Abstract
Summary: CellDesigner provides a user-friendly interface for graphical biochemical pathway description. Many pathway databases are not directly exportable to CellDesigner models. PathwayAccess is an extensible suite of CellDesigner plugins, which connect CellDesigner directly to pathway databases using respective Java application programming interfaces. The process is streamlined for creating new PathwayAccess plugins for specific pathway databases. Three PathwayAccess plugins, MetNetAccess, BioCycAccess and ReactomeAccess, directly connect CellDesigner to the pathway databases MetNetDB, BioCyc and Reactome. PathwayAccess plugins enable CellDesigner users to expose pathway data to analytical CellDesigner functions, curate their pathway databases and visually integrate pathway data from different databases using standard Systems Biology Markup Language and Systems Biology Graphical Notation.
Availability: Implemented in Java, PathwayAccess plugins run with CellDesigner version 4.0.1 and were tested on Ubuntu Linux, Windows XP and 7, and MacOSX. Source code, binaries, documentation and video walkthroughs are freely available at http://vrac.iastate.edu/∼jlv
Contact: julied@iastate.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
CellDesigner (Funahashi et al., 2008) is a tool for graphically building biochemical pathway models, which integrate model representation by Systems Biology Markup Language (SBML; Hucka et al., 2003) with graphical representation by Systems Biology Graphical Notation (SBGN; Moodie et al., 2009). There exist many databases providing application programming interface (API) libraries enabling programmatic queries. These API libraries include many biologically meaningful objects, which carry out intuitive functions. For example, a pathway object can report the set of reaction objects it contains, a protein complex object can report the monomer objects that contsruct it and a metabolite object might report its SMILES and InChi codes. The problem is that a Pathway object in one API is not the same as a Pathway object in the API of a different database. The same biological concept is represented using independently developed in-silico representations, preventing any single application from communicating and integrating across databases.
2 FUNCTIONALITY
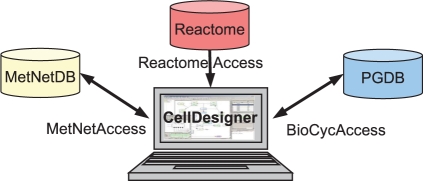
PathwayAccess plugins directly interact with pathway databases so that the user can download one or more pathways to a CellDesigner model and upload (or commit) a CellDesigner model to a database. Figure 1 shows a dataflow diagram for typical use of the PathwayAccess plugins.
Fig. 1.
Dataflow for PathwayAccess plugins. PathwayAccess plugins use respective APIs to communicate with different pathway databases and integrate data in CellDesigner. As indicated by arrows, depending on functionality supported by the datasource, dataflow is uni- or bi-directional.
The PathwayAccess plugin framework confers three major benefits, depending on whether individual database APIs support data retrieval and modification. First, the plugins make pathways stored in remote databases available to the powerful modeling and simulation functionality already provided by CellDesigner. Second, SBGN implemented by CellDesigner provides a standard representation for biologists to curate pathway databases; the user can create a pathway model and commit it to the database of his choice. A user can also download a pathway model from a database, edit it and commit it back to the database, either replacing the original pathway or creating a different version. Third, CellDesigner can be effective in visually comparing and integrating pathway data from one or many different databases; metabolic networks can be downloaded directly into CellDesigner and integrated into custom super-pathways. CellDesigner can export pathways into files for loading into other software such as Cytoscape (Shannon et al., 2003), where SBGN is an ancilliary feature to network analysis functions.
Since CellDesigner and most datasources' user interfaces provide good automatic layouts, layouts are left to the datasources and CellDesigner independently.
2.1 Pathway integration across databases
When PathwayAccess plugins download pathways, they are integrated with the growing model in memory. CellDesigner is suited to support integration because it uses the XML-based SBML data model not only for file storage, but also for objects in memory—ideal for representing annotations integrated from different sources. Among other annotations, PathwayAccess stores synonyms this way, enabling it to match integrated objects in the same subcellular compartment that may be named differently across databases. To prevent duplicate reactions in integrated pathways, a reaction hashing algorithm calculates a unique integer for every combination of reaction substrates, products and catalysts (see Supplementary Material). Each PathwayAccess plugin has a unique, but editable highlight color, which can be used to color the model objects downloaded using that plugin. Objects from multiple databases are colored by mixing the colors of the plugins that downloaded them.
2.2 Creating new PathwayAccess plugins
The PathwayAccess framework includes a core library plus one or more independent plugins. A plugin developer can easily create a new CellDesigner plugin that communicates with any pathway database providing a Java API. Simply create a new CellDesigner plugin object using the PathwayAccess library and define a set of simple database query operations, depending on whether the plugin will support download and/or saving a model to the database. To create a PathwayAccess plugin that downloads a pathway, define 18 simple functions such as to get the synonyms of an object (pathway, metabolite, gene, etc). To design a commit feature, define nine simple functions such as add substrates to a reaction in the database. With these simple operations defined for communicating with a database, PathwayAccess handles all interaction both with CellDesigner and the database, similarly to Cytoscape's Data Integration Request For Comments (Killcoyne et al., 2009), and provides a way to enrich objects beyond the annotation used for integration.
2.3 Examples
Three PathwayAccess plugins, MetNetAccess, BioCycAccess and ReactomeAccess were created. In addition to representing biological objects differently, each uses a different communication protocol: SQL, Sockets and Web Services, respectively.
2.3.1 BioCycAccess: download and commit to a Pathway Genome Database
BioCyc databases are individually deployed for specific organisms and purposes http://www.biocyc.org; Karp et al., 2005). BioCycAccess uses JavaCycO, our new library wrapped around the JavaCyc API (Krummenacker et al., 2005; Mueller et al., 2005), running in client mode to connect to a BioCyc Pathway Genome Database that is running JavaCycO in server mode. It supports both downloading and committing pathways.
2.3.2 ReactomeAccess: download from Reactome
Reactome is a large repository for pathways (Matthews et al., 2009). ReactomeAccess supports downloading pathways from Reactome directly into CellDesigner models via an API wrapped around Reactome's web services.
2.3.3 MetNetAccess: download and commit to MetNet
MetNetAccess provides CellDesigner access to the pathway database MetNetDB using MetNetAPI (Sucaet et al., 2010), which is wrapped around SQL queries. It supports both downloading and committing pathways. MetNetDB is an integrated pathway database that currently includes Arabidopsis thaliana, yeast, soybean and the grapevine. MetNetAccess has been used to curate many pathways for different organisms in MetNetDB (Wurtele et al., 2007). MetNet allows public downloading of data, but only registered curators may modify data in MetNetDB.
3 IMPACT
The PathwayAccess suite of CellDesigner plugins is a powerful tool for researchers who work with metabolic pathway data and wish to take advantage of graphical and computational CellDesigner features. By directly accessing and publishing to pathway databases, decentralized pathway integration and comparison is made possible over simply saving and loading SBML files. Although three PathwayAccess plugins have been released, the practical scope of the PathwayAccess library is as wide as the number of databases to which CellDesigner can connect because communication requires a Java API. MetNetAccess, BioCycAccess, ReactomeAccess and future PathwayAccess plugins enable CellDesigner users to expose pathway data to analytical CellDesigner functions as well as visually integrate and curate pathway data from different databases using standard SBGN—something that has been previously prevented by disparate in silico representations of biological objects.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yves Sucaet and Eve Wurtele for developing MetNetAPI and Jesse Walsh for testing JavaCycO.
Funding: National Science Foundation (grants DBI 0604755 and EEC 0813570).
Conflict of Interest: none declared.
REFERENCES
- Funahashi A, et al. Celldesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE. 2008;96:1254–1265. [Google Scholar]
- Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;9:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Karp PD, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;19:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killcoyne S, et al. Cytoscape Data Integration RFC. Request for comments, Cytoscape. 2009 Available at http://cytoscape.wodaklab.org/wiki/DataIntegration(last accessed date July 28, 2010) [Google Scholar]
- Krummenacker M, et al. Querying and computing with BioCyc databases. Bioinformatics. 2005;21:3452–3455. doi: 10.1093/bioinformatics/bti546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L, et al. Reactome knowledgebase of biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie S, et al. Systems biology graphical notation: process description language level 1. 2009 doi: 10.2390/biecoll-jib-2015-263. Available from Nature Precedings < http://dx.doi.org/10.1038/npre.2009.3721.1>. [DOI] [PubMed] [Google Scholar]
- Mueller L, et al. The SOL genomics network. a comparative resource for Solanaceae biology and beyond. Plant Physiol. 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucaet Y, et al. MetNetAPI. 2010 doi: 10.1186/1756-0500-3-312. Available at http://metnet3.vrac.iastate.edu/api(last accessed date July 28, 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele ES, et al. Concepts in Plant Metabolomics. Springer; 2007. MetNet: Systems Biology Software for Arabidopsis; pp. 145–158. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.