI] [PubMed] [Google Scholar]
- Wheelan SJ, et al. Domain size distributions can predict domain boundaries. Bioinformatics. 2000;16:613–618. doi: 10.1093/bioinformatics/16.7.613. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Xie L, Bourne PE. Detecting evolutionary relationships across existing fold space, using sequence order-independent profile-profile alignments. Proc. Natl Acad. Sci. USA. 2008;105:5441–5446. doi: 10.1073/pnas.0704422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.