Abstract
Motivation: Animal toxins operate by binding to receptors and ion channels. These proteins are short and vary in sequence, structure and function. Sporadic discoveries have also revealed endogenous toxin-like proteins in non-venomous organisms. Viral proteins are the largest group of quickly evolving proteomes. We tested the hypothesis that toxin-like proteins exist in viruses and that they act to modulate functions of their hosts.
Results: We updated and improved a classifier for compact proteins resembling short animal toxins that is based on a machine-learning method. We applied it in a large-scale setting to identify toxin-like proteins among short viral proteins. Among the ∼26 000 representatives of such short proteins, 510 sequences were positively identified. We focused on the 19 highest scoring proteins. Among them, we identified conotoxin-like proteins, growth factors receptor-like proteins and anti-bacterial peptides. Our predictor was shown to enhance annotation inference for many ‘uncharacterized’ proteins. We conclude that our protocol can expose toxin-like proteins in unexplored niches including metagenomics data and enhance the systematic discovery of novel cell modulators for drug development.
Availability: ClanTox is available at http://www.clantox.cs.huji.ac.il
Contact: michall@cc.huji.ac.il
1 INTRODUCTION
In recent years, with the expansion of genomics data and the development of ‘next-generation sequencing’ we have witnessed unprecedented growth in sequence information as reflected in the ∼20 million UniRef sequences. Several niches that remain poorly annotated are viruses, environmental metagenomics sequences and short proteins.
Short sequences are of special interest from a biotechnological and medical perspective as they are relatively easy to produce, they are often one domain proteins and more importantly, they may be used in medical research. From a bioinformatics and statistical perspective, annotation of these sequences remains challenging due to low similarity signals (often within the realm of noise). Consequently, typical automated methods cannot reliably assign such sequences to their homologous groups. As a result, short proteins constitute an uncharacterized and fragmented subset of sequence space. Another major uncharacterized group covers the world of viruses. Viruses are extremely diverse and their importance to agriculture and human health is self-evident. Nevertheless, most viral proteomes are reported as ‘translated sequences’ with a very poor annotation of their content (polymerase and envelope proteins being the exception). Co-evolution of viruses and their hosts constitutes a major evolutionary driving force (Bahir et al., 2009). As a result, key cellular functions are acquired by viruses to enhance their infectivity and replication efficiency (Woolhouse et al., 2002). Examples of host cellular components that are mimicked by viral proteins include mimicry of receptors and antigens of the immune system. In addition, sophisticated modulation by viruses that infect any taxonomical group, from bacteria to human is directed towards the most sensitive cellular function including the ubiquitin system, signal transduction cascades and the translational machinery (Marques and Carthew, 2007). These mechanisms are best studied in light of host–virus co-evolution (Sorek et al., 2008).
Animal toxins make up one of the most highly over-represented functional groups among the short proteins. Toxins are proteins that appear in animal venom and are aimed at inflicting harm to the organism on which the venom acts (Jungo and Bairoch, 2005; Tan et al., 2006). Most animal toxins are short proteins (ranging from 30 to 120 amino acids). They are extremely varied in terms of function and include ion channel inhibitors, phospholipases, protease inhibitors, disintegrins, membrane pore inducers and more (Tan et al., 2003). Such toxins are detected in sporadic species along the evolutionary tree of the animal kingdom including scorpions, snakes, bees, wasps, frogs and numerous animals living in the sea (e.g. stonefish, snail, jellyfish, cone snail and hydra). Interestingly, proteins that share a common scaffold with animal toxins have also been sequenced in non-venomous species. In recent years, an increasing number of proteins resembling animal toxins have been identified in non-venomous contexts and shown to act as natural cell modulators. These toxin-like proteins include proteases, protease inhibitors, as well as secreted proteins which resemble cell antigens, growth factors and more (Whittington et al., 2008). A strong evolutionary relation exists between animal toxins and ancestral cysteine cross-linked proteins (Fry, 2005; Fry et al., 2006; Kini, 2002). Some of the more striking examples include proteins resembling snake α-neurotoxins identified in humans and rodents (Miwa et al., 1999). To date, most of these toxin-like proteins have been discovered sporadically but the availability of rich sequence resources argues for a more systematic search strategy towards cell modulators.
We have developed a classifier for ranking protein sequences according to their toxin-like properties. The classifier is able to create a robust characterization of proteins that display characteristics of short, cysteine-based compact proteins, many of which are indeed toxin-like (Kaplan et al., 2007). Application of the classifier to the ∼10 000 predicted protein sequences from the sequenced honey bee proteome identified several such sequences, leading to the experimental confirmation of an overlooked function of a voltage-gated channel inhibitor (Kaplan et al., 2007).
Herein, we developed an improved and updated version of the classifier, called ClanTox-10 (for Classifier of Animal Toxin-2010) and applied it to the prediction of toxin-like proteins in all known viral sequences. The revised classifier is tuned to cope with genome-scale input and was refined through an updated protein training set. An improved discovery rate is achieved by combining the predictor with sequence features such as the presence of a ‘signal peptide’. We show the results of the predictor for 508 (often uncharacterized) proteins, all predicted as toxin-like proteins, ∼10% of which were classified with a very high-degree of confidence. The observation that viral proteins appear to include sequences that are candidates for growth factors, ion channel inhibitor and anti-bacterial short proteins opens new research avenues in the search for naturally evolved cell modulators. The analysis of a sample of these proteins is presented along with their postulated modulatory role.
2 METHODS
2.1 Data collection
Proteins for viruses were collected from UniProt (Apweiler et al., 2004). We included queries to the retrieval system to eliminate all ‘fragments’ and used the UniRef100 clusters to reduce the over-representation of the more heavily studied viruses. All together we started with ∼773 000 sequences (Organism: Virus). The set was reduced to 214 000 by eliminating incomplete sequences and unprocessed genomes (excluding ‘Fragment’ and ‘Polyprotein’). The short sequences (∼40 000 sequences, 10–120 amino acids) from this list were clustered according to UniRef100 to produce a filtered list of ∼26 000 proteins. Less than 5% of these are curated and represented in the SwissProt collection.
2.2 Bioinformatics analysis tools
SignalP was used for predicting signal peptides (Bendtsen et al., 2004). ClustalW and alignment viewer tools were used from EBI's (ClustalW2) server and the NCBI (Cobalt multiple sequence alignment). Multiple sequence alignment and phylogenetic analysis was based on (PHYLIP tree). PHYRE (Kelley and Sternberg, 2009) was used for fold recognition and InterProScan (Quevillon et al., 2005) was used for sequence motif detection.
2.3 Feature vector construction
Each protein sequence was represented as a vector of 545 sequence derived numerical features. In brief, the generic features include (i) amino acid frequencies (single and consecutive pair frequencies, total of 420 features); (ii) Sequence length (m, 1 feature). The rest of the features were driven by the importance of specific amino acids and their spacing, with a special consideration for cysteine. The stability of short animal toxins is often mediated by the multiple disulfide bridges of paired cysteines. Note that the features were not restricted to cysteine-related features and were derived from all 20 amino acids: (i) cysteine binary 5-mers: The sequence is divided into 5-mers. The number of such 5-mers is m-4, where m is the protein length. For example, a protein of length 30 reports on 26 such 5-mers. Cysteines are translated into 1s, and the remaining amino acids are translated into 0 s. A 25 legitimate combinations are possible, providing 32 features. (ii) Charged-polar residues binary 5-mers: similar to cysteine binary 5-mers, except that charged and polar residues (Asp, Glu, Lys, Arg, Asn, Gln) are translated into 1 and the rest are translated into 0. As above, it provides 32 features. Note that the binary descriptors replace any explicit information on the hydrophilic or polarity index of amino acids. (iii) Amino acid entropy: a quantitative measure of how much each amino acid type is dispersed throughout the sequence (20 features). For a given amino acid type c, we denote its positions in the sequence as p1,…, pk its positions in the sequence. We define p0 = 0 and pk+1 = m + 1. We define the entropy of c to be:
| (1) |
(iv) Circular center of mass (CM): we added a quantitative measure that encodes the relative location and distribution of each amino acid type in the sequence. For a given amino acid type c, we again denote its positions in the sequence as p1,…, pk. The feature formalizes the following notion: If the sequence is spread clockwise around the 2D unit circle, we can calculate the mean of the points on the circle that match p1,…, pk and define it as the circular mean of c. This measure is supported by the observation that in short folds the two ends are relatively close in space. Formally:
| (2) |
2.4 Training and learning sets
Selection of a training set is based on careful manual evaluation due to the diversity in ‘animal toxins’ and some misuses of the term ‘toxin’. We used a combination of annotations and keywords including ‘ionic channel inhibitor’. Following exclusion of protein fragments, 924 sequences were collected and manually verified. Following a sequence similarity-based clustering (cutoff at 90% identity using global sequence alignment), we remained with a positive set of 627 sequences. We manually removed 23 sequences that are mostly proteins fused to Ion channel inhibitor (ICI) or incorrectly identified as such. Note that the protein length restriction was not applied for the training set. We selected several sets of negative instances from a random UniProtKB from which we removed ‘fragments’ or any obvious redundancy (according to UniRef90). For these sets, proteins annotated as ‘toxin’ were eliminated. We selected random sets with a size distribution that matches that of the true instances that are 2-fold larger. Six independent such sets were used. The final training set consists of the union of the non-redundant sets of positive (ion channel inhibitor non-redundant set) and negative (non-toxins) proteins. The list of the 604 sequences that were used as a positive set is found under ‘technical support’ in the ClanTox web server.
2.5 Learning algorithm
The learning algorithm that was used is a meta-classifier based on the boosted stumps algorithm. In a nutshell, the stump classifier is trained to find the best linear separation available by a single feature (i.e. tries all possible thresholds on all possible features and uses the best threshold). In the boosted stumps method, the AdaBoost algorithm is applied to the stump classifier. Subsequently, a stump classifier is trained on the instances, so that instances with higher weights are penalized more heavily. Following the training, the weights of the instances that were mispredicted are adjusted, and so on. Each such iteration is referred to as a boosting iteration. The final prediction is in the form of a linear combination of stump classifier predictions (one per boosting iteration, where the weight of each stump classifier is determined by its error rate). The prediction is a real number, rather than a binary prediction. A value >0 signifies a sequence that is toxin like (positive).
Since learning algorithms often require input of additional parameters and each choice of parameters can give different results, a parameter-tuning framework was included. In order to determine the optimal number of iterations, the classifier is evaluated on every boosting step by its area under the curve (AUC) performance (in a 3-fold cross-validation test). The parameter value which maximizes the AUC is chosen for the final classifier. The boosted stumps method performed faster and outperformed support vector machine (SVM) and boosted decision trees (data not shown).
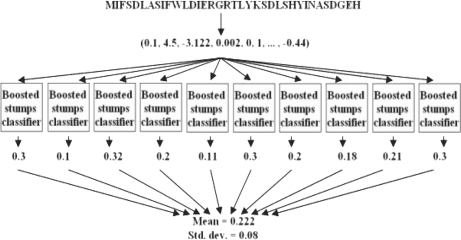
We adapted a meta-classifier so that the predictions it makes are a combination of the predictions made by 10 different boosted-stumps classifiers. For a given set of true instances, 10 separate training sets are generated. Next, a parameter-tuned boosted stump classifier is trained on each set. The outputs of all 10 classifiers are normalized by the highest positive prediction of each classifier on the training set (respective to each classifier). The final prediction of the meta-classifier is the arithmetic mean of the predictions of all 10 classifiers. Additionally, the meta-classifier provides the SD of the predictions on each sequence as a measure of robustness. Other methods of ranking and binning the results were similarly effective (data not shown). A prediction is considered to be positive prediction if the mean is greater than the SD. By this approach, the meta-classifier is able to provide a more robust hypothesis, which is minimally biased by a specific choice of false instances. An overview of the prediction scheme is show in Figure 1.
Fig. 1.
A scheme of the prediction procedure. A protein sequence is transformed into a vector of 545 features. The vector is independently sent to 10 boosted stump classifiers, each of which produces a numerical result. The mean and the standard deviation of the 10 results are reported.
A formal detailed outline of the learning scheme is presented under ‘technical support’ in Clantox web server.
2.6 Cross-validation
Cross-validation was performed and evaluated by the area under the curve (AUC). The receiver operating characteristic (ROC) curve formed by the predictions of the classifier shows the TP rate and FP rate at all possible thresholds (i.e. each possible threshold is plotted as a point). The AUC is measured as the area under the ROC curve formed by the predictions.
3 RESULTS
3.1 Prediction performance of ClanTox-10
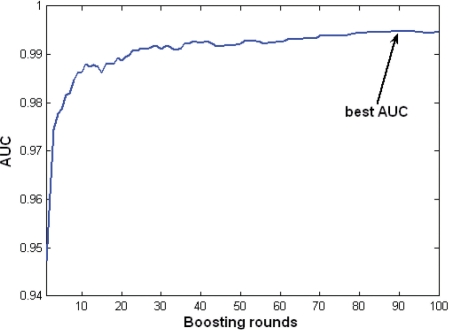
An evaluation of the predictor (presented initially in Kaplan et al., 2007) was performed to test the impact of updating the training sets (UniProtKB release 15.13). In ClanTox-10, negative instances were selected differently than in the original version of the classifier (Kaplan et al., 2007). The classifier showed a high level of success, with a mean AUC of >0.99 (Fig. 2). Note that the value calculated is equal to the Mann–Whitney–Wilcoxon non-parametric test. From a list of >500 features (see Section 2), the most dominant features repeatedly identified by the classifier are the density (Entropy) and the spacing of cysteines (CM, see Section 2). Both feature types are crucial to structural properties underlying the fold stability. The high performance in the cross-validation test indicates that the classifier captures a robust phenomenon. However, the success of AUC test is not necessarily a direct measure for the success of the classifier towards an unseen data set.
Fig. 2.
The classifier showed a mean AUC of 0.9936 (SD = 0.003). The graph shows the 3-fold cross-validation AUC of one of the boosted stumps classifiers, as a function of the boosting rounds.
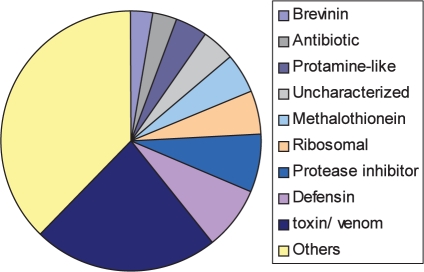
We tested the performance on an updated set of SwissProt that covers 500 000 proteins (a set that is 2-fold larger than was initially tested). We characterized the performance of the classifier towards short proteins (67 900 proteins in the range of 10–120 amino acids, represented by 36 000 non-redundant representatives). The classifier successfully identified among the positive predictions hundreds of proteins enriched in secretory proteins. This is an intriguing finding as no explicit features support this property. In addition, significant enrichment was associated with multiple general categories (Fig. 3). The classifier positively predicted ∼900 sequences that are enriched in proteins from toxin/venom, defensin and brevenin, anti-fungal and anti-bacterial sequences, uncharacterized proteins and more. Manual inspection of the results indicated that most of the positive predictions do not belong to genuine ICI proteins (recall that all ICI annotated set was eliminated for such test). Instead, enrichment in proteins that act as cell modulators in diverse biological contexts become evident (Fig. 3). For example, defensins and brevenins and short secreted peptides from the skin of mammals and amphibians, respectively. These proteins are active in host-defense mechanism against invading organisms and thus are also considered antimicrobial peptides. Interestingly, ribosomal proteins and protamine-like proteins were positively identified yet with a reduced confidence score. These finding suggests that the classifier is able to identify modulatory functions beyond the ‘ion channel inhibitors’ for which the system has been trained for.
Fig. 3.
Partition of the protein categories that were positively predicted by ClanTox-10. The classifier was applied to ∼36 000 non-redundant sequences from SwissProt ranging between 10 and 120 amino acids in length. The major broad categories (> 25 proteins each) are indicated.
We then tested all the keyword annotations (from UniProt) that are most statistically significant according to their P-value (P-value is calculated according to the hypergeometric distribution and corrected by Bonferroni correction for multiple hypothesis testing). Among the most significant annotations (P < E-100) are for snake toxins, secreted proteins (based on annotation of signal peptides), Phospholipase A2, Knottin, protease inhibitor and defensin (ranges at E-70–E-60). A detailed list can be found in technical ‘help’ on the webserver (Naamati et al., 2009). Functional enrichment terms also included immune recognition elements and disintegrins (P < E-30).
3.2 Viral proteins are fragmented, poorly annotated and redundant
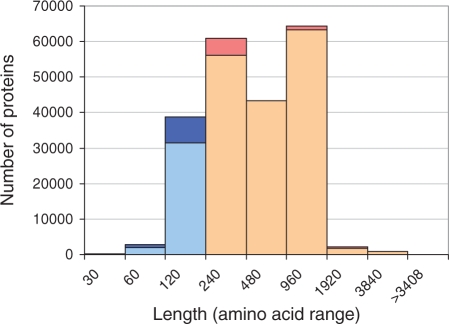
Viral proteins are very biased and overrepresented by few viral families (Bahir et al., 2009). Currently, ∼8% of all sequences in the UniProtKB database (Apweiler et al., 2004) are viral proteins (773 000 proteins). The number of viral proteins that are full-length proteins (without polyproteins) drops to 214 000 sequences. The length distribution of viral proteins is also different from that of the other kingdoms. We limited the analysis for proteins of length 10–120 amino acids that comprise ∼40 000 sequences that are clustered into ∼26 000 representatives (according to UniRef100, Fig. 4). While the number of ‘uncharacterized’ and ‘putative’ viral proteins (excluding ‘fragment’) is 6.8%, for proteins shorter than 120 amino acids this fraction reaches 22%.
Fig. 4.
Length distribution of viral representative proteins from UniProtKB. Total number of UniProtKB viral proteins according to their length. The list includes only full-length proteins. The x-axis values indicate the range (240 are proteins with a length of 120–240 amino acids). In blue, the fraction of viral proteins analyzed in this study. Dark blue and dark pink indicate proteins marked as ‘uncharacterized’. This fraction is maximal for short proteins <120 in length.
3.3 Positive prediction of toxin-like cell modulators in virus proteome
We used the 25 982 short proteins as input for ClanTox. Only 2% (508 proteins) of these sequences were predicted as toxin-like proteins (Table 1).
Table 1.
Partition of the prediction according to the length
| Very | High | Mod. | Neg. | Total | Total | |
|---|---|---|---|---|---|---|
| high | Seq. | clusters | ||||
| Viruses: all | 19 | 32 | 457 | 25 474 | 39 774 | 25 982 |
| 10–80 aa | 16 | 26 | 235 | 5727 | 7732 | 6004 |
| 81–100 aa | 3 | 6 | 167 | 12 928 | 24 098 | 13 104 |
| 101–120 aa | 0 | 3 | 55 | 6960 | 7844 | 7018 |
Prediction confidence is marked as Very high, High, Moderate (Mod.) and Negative (Neg.) based on the mean score and SD from ClanTox.
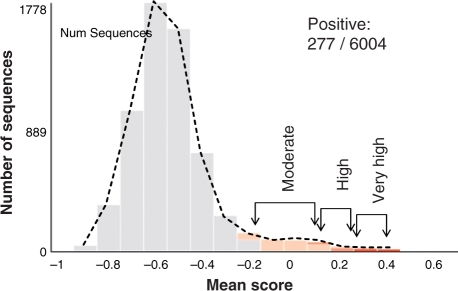
From a biotechnological and medical perspective, the most attractive proteins are short ones. Driven by this preference, we tested the viral sequences according to their length partition. The results of the 6004 proteins whose sizes are in the range of 10–80 amino acids are shown in Figure 5. This set contributes to ∼50% of all positive predictions and ∼80% of the most confident predictions (class ‘High’ and ‘Very high’). The discovery rate drops drastically in sets composed of proteins that are longer (Table 1).
Fig. 5.
Histogram of the predictor mean score for 6004 viral proteins (length 10–80 amino acids). The tail of the distribution is marked according to the prediction quality categories. A total of 277 positive predictions are reported.
3.4 Conotoxin-like proteins are prevalent among the highest rank predicted proteins
Fifty-one proteins were identified at the highest scores (Table 1), of which 12 are marked as ‘uncharacterized’. A sample of the top-ranked predicted proteins is shown, alongside the distribution of cysteine residues (Fig. 6, left). Surprisingly, among the top listed proteins, we identified several proteins annotated as conotoxin-like proteins. Conotoxins are one of the largest groups of neurotoxic peptides isolated from the venom of the marine cone snail. Conotoxins mostly act to modulate the activity of ion channels.
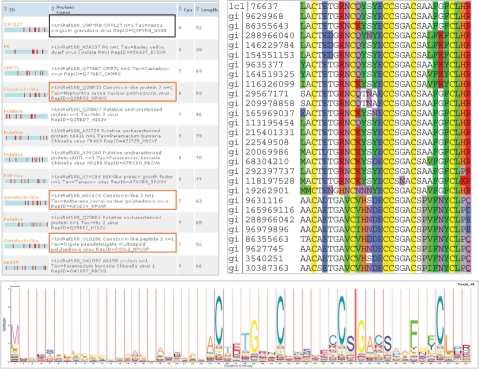
Fig. 6.
Top predicted proteins from short viral proteins. Left: 12 proteins predicted by ClanTox. The three top scoring conotoxin-like proteins are framed in orange; an overlooked homologue to conotoxin-like protein is framed in black. Right: multiple sequence alignment based on BLAST nr search. Twenty-six out of 28 representative proteins are viral proteins that are present in 21 different viruses. Bottom: an HMM logo of Pfam family PF08087 that is associated with the Conotoxin O-superfamily (see text).
We performed a BLAST search and a multiple sequence alignment (MSA) analysis for the proteins listed as conotoxin-like proteins (Fig. 6, Framed). This family was further expanded by including all (non-identical) sequences supported by NCBI. A total of 67 proteins were identified in 26 taxa. A total of 24/26 organisms are found in dsDNA viruses. The other two are found in fungi and sea anemone proteomes. The MSA is composed of 28 viral proteins that are reported by UniProtKB (Fig. 6, right).
Several observations strengthen the relevance of these top predicted viral proteins as genuine conotoxins: (i) all 65 viral conotoxin-like proteins have a signal peptide as predicted by Signal P (Bendtsen et al., 2004). (ii) The MSA of the signal peptide is weakly conserved relative to the ‘mature’, apparently active, peptide (Fig. 6, right). (iii) Like many of the conotoxins, six cysteines (Fig. 6, right, marked in yellow) that form three disulfide bonds are conserved throughout. (iv) Key residues that are essential in the active conotoxin peptides are shown by the MSA. These residues include the conserved Gly, two conserved Glu and a hydrophobic residue (either Phe, Tyr). (v) Structural modeling (Kelley and Sternberg, 2009) of the sequence of the top predicted conotoxin-like proteins overlap the template of the conotoxin fold (solved by NMR).
Most viral conotoxin-like proteins that are predicted by ClanTox-10 match the PFAM PF08087, Toxin_18, Conotoxin O-superfamily. The HMM based logo of these sequences is presented in Figure 6 (bottom). The HMM-logo based on Pfam covers mostly Baculoviridae and sequences from few insects (Aphid and Mosquitos). Based on the MSA using the ClanTox-10 conotoxin-like proteins as a seed we were able to expand the family and to include sequences from Fungi and Sea anemone. Note that sequences presented in the MSA include all six conserved cysteines (as opposed to four cysteines in PF08087, Fig. 6, bottom).
Among the viral conotoxin-like proteins, two major subfamiles are evident (Fig. 6, right). The 20 protein sequences in the top of the MSA belong to one subfamily while the other eight sequences comprise the second subfamily. Whether the proteins that belong to the two subfamilies differ in their activity towards specific ion channel targets is unknown and calls for experimental testing.
3.5 Knottin fold is abundant among positively predicted proteins
Most top predicted proteins (classified with either ‘Very high’ or ‘High’ confidence, Table 1) lack similarity to other proteins. Structural modeling (Kelley and Sternberg, 2009) was used to propose structural resemblance. For example, the 70 amino acids of UniProt: Q5QC64 from Monkeypox virus (predicted as ‘High’) was modeled as a knottin-related structure (using PDB 2B68 as a template). Best fold prediction suggested the defensin and to a lesser extent a scorpion toxin which blocks calcium-activated potassium channels (PDB 1DU9). Knottin fold proteins (Hartig et al., 2005) share a variety of cell modulatory functions such as immune defense, anti-bacterial activity, receptor binding domains and as insecticides. Among the positively identified viral proteins, we identified tumor necrosis factor (TNF)-like receptor and epidermal growth factor (EGF)-like growth factor (similar to neuronal neuregulin).
3.6 Inference annotation task
Most positively predicted proteins were assigned with moderate confidence (Fig. 5, Table 1). Although we set the predictor threshold to be rather conservative, the quality of such predictions is questionable. Nonetheless, we suggest that many of the ‘moderate’ predictions are valid as potential cell modulators: (i) testing proteins of the venomous snake proteins indicated many validated toxins to be assigned to the ‘moderate’ class (Naamati et al., 2009). (ii) Bioinformatics inspection of the listed 457 proteins (Table 1) reveals that 30.5% contains a signal peptide (using SignalP), but only 5% are annotated as such in the entire input set. Recall that all toxin-like proteins are expected to be secreted. (iii) Applying the classifier to the entire set of viral proteins that are annotated as ‘secreted proteins’ (total of 628 proteins represented by 537 clusters, UniRef100), identified 20 proteins in accord with the view that the signal peptide per se does not convey the predictive power of our classifier.
We were able to suggest new annotation for a number of positively predicted proteins. For example, UniProt P24284 (VB05_VACC8) named ‘truncated plaque-size/host range protein’ from Vaccinia virus is a short protein (92 amino acids). This protein contains a classical signal peptide and a Sushi domain. This gene product was shown to affect plaque size (Takahashi-Nishimaki et al., 1991). The Sushi domain is found in many adhesion proteins. Proteins with this domain (large proteins) were identified in other viruses such as Epstein Barr virus (EBV), where the protein is a glycoprotein receptor. We propose that the P24284 is a non-functional mimic of an adhesion unit and potentially affects virus infectivity. This hypothesis calls for further research.
3.7 ‘Toxin-like’ viral proteins under-represented in domain and family annotations
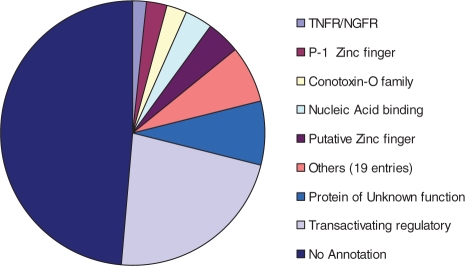
ClanTox exposed 508 sequences that are candidate toxin-like proteins. We tested the overlap between this set and several domain and family expert systems. Pfam and InterPro provide the most extensive cover for domains and families. However, we found that this ‘toxin-like set’ is poorly covered by such systems. Among the 508 sequences, about half (248) have no annotations by Pfam or InterPro. All together, this list is associated with 26 Pfam entries including entries that provide little information such as ‘Uncharacterized protein family’ (39). Additional Pfam entries include conotoxin-like proteins, growth factors and their receptors, adhesion-like domains and putative zinc finger. Interestingly, the largest group of proteins that were annotated are the TAT transactivation regulators (Fig. 7). While TAT function as RNA binding proteins, they are considered to be multifunctional virus-encoded cytokine-like protein factor. As such it displays the activities of several growth factors and chemokines. TAT proteins have a unique stability and pleotropic function on cells activation, growth, and transformation even in the absence of other viral structures. This overlap between our prediction and the large body of TAT co-activators strongly support the notion that other cell modulators are among the list of ∼500 identified proteins. A list of 20 viral proteins that belong to this diverse set of viruses that infect human is shown (Table 2).
Fig. 7.
Partition of the 508 toxin-like proteins according to coverage by Pfam entry. A total of 248 proteins have no domain and family annotation by Pfam or InterPro. All together, 26 Pfam entries are included. Only Pfam entries covering more than eight proteins are indicated.
Table 2.
A list of 20 proteins belonging to diverse set of viruses that infect human
| Accession | Protein names | Organism | aa | Virus hosts |
|---|---|---|---|---|
| Q9DHU7 | 15L protein | YLDV | 77 | Human and others |
| Q9PX43 | B3 | HHV-6 variant B | 59 | Human |
| Q8QN46 | CD30 (VCD30) | CPV | 110 | Human and others |
| O57094 | Crm-B secreted TNF-α-receptor-like | CPV | 81 | Human and others |
| Q779N0 | CrmE protein | CPV | 81 | Human and others |
| P87599 | D13L protein | CPV | 111 | Human and others |
| Q0GP92 | HSPV010 | HSPV | 111 | Human and others |
| Q5QC64 | K1R, TNF-α-receptor-like | Monkeypox virus | 70 | Human and others |
| Q98181 | MC010R | MCVI | 73 | Human |
| Q98194 | MC026L | MCVI | 83 | Human |
| Q98220 | MC052R | MCVI | 100 | Human |
| Q6TW86 | ORF014 modified RING finger | ORFV | 93 | Human and others |
| D3J321 | Phosphoprotein | HEV | 102 | Human |
| Q2F8G1 | RING-H2 motif protein | ORFV | 93 | Human and others |
| Q75005 | Tat (transactivating regulatory) | HIV-1, Isolate ETH2220 | 42 | Human |
| Q0NQ67 | TNF-α-receptor-like | CPV | 109 | Human and others |
| P24284 | Truncated plaque-size/host range, B5 | VACV, strain LC16m8 | 92 | Human |
| P20530 | Uncharacterized 8.8 kDa | VACV, strain Copenhagen | 72 | Human |
| A2RQH9 | Z protein | GTOV, NH-95551 isolate | 95 | Human and others |
| A2RQH8 | Z protein | GTOV, NH-95551 isolate | 95 | Human and others |
4 CONCLUSIONS AND DISCUSSION
Applying ClanTox-10 to viral proteomes has become possible through the use of an implementation can cope with genomic-scale input (Naamati et al., 2009). Using ClanTox as a ‘microscope’ for overlooked modulators predicted to be toxin-like proteins was proposed. Our approach can be extended to other sequencing resources and transcriptomic data. An attractive resource is that of the metagenomics data originating from environmental samples. The number of phages and viral sequences is very high and at present, most lack annotation schemes (Edwards and Rohwer, 2005), while the relevance of metagenomics to human health has become increasingly appreciated.
The finding of a large number of conotoxin-like peptides in viruses is intriguing. Two scenarios may account for this observation: (i) fast evolving convergent evolution of short secreted proteins as proposed for many of the cysteine-rich proteins (Fry, 2005). (ii) Genetic material exchange from hosts to viruses. Such a scenario was suggested in some instances. A cysteine-rich encoding region was shown to transfer from the endoparasitic wasp Campoletis sonorensis to the symbiotic polydnavirus (CsPDV) (Cui and Webb, 1996). The role of such toxin-like proteins to the life cycle of the viruses is unknown. Interestingly, a report on a conotoxin-like peptide in baculovirus from 20 years ago showed that deletion of the gene (called ctl) had no effect on the kinetics and virulence of infection (Eldridge et al., 1992). For over half of the 508 proteins that were positively identified by the classifier, no significant similarity can be found. It is an expected outcome of the fast evolution of viral genomes (Fig. 7). However, in instances where a significant similarity can be traced (using PSi-BLAST, IntrProScan (Quevillon et al., 2005), ProtoNet cluster assignment (Kaplan et al., 2005) and annotations based on Pfam, an over-representation of insect genomes is apparent. An example is the protein B6S6X8 (113 aa) from Betabaculovirus that is similar to many short peptides in Drosophilae proteomes.
The cross-talk of the viral proteins as candidates for cell modulatory function in view of the specific host is currently being investigated. The extreme underrepresentation of toxin-like viral proteins for infected bacteria may reflect the inability to support disulfide bridges in most bacteria.
The systematic discovery of short peptides as modulators is of great interest for peptide therapy. We expect to use our procedure to find such potential candidate sequences. Currently, several conotoxins were proven successful in treatment of acute pain in animal models (Scott et al., 2002). Some pain managing drugs that are based on a conotoxin scaffold were approved for clinical use (Alonso et al., 2003).
In summary, the ClanTox can identify short proteins that function as non-toxin cell modulators that are beyond the function of toxins. Among the ∼500 viral positive predictions, we identified a surprising number of conotoxin-like proteins, growth factors and their receptors and several sequences resembling adhesion units. About 34% of the toxin-like proteins for which the host is known are from viruses that infect human and primate. Therefore, we postulate that some of the sequences identified in this study carry a potential for drug development and for manipulating virus–host interaction.
ACKNOWLEDGEMENTS
We would like to thank Solange Karsenty for design and managing ClanTox and Menachem Fromer for a critical reading. We specifically thank Noam Kaplan that initiated the project and set the foundation for the classifier.
Funding: Israel Science Foundation (ISF 592/07); the US-Israel Binational Science Foundation (BSF 07219); Sudarsky Center for Computational Biology (SCCB to G.N.).
Conflict of Interest: none declared.
REFERENCES
- Alonso D, et al. Drugs from the sea: conotoxins as drug leads for neuropathic pain and other neurological conditions. Mini Rev. Med. Chem. 2003;3:785–787. doi: 10.2174/1389557033487746. [DOI] [PubMed] [Google Scholar]
- Apweiler R, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahir I, et al. Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Mol. Syst. Biol. 2009;5:311. doi: 10.1038/msb.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, et al. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Cui L, Webb BA. Isolation and characterization of a member of the cysteine-rich gene family from Campoletis sonorensis polydnavirus. J. Gen. Virol. 1996;77(Pt 4):797–809. doi: 10.1099/0022-1317-77-4-797. [DOI] [PubMed] [Google Scholar]
- Edwards RA, Rohwer F. Viral metagenomics. Nat. Rev. Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- Eldridge R, et al. Characterization of a baculovirus gene encoding a small conotoxinlike polypeptide. J. Virol. 1992;66:6563–6571. doi: 10.1128/jvi.66.11.6563-6571.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG. From genome to ‘venome’: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15:403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, et al. Early evolution of the venom system in lizards and snakes. Nature. 2006;439:584–588. doi: 10.1038/nature04328. [DOI] [PubMed] [Google Scholar]
- Hartig GR, et al. Intramolecular disulphide bond arrangements in nonhomologous proteins. Protein Sci. 2005;14:474–482. doi: 10.1110/ps.04923305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungo F, Bairoch A. Tox-Prot, the toxin protein annotation program of the Swiss-Prot protein knowledgebase. Toxicon. 2005;45:293–301. doi: 10.1016/j.toxicon.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Kaplan N, et al. Novel families of toxin-like peptides in insects and mammals: a computational approach. J. Mol. Biol. 2007;369:553–566. doi: 10.1016/j.jmb.2007.02.106. [DOI] [PubMed] [Google Scholar]
- Kaplan N, et al. ProtoNet 4.0: a hierarchical classification of one million protein sequences. Nucleic Acids Res. 2005;33:D216–D218. doi: 10.1093/nar/gki007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kini RM. Molecular moulds with multiple missions: functional sites in three-finger toxins. Clin. Exp. Pharmacol. Physiol. 2002;29:815–822. doi: 10.1046/j.1440-1681.2002.03725.x. [DOI] [PubMed] [Google Scholar]
- Marques JT, Carthew RW. A call to arms: coevolution of animal viruses and host innate immune responses. Trends Genet. 2007;23:359–364. doi: 10.1016/j.tig.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Miwa JM, et al. lynx1, an endogenous toxin-like modulator of nicotinic acetylcholine receptors in the mammalian CNS. Neuron. 1999;23:105–114. doi: 10.1016/s0896-6273(00)80757-6. [DOI] [PubMed] [Google Scholar]
- Naamati G, et al. ClanTox: a classifier of short animal toxins. Nucleic Acids Res. 2009;37:W363–W368. doi: 10.1093/nar/gkp299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA, et al. Actions of intrathecal omega-conotoxins CVID, GVIA, MVIIA, and morphine in acute and neuropathic pain in the rat. Eur. J. Pharmacol. 2002;451:279–286. doi: 10.1016/s0014-2999(02)02247-1. [DOI] [PubMed] [Google Scholar]
- Sorek R, et al. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Takahashi-Nishimaki F, et al. Regulation of plaque size and host range by a vaccinia virus gene related to complement system proteins. Virology. 1991;181:158–164. doi: 10.1016/0042-6822(91)90480-y. [DOI] [PubMed] [Google Scholar]
- Tan PT, et al. Bioinformatics for venom and toxin sciences. Brief Bioinform. 2003;4:53–62. doi: 10.1093/bib/4.1.53. [DOI] [PubMed] [Google Scholar]
- Tan PT, et al. SCORPION2: a database for structure-function analysis of scorpion toxins. Toxicon. 2006;47:356–363. doi: 10.1016/j.toxicon.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Whittington CM, et al. Defensins and the convergent evolution of platypus and reptile venom genes. Genome Res. 2008;18:986–994. doi: 10.1101/gr.7149808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse ME, et al. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]