Abstract
Motivation: Detecting human proteins that are involved in virus entry and replication is facilitated by modern high-throughput RNAi screening technology. However, hit lists from different laboratories have shown only little consistency. This may be caused by not only experimental discrepancies, but also not fully explored possibilities of the data analysis. We wanted to improve reliability of such screens by combining a population analysis of infected cells with an established dye intensity readout.
Results: Viral infection is mainly spread by cell–cell contacts and clustering of infected cells can be observed during spreading of the infection in situ and in vivo. We employed this clustering feature to define knockdowns which harm viral infection efficiency of human Hepatitis C Virus. Images of knocked down cells for 719 human kinase genes were analyzed with an established point pattern analysis method (Ripley's K-function) to detect knockdowns in which virally infected cells did not show any clustering and therefore were hindered to spread their infection to their neighboring cells. The results were compared with a statistical analysis using a common intensity readout of the GFP-expressing viruses and a luciferase-based secondary screen yielding five promising host factors which may suit as potential targets for drug therapy.
Conclusion: We report of an alternative method for high-throughput imaging methods to detect host factors being relevant for the infection efficiency of viruses. The method is generic and has the potential to be used for a large variety of different viruses and treatments being screened by imaging techniques.
Contact: r.eils@dkfz.de; r.koenig@dkfz.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Despite many remarkable discoveries in virology, viruses are still a major cause of severe diseases including Dengue fever, hepatitis, immune deficiency and severe influenza. Viruses employ specific human host proteins (host factors) for each step of their ‘life’ cycle (Carter and Ehrlich, 2008; Malim and Emerman, 2008; Martin and Sattentau, 2009). Discovering these host factors may not only unravel fundamental principals of viral modes of operation, like their replication, but also, notably, may lead to promising drug therapies which are not affected by the high mutational variability in viral populations. Fluorescence microscopy imaging of RNA interference (RNAi) knockdown screens has become a major method of choice to identify the function of the proteins corresponding to the silenced genes and specifically to detect potential drug targets. Typically, these screens are based on endpoint assays of transfected cells with a direct intensity readout (Boutros et al., 2004). More recently, large-scale imaging has been used to study the transfected cells (Neumann et al., 2006; Nir et al., 2010). Image analysis software was developed to segment cells and extract cellular texture features enabling machine learning methods to identify subcellular location (Conrad et al., 2004; Peng et al., 2010) and to classify the mitotic phase of imaged cells (Carpenter et al., 2006; Harder et al., 2006; Jones et al., 2008; Lamprecht et al., 2007; Neumann et al., 2010; Vokes and Carpenter, 2008). For HIV, three such genome wide knock-down studies have been performed (Brass et al., 2008; Konig et al., 2008; Zhou et al., 2008). However, there was only little overlap in the predicted host factors reducing the infection. This discrepancy might be due to differences in the experimental conditions like using different viral strains, investigating different time intervals or using different silencing sequences. In addition, it may have also resulted from incomplete data analysis. Therefore, we developed an alternative approach to detect host factors with such a screening method.
Viruses can spread within the host by release of cell-free virions or direct passage between infected and non-infected cells. In general, direct cell–cell transfer is considerably more efficient than a cell-free transfer (Timpe et al., 2008) and is supported by filopodial bridges (Sherer et al., 2007). As a consequence of such a viral cell–cell spreading, clusters of infected cells may be formed. It was reported recently that spatial distribution of cells can influence the infection behavior. Snijder and co-workers observed intriguing relationships between virus species, the spatial distribution of cells and the infection rate. While the infection efficiency of a rotavirus was considerably increased in sparse populations, Dengue viruses mainly employed cells located at edges of islets, and murine hepatitis viruses were preferably found in dense cell populations (Snijder et al., 2009). To analyze such clustering patterns systematically, statistical methods for point pattern analysis can be employed. Ripley's K-function is an established measure for defining the degree of clustering. It evaluates all interparticle distances over the studied area and compares the observed distribution with a random distribution of spots. Ripley's K-function has been used in ecology, epidemiology and geography (Ersboll and Ersboll, 2009). In cell biology, it was applied to study integrin-sensing extracellular matrix properties (Paszek et al., 2009) and to analyze lipid rafts by observing clustering of RAS proteins (Prior et al., 2003).
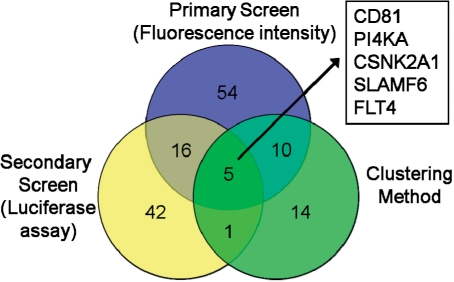
In our study, we investigated HCV infection in a human hepatoma cell line to detect human host factors that are necessary for virus replication. We employed the RNA interference technology and screened a comprehensive set of 719 kinases expressing genes. We tracked the infection efficiency by fluorescence imaging of cells infected with GFP-expressing viruses. Three bioinformatics approaches were applied to yield host factors that significantly reduce infection efficiency. We employed (i) a statistical method described recently using B-score and Z-score normalization of intensity read-outs of segmented cell images (Brideau et al., 2003), (ii) intensity read-outs of a luciferase based secondary screen and (iii) our new application of the point pattern analysis method. The idea of this approach bases on the observation that reduced virus replication results in a reduced grouping (clustering) of infected cells. For each knockdown, we compared the clustering of infected cells and non-infected cells and estimated a reduction of clustering of the infected cells. We yielded 30 promising candidates suiting as potential host factors for therapeutical drug targeting, five out of which were found with all three methods comprising CD81, PI4KA, CSNK2A1, SLAMF6 and FLT4.
2 METHODS
2.1 Experimental setup
The siRNA library used for the primary screen in this study was purchased from Ambion (Silencer® Human Kinase siRNA Library V3 (AM80010V3)). Reverse transfection of siRNAs into Huh7.5 cells (Blight et al., 2002) in a LabTek format was optimized according to a previously described protocol (Erfle et al., 2007). Overall, 2157 siRNAs targeting 719 human kinase genes plus positive controls targeting the entry receptor CD81 or the viral genome itself (HCV321 and HCV138) and four different negative controls (non-silencing siRNA) were spotted in transfection mixture onto LabTeks. After seeding of Huh7.5 cells we allowed siRNA silencing for 36 h. Cells were infected with a HCV GFP reporter virus, fixed 36 h later and immunostained with a GFP-specific antibody. Cell arrays were imaged with a scanning microscope (ScanˆR, Olympus Biosystems) using 10× objective (Olympus, cat. no. UPSLAPO 10×) and images were analyzed with an image analysis method (see Section 2.2). The primary screen was conducted in 12 repetitions. All images with less than 125 or more than 500 cells within siRNA spots were excluded from the analysis. As an additional quality control for staining artefacts all images were analyzed by eye resulting in an overall exclusion of 15% of the images. Then statistical analysis was performed to compute a mean z-score and a P-value for each gene (see Section 2.5). During validation of the 178 gene candidates selected from the primary screen, three independent siRNAs per gene were used to minimize the number of potential off-target hits. In addition, the format of the assay was changed to a statistically more robust 96-well plate format to increase the number of transfected cells per siRNA and thus statistical power (about 300 cells in the LabTek format but about 10 000 in this well-based assay). The method of solid phase reverse siRNA transfection was adapted to the 96-well plate format as described elsewhere (Erfle et al., 2008). This assay format allowed to use a luciferase reporter virus facilitating the analysis of the screen. To validate effects of kinase knockdowns on HCV entry and replication 5 × 103 Huh7.5FLuc cells (stably expressing firefly luciferase) were seeded per siRNA-coated well of a 96-well plate. After 36 h, cells were infected with a HCV renilla luciferase reporter virus. Forty-eight hours post-infection cells were harvested and the firefly luciferase and renilla luciferase activities measured. The secondary screen was performed twice in duplicates and statistically analyzed (see Section 2.5).
2.2 Image analysis and quality assessment
To analyze the images of the siRNA screen, an automated system was employed which was described in detail recently (Matula et al., 2009). Briefly, the inputs of this system consisted of two dye channel images from a chamber plate with printed siRNA spots. The fluorescence signals originated from DAPI stained cell nuclei (1st channel) and GFP incorporated into the viral strain (2nd channel). In the DAPI channel, single-cell nuclei were segmented using an edge-based approach based on combining responses of the gradient magnitude and the Laplacian of Gaussian filters with morphological closing and hole filling operators. Cell nuclei were identified among the segmented objects by applying size, intensity and circularity criteria. The viral protein production level (virus signal) of each cell was computed by the mean intensity in channel 2 inside the nucleus neighborhood. Positive and negative controls had been spotted on each plate. In positive controls, the siRNAs hindered viral protein production resulting in a low virus signal, whereas in negative controls virus replication was not altered. According to the virus signal, cells were classified as infected and non-infected using a threshold. Cells with a virus signal less than the threshold were classified as non-infected, otherwise as infected. The threshold was defined by maximizing the difference in infection rates between positive and negative controls. Quality filtering was performed eliminating out-of-focus images and image artifacts. On the single image level, images were automatically classified as low quality if they contained too few or too many cells or if they were out-of-focus. On the whole plate level, the percentage of saturated pixels in channel 2 was computed. Plates which showed over-exposure were scanned again with decreased exposure times (Matula et al., 2009).
2.3 Ripley's K-function
The distribution of cells on fluorescence microscopy images was represented as a spatial pattern of spots. Spots (cells) were classified as infected and non-infected and their respective clustering behavior studied using the K-function as described elsewhere (Ripley, 1977). K is calculated by
| (1) |
for a given radius parameter r > 0. N is the number of spots in the observed area A (whole image), λ is the intensity of spots which can be estimated by N/A, dij is the (Euclidean) distance between spot i and j. Ir (dij) equals to one if dij < r and is zero otherwise. The weighting factor w(xi, dij) copes for edge effects and is the proportion of the circumference of a circle with center xi and distance dij that falls in the studied area. If the circle is entirely inside the studied area, it equals to one.
Ripley's K-function is used to compare the observed spot distribution with a random distribution. The given spot distribution is tested against the null hypothesis that the spots are randomly distributed. For clustering distributions, the expected value of K(r) is larger than the value of a random distribution, for regular patterns it is less than for a random distribution (examples are given in the Supplementary Material S1). To cope for biases caused by clustering of proliferating cells, we derived the random distribution by using the actual positions of the spots of infected and non-infected cells. The sth simulated null-hypothesis of the K-function was estimated by randomly drawing Nc spots from all spots (infected and non-infected cells) and applying them to the K-function. The final null-hypothesis was calculated from the mean value of these simulated K-functions (s = 1…100). Applying Ripley's K-function to spot distributions with local spatial variation (independent from their clustering), the inhomogeneous K-function was defined by Baddeley and co-workers (Baddeley et al., 2000) which we used for our study. It is given by
| (2) |
|A| denotes the observation area (distance ≤ r), eij is the edge-correction factor calculated by the border method (Ripley, 1981). λ(yi) and λ(yj) are estimated intensities at spots yi and yj. They were estimated by a Gaussian kernel smoother using the intensity surface model (Baddeley et al., 2000). The maximum ranges of the radius r we investigated were 25%, 30%, 35% and 40% of the shorter side of the whole image. To get the clustering score, the area between the curves of the inhomogeneous K-function and a simulated random distribution was calculated. The score was positive if the curve for the inhomogeneous K-function was mainly above the curve of the simulated random distribution (tendency for clustering), and negative otherwise. This score was calculated for infected and non-infected cells, respectively, using the function Kinhom from the library Spatstat of the R package (www.r-project.org, version 2.8.0). To obtain the final clustering score for estimating the infection rate, the score of the infected cells was subtracted by the score of the non-infected cells.
2.4 Quadrat analysis
Quadrat analysis observes the frequency distribution of cells within a set of grid squares (quadrat) (Wong and Lee, 2005). The mean number of cells per quadrat is estimated and its variance computed to obtain the variance–mean ratio (VMR) as a measure for clustering of points, i.e.
| (3) |
| (4) |
m is the number of quadrats, xi is the number of points in quadrat i and  is the mean of the number of points per quadrat. VMR greater than one indicates a clustered distribution, VMR less than one indicates a random distribution and VMR = 0 a uniform distribution. To obtain the final clustering score, we subtracted VMR of the non-infected cells from VMR of the infected cells. The clustering score was calculated for all knocked down genes and the controls and a z-normalization was performed.
is the mean of the number of points per quadrat. VMR greater than one indicates a clustered distribution, VMR less than one indicates a random distribution and VMR = 0 a uniform distribution. To obtain the final clustering score, we subtracted VMR of the non-infected cells from VMR of the infected cells. The clustering score was calculated for all knocked down genes and the controls and a z-normalization was performed.
2.5 Identification of host siRNA hits
Statistical analysis of processed imaging data was carried out in R using the Bioconductor packages RNAither (Rieber et al., 2009) and cellHTS (Boutros et al., 2006). For the primary screen, we excluded wells with less than 125 and more than 500 cells. For the secondary screen, wells showing the lowest 5% and highest 5% of firefly reporter activity (correlated to the number of viable cells) were excluded. Those wells were excluded to eliminate possible interference of cytostatic or cytotoxic effects or high variability in cell number with the readout of viral replication. As well, in some wells, cells may have grown densely and possible incorrect segmentation of images may have occurred (Börner et al., 2009). Virus-specific signal intensities per siRNA were normalized for effects of differing cell counts using locally weighted scatterplot smoothing (Cleveland, 1979). B-score normalization was used to remove spatial effects within individual LabTeks (Brideau et al., 2003). Variability between plates was addressed by subtracting the plate median from each measurement per siRNA and dividing by the plate median absolute deviation (1σ) resulting in one z-score per siRNA per LabTek. Replicates were summarized using the mean z-score; furthermore, Student's t-tests were carried out to determine whether siRNA effects differed significantly from zero. Only hits with negative z-scores were taken. For all three analyses (primary screen, secondary screen and clustering analysis), hits were selected if their P-values were <0.05.
3 RESULTS
3.1 Parameter optimization, choice of the most suitable clustering analysis method and assembling significant hits
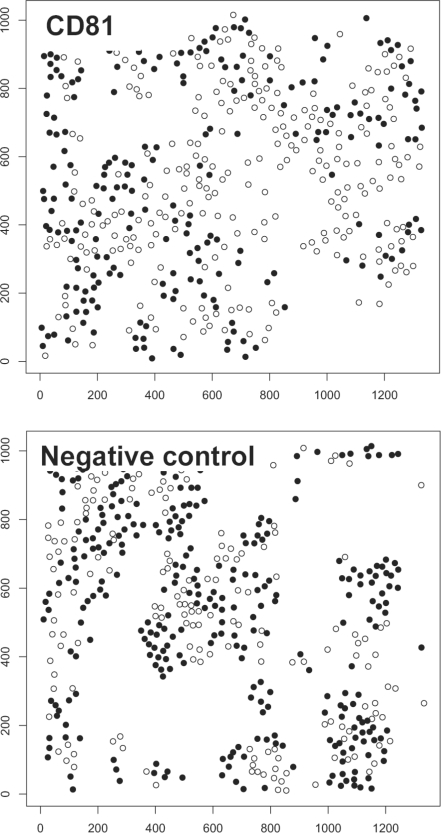
We identified cellular protein kinases involved in HCV replication by observing replication and clustering of the infected cells upon silencing of protein kinases (2157 siRNAs targeted 719 human protein kinase genes). Virus-infected cells were identified by viral GFP expression observed with fluorescence microscopy analysis. Host siRNA hits were identified by three different approaches, (i) using viral GFP fluorescence intensity of the primary screen, (ii) luciferase intensity of the secondary screen and (iii) the clustering analysis method. For the clustering analysis method, we computed a z-transformed clustering score for all knockdowns. We analyzed the clustering of infected cells using the DAPI channel (nucleus staining) for defining the center of mass and the viral GFP signal for labeling the cells as infected and non-infected. Low clustering scores were yielded if the infected cells did not cluster, while high values resulted if specifically the infected cells showed a high clustering. This is demonstrated exemplarily in Figure 1. For Ripley's K-function, we optimized the performance by varying the range of radius. As the objective function, we analyzed the correlation of the z-scores from Ripley's K-function for all knocked down genes with the z-scores from the intensity readout of the primary screen and secondary screen. Table 1 shows the results. The best correlation to the primary screen was 0.55 using a radius range of 35%. We investigated the performance of a well established clustering analysis method, the Quadrat Analysis (Wong and Lee, 2005). However, the method showed less correlation to the intensity readouts (Supplementary Table S2 shows the results for several parameter settings).
Fig. 1.
Images of a positive (knockdown of CD81) and a negative control (non-silencing siRNA). Knocking down CD81 resulted in a rather random distribution of infected cells (black dots), while infected cells were highly clustered when no gene was silenced (unhindered viral replication).
Table 1.
Pearson's correlation coefficients of intensity values of the scores from Ripley's K-function and the standard readouts (intensity values of the primary and secondary screen)
| K-function | K-function | ||||
|---|---|---|---|---|---|
| (Inhomogeneous) |
(Homogeneous) |
||||
| 40% | 35% | 30% | 25% | 35% | |
| Correlation with intensities of the primary screen | 0.51 | 0.55 | 0.49 | 0.36 | 0.36 |
| Correlation with intensities of the secondary screen | 0.31 | 0.32 | 0.34 | 0.28 | 0.23 |
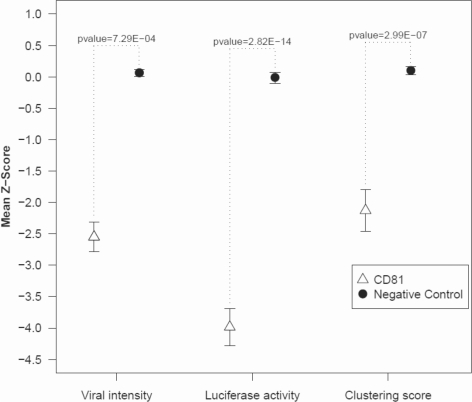
Also, the homogeneous K-function was inferior to the inhomogenous K-function (result with the best range of radius is given in Table 1). In the following, we report results using the inhomogenous K-function with the optimized parameter (radius range = 35%). Knockdown of gene CD81 (positive control) resulted in low clustering of the infected cells, while the negative control (non-silencing siRNAs) showed a comparably high tendency for infected cells (black dots) to cluster. The clustering scores were −2.3 and 2.2 for CD81 and the negative control, respectively. For the primary screen, mean intensities of viral GFP was calculated for each knockdown and replicate (12 replicates), their z-scores computed in respect to the bulk of the data, and genes with significant low z-scores selected (P < 0.05). Similarly, significant genes were defined from the secondary screen. The difference of z-score distributions of the positive control (CD81) and the negative controls is shown in Figure 2. The separation of distributions shows CD81 as a significant down regulator in all three approaches (primary screen, secondary screen and clustering analysis method). The numbers of significant hits and their intersections are summarized in Figure 3. Observing viral signal intensities in the primary screen yielded 85 significant genes. A total of 178 genes selected from the primary screen were observed with the secondary screen yielding 64 significant genes. The clustering analysis method yielded 30 genes (shown in Supplementary Table S3). All three positive controls showed significantly low clustering scores (CD81: P = 6.61E-07; HCV-321: P = 1.53E-13; HCV-138: P = 1.20E-10). Five genes were significant in all three methods comprising CD81, PI4KA, CSNK2A1, SLAMF6 and FLT-4 (Table 2). Note that the positive controls HCV-321 and HCV-138 were not used in the secondary screen. CD81 was used as a positive control. It is well known as a viral receptor of HCV (Zhang et al., 2004) and involved in HCV entry (Randall et al., 2007).
Fig. 2.
Comparison of the scores of the positive control CD81 and the negative controls (non-silencing siRNA) for all applied methods. The P-values were calculated for the two distributions of CD81 and the negative controls using a Student's t-test.
Fig. 3.
Venn diagram of the hits of all three used methods.
Table 2.
Host factors detected with all three analysis methods
| Entrez gene ID | Gene symbol | Gene name | P-value (Clustering method) |
|---|---|---|---|
| 975 | CD81 | CD81 molecule | 6.61E-07 |
| 5297 | PI4KA | Phosphatidylinositol 4-kinase, catalytic, alpha | 0.0019 |
| 1457 | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide | 0.0274 |
| 114 836 | SLAMF6 | SLAM family member 6 | 0.0345 |
| 2324 | FLT4 | fms-related tyrosine kinase-4 | 0.0445 |
3.2 Functional interpretation of the results
Besides CD81 we detected four host factors being significant in all three analysis approaches (PI4KA, CSNK2A1, SLAMF-6 and FLT-4). Phosphatidylinositol 4-kinase-α (PI4KA) is well known to be required for HCV replication (Berger et al., 2009; Borawski et al., 2009; Li et al., 2009; Tai et al., 2009; Trotard et al., 2009; Vaillancourt et al., 2009). It was shown in vitro that Casein kinase II (CSNK2A1 is coding its subunit alpha) phosphorylates the non-structural HCV protein NS5A (Kim et al., 1999). Fms-related tyrosine kinase 4 (FLT-4) is also known as vascular endothelial growth factor receptor 3 (VEGFR-3). It is a member of the tyrosine kinase receptor family. Over-expression of the short splice variant of VEGFR-3 stimulated cell growth in HepG2 cells (Lian et al., 2007) which may advantage infectious spreading of the virus. Interestingly, a retrovirus was found to be integrated into an intron of FLT-4 in the genome which may have resulted in an evolutionary advantage of this virus (Hughes, 2001). SLAMF-6 belongs to the signaling lymphocytic activation molecule family and is a transmembrane receptor mainly expressed in natural killer (NKT) cells. The receptor serves as a docking site for several signaling molecules (Engel et al., 2003; Veillette, 2006). It was shown that SLAMF-1 and SLAMF-6 critically control the characteristic expansion and differentiation of NKT cells after thymic selection (Griewank et al., 2007). SLAMF-6 may suit as an interesting candidate for investigating uptake and signal propagation of the virus during its entry into the host cell.
3.3 Comparing the clustering behavior of HCV and the dengue virus infection
The same experimental set-up as for HCV was also applied to observe cells infected with the dengue virus (DV) (Matula et al., 2009). It is known that DV infects edges of islets of cell populations rather than forming clusters of infections (Snijder et al., 2009). We observed this behavior also in our data which is shown exemplarily in the Supplementary Material (Supplementary Fig. S4). We compared the clustering scores for non-silencing siRNA images of both datasets and observed significantly higher clustering scores for cells with HCV infection (P = 4.8E-4, Wilcoxon test, see Supplementary Fig. S4 for the distribution of all scores for both data sets).
4 DISCUSSION
We applied an image processing analysis, a clustering analysis method and statistical analyses of intensity readouts to detect host factors involved in HCV infection. Instead of observing knockdowns of viral components, we focused on specific proteins in the host cell. Targeting host factors which are relevant to viral replication showed distinct lower clustering of the infected cells. Specifically, all three positive controls showed significantly low clustering scores. Additionally, we got hits having significantly low viral GFP intensities observed in the primary screen and hits from a secondary screen based on a luciferase read-out. Computing the intersection of hits from all three approaches yielded five genes to be considered as attractive targets against HCV infection.
Besides two well-known host factors being relevant for HCV replication (CD81 and PI4KA) and one host factor which has been described to phosphorylate an HCV protein, we also found two new challenging candidates (FLT-4 and SLAMF-6). FLT-4 has interesting characteristics. It was observed that it suited for a retrovirus to be genomically incorporated (Hughes, 2001). Even though known virulence principles of HCV and retroviruses are very different, such a mechanism may have similar advantages for replication of HCV as for the evolutionary benefits of the retrovirus. To measure clustering, we used the inhomogeneous Ripley's K-function which has been used in a broad variety of scientific applications ranging from the clustering behavior of infected habitants in a country (Ersboll and Ersboll, 2009) to cell biological concerns as e.g. studying the clustering of integrins when cells sense the extra cellular matrix (Paszek et al., 2009). We used Ripley's K-function now for observing the clustering behavior of individual infected cells in a cellular in vitro assay. With such a clustering analysis method we were able to track infection populations in a systematic way and used it to support finding crucial host factors for viral replication. Besides applying Ripley's K-function to detect relevant host factors as shown in this study, it additionally may be applied to systematically investigate the infection behavior of different virus families. Snijder and co-workers observed principal differences of virus entities to populate cell samples (Snijder et al., 2009). Ripley's K-function may be used to follow up this study by a quantitative clustering analysis supporting putting up a taxonomy for virus strains based on their population characteristics in the host. It is known e.g. that the Dengue virus infects edges of islets in cell colonies and therefore does not exhibit such a clustering tendency as HCV (Snijder et al., 2009). In an initial trial, we observed distinct higher clustering scores for cells infected by HCV in comparison to cells infected by the Dengue virus.
Applying a clustering analysis method for estimating the virulence in cellular assays is general and can be used for other screens to observe infectious propagation in cellular populations. It may also be used for a quantitative and systematic analysis of the specific spreading and populating behavior of distinct virus families which may also have an impact on the discovery of their specific use of host factors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rolf Kabbe, Karlheinz Groß and Marc Hemberger for IT support, and Maik Lehmann for fruitful discussions.
Funding: BMBF-FORSYS Consortium, Viroquant (#0313923); the Landesstiftung Baden-Württemberg (research program RNS/RNAi, contract no. P-LS-RNS30); the Helmholtz Alliance on Systems Biology of Signaling in Cancer; the Nationales Genom-Forschungs-Netz (NGFN+) for the neuroblastoma project ENGINE; the Deutscher Akademischer Auslandsdienst (DAAD).
Conflict of Interest: none declared.
REFERENCES
- Baddeley AJ, et al. Non- and semi-parametric estimation of interaction in inhomogeneous point patterns. Stat. Neerlandica. 2000;54:329–350. [Google Scholar]
- Berger KL, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl Acad. Sci. USA. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, et al. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borawski J, et al. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner K, et al. From experimental setup to bioinformatics: an RNAi screening platform to identify host factors involved in HIV-1 replication. Biotechnol. J. 2009;5:39–49. doi: 10.1002/biot.200900226. [DOI] [PubMed] [Google Scholar]
- Boutros M, et al. Analysis of cell-based RNAi screens. Genome Biol. 2006;7:R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Brideau C, et al. Improved statistical methods for hit selection in high-throughput screening. J. Biomol. Screen. 2003;8:634–647. doi: 10.1177/1087057103258285. [DOI] [PubMed] [Google Scholar]
- Carter CA, Ehrlich LS. Cell biology of HIV-1 infection of macrophages. Annu. Rev. Microbiol. 2008;62:425–443. doi: 10.1146/annurev.micro.62.081307.162758. [DOI] [PubMed] [Google Scholar]
- Cleveland WS. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 1979;74:829–836. [Google Scholar]
- Conrad C, et al. Automatic identification of subcellular phenotypes on human cell arrays. Genome Res. 2004;14:1130–1136. doi: 10.1101/gr.2383804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P, et al. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat. Rev. Immunol. 2003;3:813–821. doi: 10.1038/nri1202. [DOI] [PubMed] [Google Scholar]
- Erfle H, et al. Reverse transfection on cell arrays for high content screening microscopy. Nat. Protocols. 2007;2:392–399. doi: 10.1038/nprot.2006.483. [DOI] [PubMed] [Google Scholar]
- Erfle H, et al. Work flow for multiplexing siRNA assays by solid-phase reverse transfection in multiwell plates. J. Biomol. Screen. 2008;13:575–580. doi: 10.1177/1087057108320133. [DOI] [PubMed] [Google Scholar]
- Ersboll AK, Ersboll BK. Simulation of the K-function in the analysis of spatial clustering for non-randomly distributed locations–exemplified by bovine virus diarrhoea virus (BVDV) infection in Denmark. Prev. Vet. Med. 2009;91:64–71. doi: 10.1016/j.prevetmed.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Griewank K, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DC. Alternative splicing of the human VEGFGR-3/FLT4 gene as a consequence of an integrated human endogenous retrovirus. J. Mol. Evol. 2001;53:77–79. doi: 10.1007/s002390010195. [DOI] [PubMed] [Google Scholar]
- Kim J, et al. Hepatitis C virus NS5A protein is phosphorylated by casein kinase II. Biochem. Biophys. Res. Commun. 1999;257:777–781. doi: 10.1006/bbrc.1999.0460. [DOI] [PubMed] [Google Scholar]
- Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl Acad. Sci. USA. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Z, et al. Hepatitis B x antigen up-regulates vascular endothelial growth factor receptor 3 in hepatocarcinogenesis. Hepatology. 2007;45:1390–1399. doi: 10.1002/hep.21610. [DOI] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Martin N, Sattentau Q. Cell-to-cell HIV-1 spread and its implications for immune evasion. Curr. Opin. HIV AIDS. 2009;4:143–149. doi: 10.1097/COH.0b013e328322f94a. [DOI] [PubMed] [Google Scholar]
- Matula P, et al. Single-cell-based image analysis of high-throughput cell array screens for quantification of viral infection. Cytometry A. 2009;75:309–318. doi: 10.1002/cyto.a.20662. [DOI] [PubMed] [Google Scholar]
- Neumann B, et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- Nir O, et al. Inference of RhoGAP/GTPase regulation using single-cell morphological data from a combinatorial RNAi screen. Genome Res. 2010;20:372–380. doi: 10.1101/gr.100248.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, et al. Integrin clustering is driven by mechanical resistance from the glycocalyx and the substrate. PLoS Comput. Biol. 2009;5:e1000604. doi: 10.1371/journal.pcbi.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, et al. Determining the distribution of probes between different subcellular locations through automated unmixing of subcellular patterns. Proc. Natl Acad. Sci. USA. 2010;107:2944–2949. doi: 10.1073/pnas.0912090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, et al. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl Acad. Sci. USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieber N, et al. RNAither, an automated pipeline for the statistical analysis of high-throughput RNAi screens. Bioinformatics. 2009;25:678–679. doi: 10.1093/bioinformatics/btp014. [DOI] [PubMed] [Google Scholar]
- Ripley BD. Modelling spatial patterns. J. Roy. Stat. Soc. Series B Stat. Methodol. 1977;39:172–192. [Google Scholar]
- Ripley BD. Spatial Statistics. New York; Chichester: Wiley; 1981. [Google Scholar]
- Sherer NM, et al. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder B, et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- Tai AW, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpe JM, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47:17–24. doi: 10.1002/hep.21959. [DOI] [PubMed] [Google Scholar]
- Trotard M, et al. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 2009;23:3780–3789. doi: 10.1096/fj.09-131920. [DOI] [PubMed] [Google Scholar]
- Vaillancourt FH, et al. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat. Rev. Immunol. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- Wong DWS, Lee J. Statistical Analysis of Geographic Information with Arcview GIS and ArcGIS. New York: Wiley; 2005. [Google Scholar]
- Zhang J, et al. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 2004;78:1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.