Abstract
Motivation: Peaks are the key information in mass spectrometry (MS) which has been increasingly used to discover diseases-related proteomic patterns. Peak detection is an essential step for MS-based proteomic data analysis. Recently, several peak detection algorithms have been proposed. However, in these algorithms, there are three major deficiencies: (i) because the noise is often removed, the true signal could also be removed; (ii) baseline removal step may get rid of true peaks and create new false peaks; (iii) in peak quantification step, a threshold of signal-to-noise ratio (SNR) is usually used to remove false peaks; however, noise estimations in SNR calculation are often inaccurate in either time or wavelet domain. In this article, we propose new algorithms to solve these problems. First, we use bivariate shrinkage estimator in stationary wavelet domain to avoid removing true peaks in denoising step. Second, without baseline removal, zero-crossing lines in multi-scale of derivative Gaussian wavelets are investigated with mixture of Gaussian to estimate discriminative parameters of peaks. Third, in quantification step, the frequency, SD, height and rank of peaks are used to detect both high and small energy peaks with robustness to noise.
Results: We propose a novel Gaussian Derivative Wavelet (GDWavelet) method to more accurately detect true peaks with a lower false discovery rate than existing methods. The proposed GDWavelet method has been performed on the real Surface-Enhanced Laser Desorption/Ionization Time-Of-Flight (SELDI-TOF) spectrum with known polypeptide positions and on two synthetic data with Gaussian and real noise. All experimental results demonstrate that our method outperforms other commonly used methods. The standard receiver operating characteristic (ROC) curves are used to evaluate the experimental results.
Availability: http://ranger.uta.edu/∼heng/MS/GDWavelet.html or http://www.naaan.org/nhanguyen/archive.htm
Contact: heng@uta.edu
1 INTRODUCTION
Mass spectrometry (MS) is a crucial analytical tool in proteomics research to provide tremendous information for disease proteomics study and drug targets identification at the protein/peptide level. Due to measurement error, chemical and other background noise, MS usually contains high-frequency noise and consequently a multitude of misleading peaks. Peak detection is one of the most important steps in MS data analysis because its performance directly effects the final proteomics study results.
Because the noise in MS comes from different resources and cannot be estimated, false positive peak detection results are unavoidable. This makes peak detection as a challenging problem. In recent years, several peak detection methods have been proposed (Coombes et al., 2005; Du et al., 2006; Morris et al., 2005; Nguyen et al., 2009). Most previous algorithms have four common preprocessing steps: denoising, baseline correction, alignment of spectrograms and normalization. After preprocessing, local maxima is usually used to detect peak positions and design rules to quantify peaks. In this article, we will explore the limitations of existing peak detection methods and propose several new algorithms to solve them.
Most peak detection methods employed denoising step by removing noise in each scale of wavelet, such as commonly used Cromwell (Coombes et al., 2005; Morris et al., 2005) and continuous wavelet transform (CWT) (Du et al., 2006) methods. However, true peaks in MS could have large frequency response and be removed by denoising step. As a result, these true peaks cannot be detected. We propose using bivariate shrinkage model, which considers relationship of two neighbor scales, to remove noise in stationary wavelet domain. Because utilizing relationship between two neighbor coefficients or two scales of wavelets can keep high-frequency true signal (Selesnick et al., 2001). Stationary wavelet transform (SWT) utilizes spatial information of signals and suppress artifacts by redundant representations.
Baseline removal step was widely used in peak detection methods, but it often got rid of true peaks and created new false peaks. To avoid removing baseline, the CWT-based pattern-matching algorithm was introduced in study by Du et al. (2006). Using Mexican Hat wavelet in multi-scale, this method gave good results in peak detection with high sensitivity and low false discovery rate (FDR). However, the more important property of multi-scale in wavelet domain was not used in this method (Mallat, 2009). Instead of considering peaks as the sum of delta functions, more generally, we consider MS peaks as a mixture of Gaussian in which each peak corresponds to one Gaussian. We propose to use Gaussian derivative wavelet, instead of Mexican Hat wavelet which is only the second derivative of Gaussian wavelet. Zero-crossing lines which are robust to noise are also introduced to replace Ridge-lines in Du et al. (2006). We study the zero-crossing lines in multi-scale wavelet and provide new theoretical analysis.
In most peak detection methods, signal-to-noise ratio (SNR) was used to remove the small energy peaks with SNR values less than a threshold. But MS noise cannot be correctly estimated in either time domain or wavelet domain. Thus, in this article, instead of SNR, frequency response, height and SD of Gaussian peaks calculated by zero-crossing in Gaussian derivative wavelet domain are used to remove false peaks. In order to improve sensitivity, the Envelope analysis (Nguyen et al., 2009) is also used to save some important peaks with small energy.
In this article, we propose a new Gaussian derivative wavelet-based peak detection method (GDWavelet) for Surface-Enhanced Laser Desorption/Ionization Time-Of-Flight (SELDI-TOF) spectrum. Both simulated and real spectrum with known polypeptide positions and compositions are used to evaluate our method. With simulated data, we compare different peak detection algorithms by both Gaussian and real noise. All experimental results show that our new approach can detect more peaks (in both high and low amplitude) with a lower FDR than state-of-the-art methods.
2 METHODS
In this section, our new GDWavelet method will be introduced. In GDWavelet, we utilize bivariate smoothing model, Gaussian derivative wavelet and envelope analysis. First, bivariate shrinkage estimator in SWT domain will be used to reduce noise and to keep whole true signal. Second, we will introduce how to detect peaks using Gaussian derivative wavelet through peak properties such as frequency response, SD and height. Finally, envelope analysis is performed to save true small energy peaks which will be missed if only peak properties are used.
2.1 Smoothing by bivariate shrinkage function
Noise smoothing in MS is an important step which should remove noise and keep true peaks. In Myers et al. (2004), they tried to remove noise as much as possible, hence some true peaks were also removed. We propose to utilize bivariate shrinkage estimator in SWT domain to reduce noise and keep whole true signal. More precisely, we decrease the noise level without removing most of them. SWT is chosen due to its fast speed and redundant representations. The later step will further handle the remaining noise.
To estimate wavelet coefficients, the most well-known rules are universal thresholding and soft thresholding (Donoho et al., 1995) which was applied to Cromwell method (Coombes et al., 2005; Morris et al., 2005). These algorithms assume that wavelet coefficients are independent. Unfortunately, frequency response of peak is rather wide. Hard or soft thresholding only considers coefficients in a sub-band with narrow frequency. Recent research shows that algorithms utilizing the dependency between coefficients can give better results than those using the independency assumption (Sendur et al., 2002). Sendur et al. exploited this dependency between coefficients and proposed a non-Gaussian bivariate pdf for the child coefficient w1 and its parent w2 as follows
| (1) |
The marginal variance σ2 is dependent on the coefficients index k. By this bivariate pdf and the Bayesian estimation theory, the MAP estimator of w1 (Sendur et al., 2002) is derived as
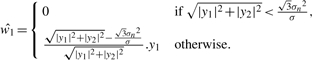 |
(2) |
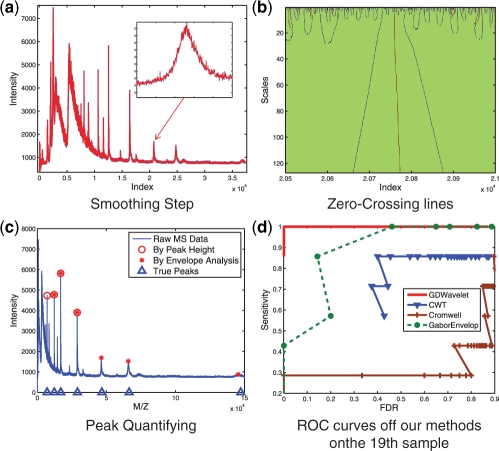
where y1 is child noisy coefficient, y2 is parent noisy coefficient. This estimator is a bivariate shrinkage function. It has been used to smooth many kinds of signals such as image (Sendur et al., 2002), DNA copy number (Huang et al., 2008; Nguyen et al., 2010), etc. In this article, bivariate shrinkage estimator is used to smooth MS signals. An example of denoising result is shown in Figure 3a. This example will be discussed in Section 2.4.
Fig. 3.
An Illustration of GDWavelet. The 19-th sample of (CAMDA, 2006) dataset is selected to illustrate how GDWavelet method detects MS peaks. (a) Blue signal is raw signal and red one is signal smoothed by bivariate shrinkage estimator in wavelet domain. (b) Parameters of peaks are estimated by zero-crossing lines. This figure shows zero-crossing lines of one zoomed peak in (a). (c) Peaks are quantified by peak height and PR. Union results include peaks with heights larger than a threshold or with PRs as one. (d) ROC curves of four methods' performance on the 19-th sample of CAMDA (2006) dataset. GDWavlet gives the best performance.
2.2 Peak detection by Gaussian derivative wavelet
In previous works (Coombes et al., 2005; Morris et al., 2005), MS peaks were considered as the sum of delta functions. That means only heights of peaks have been used for peak detection throughout SNR. Du et al. (2006) utilized width of peaks to improve peak detection results a lot. We consider MS peaks as a mixture of Gaussian in which each peak corresponds to one Gaussian:
| (3) |
With this assumption, four parameters providing intrinsic differences between true peaks and noise are peak position, SD, height and frequency response of peak. To find these parameters of a peak, we use zero-crossing lines in multi-scale of Gaussian derivative wavelet instead of ridge-lines in multi-scale of Mexican hat wavelet that was used by Du et al. (2006).
2.2.1 Theory of zero-crossing lines in multi-scale
Scaling theory for zero-crossings has been studied and applied to many applications. Yuille et al. (1986) assumed that signal is the sum of delta functions. Another similar assumption of signal, bandlimited signal, has been studied in Vo et al. (1996). However, studying zero-crossing of signals with Gaussian mixture assumption still is a new and challenging problem. We will build new theory of zero-crossing lines in multi-scale in following sections. Through our theory, parameters (position, SD, height and frequency response) of a Gaussian peak can be accurately estimated.
We use the first derivative of fi(t) to locate local maxima corresponding peak position: fi′(t0) = 0 with t0 = μi. We continue using the second derivative and third derivative of fi(t) to estimate height and SD of Gaussian peak: fi′′(t0) = 0 with t0 = μi±σi, fi′′(t0) = 0 with t0 = μi and 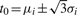 .
.
Since smoothing performed in denoising step only reduces noise and keeps small true peaks, multi-scales of Gaussian derivative wavelet are used to make local maxima and minima more robust to noise instead of only one Gaussian filter in Nguyen et al. (2009). The wavelet transform can be written as convolution product in (4):
| (4) |
According to Chapter 6 in Mallat (2009), the wavelet transform in (4) can be rewritten as a multi-scale differential operator in (5)
| (5) |
In this article, the Gaussian wavelet is used. So,  can be followed as (6):
can be followed as (6):
| (6) |
If convoluting fi(t) and 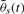 , we get result in (7)
, we get result in (7)
| (7) |
where 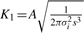 and
and 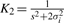 .
.
Remark. —
The zero-crossing of W1f(u, s) and W2f(u, s) belong to connected curves that are never interrupted when the scale decreases.
Proof. —
With the first derivative, (5) can be rewritten as (8)
(8) If W1f(u, s) = 0, we got u0 = μi and u0(s+1)−u0(s) = 0 with any scale s. With the second derivative, (5) can be rewritten as (9):
(9) If W2f(u, s) =0 we get
, then 0 < u0(s+1)−u0(s) ≤ 1 with any scale s.
With the third derivative, (5) can be rewritten as (10)
(10) If W3f(u, s) = 0, we get
. If we select s = 100 and σi = 0.1, then u0(100+1)−u0(100)=1.2247. In conclusion, 0≤u0(s+1)−u0(s)≤1 with the first and second derivative and zero-crossing lines belong to connected curves. Another conclusion is that zero-crossing lines is discontinuous lines if the third derivative Gaussian wavelet is used. Thus, only the first and second derivative Gaussian wavelets should be used in peak detection.
If fi is a discrete signal, (4) can be rewritten as follows:
(11) We get f(k) by sampling fi(t) with Ts:
(12) If W2f(u, s) = 0, we get
. if W3f(u, s) = 0, we get
.
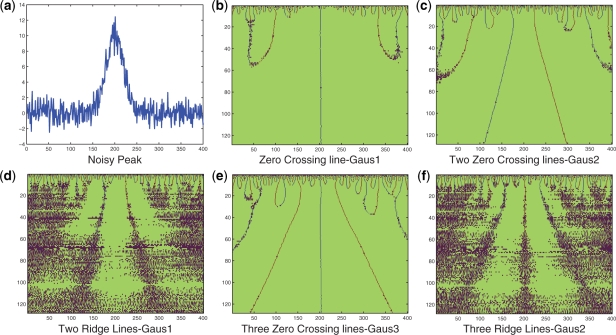
Note: zero-crossing line is more robust to noise than ridge line. This conclusion is illustrated by an example in Figure 1. Figure 1c and e show that zero-crossing lines are easier to detect than ridge lines linking local maxima or local minima points.
Fig. 1.
An illustration of zero-crossing lines and ridge lines comparison. (a) A peak sample with shape followed (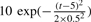 ) and Gaussian noise (SD = 1); (b) Using Gaus1, the zero-crossing line corresponds to peak position, t = 5; (c) Using Gaus2, two zero-crossing lines correspond to two peak edges whose distances to peak position are σi = 0.5; (d) Using Gaus1, two ridge lines are corresponding to two peak edges whose distances to peak position are σi = 0.5; (e) Using Gaus3, three zero-crossing lines are corresponding to one peak position and two peak edges whose distances to peak position are
) and Gaussian noise (SD = 1); (b) Using Gaus1, the zero-crossing line corresponds to peak position, t = 5; (c) Using Gaus2, two zero-crossing lines correspond to two peak edges whose distances to peak position are σi = 0.5; (d) Using Gaus1, two ridge lines are corresponding to two peak edges whose distances to peak position are σi = 0.5; (e) Using Gaus3, three zero-crossing lines are corresponding to one peak position and two peak edges whose distances to peak position are 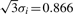 ; (f) Using Gaus2, three ridge lines are corresponding to one peak position and two peak edges whose distances to peak position are
; (f) Using Gaus2, three ridge lines are corresponding to one peak position and two peak edges whose distances to peak position are 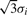 .
.
2.2.2 Applying zero-crossing to peak detection
From Section 2.2.1, parameters of a Gaussian peak could be estimated as follows:
Estimation of peak position: there are three ways to estimate peak positions throughout zero-crossing of three kind Gaussian derivative wavelets.
- The first Gaussian Derivative Wavelet (Gaus1): zero-crossing line corresponds peak position. In multi-scale, this zero-crossing line is a continuous line with length N. Peak position should be estimated by
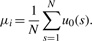
(13) - The second Gaussian Derivative Wavelet (Gaus2): there are two zero-crossing lines that correspond two edges of Gaussian peak. They are u0left and u0right. Because two zero-crossing lines are symmetric at peak position, peak position should be estimated by
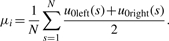
(14) - The Third Gaussian Derivative Wavelet (Gaus3): there are three zero-crossing lines if using the third GD Wavelet. They are u0left, u0middle and u0right. Because u0left and u0right are non-continuous lines, they should not be used to estimate peak position. From u0middle, we can find peak position by
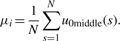
(15)
Estimation of peak's SD: Another parameter of Gaussian peak is SD σi. There are two ways to estimate σi as follows
-
The second Gaussian Derivative Wavelet (Gaus2): from Remark, σi at scale s could be calculated by
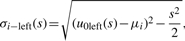
(16) 
(17) After calculating σi−left(s) and σi−right(s) at all scales, σi should be estimated by
where Nl and Nr are length of left and right zero-crossing lines.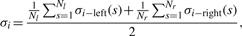
(18) - The third Gaussian Derivative Wavelet (Gaus3): from Remark, σi at scale s could be calculated by
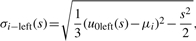
(19) 
(20)
After calculating σi−left(s) and σi−right(s) at all scales, σi should be estimated by
| (21) |
where Nl and Nr are length of left and right zero-crossing lines. However, zero-crossing lines at left and right sides of the third Gaussian derivative wavelet are disconnected lines, so it is not easy to estimate σi through (19, 20, 21).
Estimation of peak height: finally, we develop a way to estimate real height of Gaussian peak. With Gaussian peak 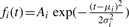 , we have
, we have
| (22) |
We can use (22) to estimate height of Gaussian peak after knowing μi and σi.
An Example: to demonstrate the above theory, we assume we have a Gaussian peak as follows:
| (23) |
where Ax = 10, μx = 5 and σx = 0.5. This peak is added Gaussian noise and baseline as follows:
| (24) |
where b is constant, a representation of baseline, μ = 0 and σ = [0.25; 0.5; 0.75; 1]. With each σ value, 200 signals f(s) have been created. One sample f(t) is shown in Figure 1a. We will estimate μx, σx and Ax using above zero-crossing theory. Error rate which is defined in (25) will be used to compare accuracy of different estimation methods:
| (25) |
Figure 1b, c and e show zero-crossing lines in 128 scales using Gaus1, Gaus2 and Gaus3. These zero-crossing lines will be used to estimate μx, σx and Ax. Table 1 lists error rates of four methods to estimate peak position μx. With Gaus1, Gaus2, and Gaus3 methods, μx values are calculated by (13, 14, 15) correspondingly. The term ‘with denoise’ means bivariate shrinkage estimator is used to denoise Gaussian noise in signal f(t). The Mexh, Mexican hat wavelet corresponding to Gaus2, is used as core part to detect peak in CWT method (Du et al., 2006) and peak tree method (Zhang et al., submitted for publication). Based on result's in Table 1, the error rate when using Mexh wavelet (Du et al., 2006) is the largest. We note that we use package ‘MassSpecWavelet’ (Du et al., 2009) which uses denoising with DWT and finds peak position using ridge lines (Du et al., 2006) with Mexh wavelet. ‘Gaus1 with denoise’ has the smallest error rate. However, error rates in Gaus1 without denoising and in Gaus2 are still acceptable and much better than in Mexh wavelet.
Table 1.
Error of peak position estimation
| σ in (24) | Gaus1 | Gaus1 | Gaus2 | Gaus2 | Gaus3 | Gaus3 | Mexh |
|---|---|---|---|---|---|---|---|
| without denoise | with denoise | without denoise | with denoise | without denoise | with denoise | (Du et al., 2006) | |
| 0.25 | 0.0519 | 0.0365 | 0.1533 | 0.1434 | 0.4890 | 0.2652 | 1.979 |
| 0.50 | 0.1319 | 0.0809 | 0.2253 | 0.1943 | 0.6918 | 0.3851 | 2.0170 |
| 0.75 | 0.1658 | 0.1034 | 0.3382 | 0.2353 | 0.7008 | 0.4855 | 2.1137 |
| 1.00 | 0.2118 | 0.1469 | 0.4630 | 0.2672 | 0.8681 | 0.5874 | 2.1618 |
Using zero-crossing lines in multi-scale of Gaussian derivative wavelet, there are three ways to estimate peak position as in (13, 14, 15). We compare errors of these estimations and CWT's estimation (Du et al., 2006). The error rate is defined by (25). In each Gaussian noise level, σ, we created 200 signals. Error value shown in this table is average value.
We can estimate σx by (18) or (21). However, with Gaus3, zero-crossing lines are not continuous lines (see Remark in Section 2.2.1). Thus, estimation of zero-crossing in 128 scales of Gaus3 is a problem. This problem causes a larger error in calculating the σx. From result of Table 2, we can conclude that Gaus2 with denoising should be used to estimate σx because its error rate is the smallest.
Table 2.
Error of peak's SD Estimation
| σ in | Gaus2 | Gaus2 | Gaus3 |
|---|---|---|---|
| (24) | without denoise | with denoise | without denoise |
| 0.25 | 1.6560 | 1.3829 | 2.3371 |
| 0.50 | 2.5626 | 2.3392 | 3.7318 |
| 0.75 | 3.3841 | 2.5087 | 4.7881 |
| 1.00 | 3.9726 | 2.8529 | 5.9220 |
By using (22) with zero-crossing lines of both Gaus2 and Gaus3, the height of Gaussian peak is estimated. In this case, baseline b which is used in (24) is a constant. From Table 3, Gaus2 with denoising gives the smallest error rate and should be used to calculate Ax.
Table 3.
Error of peak's height estimation
| σ in (24) | Gaus2 | Gaus2 | Gaus3 |
|---|---|---|---|
| without denoise | with denoise | without denoise | |
| 0.25 | 4.1032 | 1.7544 | 4.8886 |
| 0.50 | 7.8084 | 2.6869 | 8.2126 |
| 0.75 | 11.0612 | 2.8954 | 14.3860 |
| 1.00 | 13.6141 | 3.0502 | 16.9405 |
From above example, the best way to estimate peak position μx is from the first Gaussian derivative wavelet, Gaus1. The second Gaussian derivative wavelet, Gaus2, should be used to estimate SD σx and height Ax of a Gaussian peak. Figure 1d and f shows Ridge lines which correspond to zero-crossing lines in Figure 1c and e. It is clearly that detecting Ridge lines is more difficult than detecting zero-crossing lines. Ridge lines in Du et al. (2006) are similar to Ridge lines in Figure 1f, corresponding to zero-crossing line in Gaus3 which should not be used because of its high error in calculating parameters of peaks.
2.3 Saving small energy peaks by Envelope analysis
Envelope analysis was introduced by Nguyen et al. (2009). Any finite energy signal y(t) can be analyzed into three envelope signals including MAX, MIN and MED envelops at the first level. Each of these envelops can be considered as a signal and will be decomposed into three envelopes. In this article, we use MAX and MED envelops to detect peaks because MIN envelops contain no peak. We decompose the original signal into one MAX envelop at level 1, one MAX and one MED envelops at level 2 and four envelops which comprise two MAX envelops and two MED envelops at level n > 2. Empirically, 5–7 are recommended as the number of levels to get significant peaks.
2.4 Proposed GDWavelet method
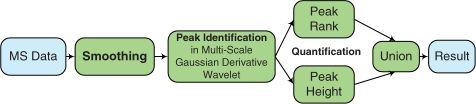
The framework of our proposed GDWavelet method is shown in Figure 2. First, raw MS data is smoothed by bivariate shrinkage estimator (2) in SWT domain to keep true signal and reduce noise. Note that, the lowest frequency detail scale and approximate scale which may include true signal should not be applied with any estimator, so that true signal is not removed. As a result, noise in signal is reduced and smoothed signal still has a little noise. Second, without applying baseline removal that often discards true peaks and creates new peaks, smoothed signal is used to estimate frequency response, position, height and SD of Gaussian peak by zero-crossing lines in multi-scale Gaussian derivative wavelet domain. Frequency response of Gaussian peak is proportional to the length of zero-crossing line if the first derivative Gaussian (Gaus1) is used. Peak position, μi, is estimated by (13). SD, σi, of Gaussian peak is calculated by (18). Result of (22) with Gaus2 gives heights of peaks. Using the first and the second derivative Gaussian wavelet, we can estimate all parameters of a Gaussian peak. After removing peaks with frequency response and SD less than a threshold, we get all peak candidates. Third, in peak quantification step, we use two rules to remove false peaks: (i) all peak candidates are quantified by peak rank (PR; Nguyen et al., 2009) in Envelop analysis. Peaks with PR = 1, even small peaks, are important peaks. (ii) Peak height is used to remove peaks with height smaller than threshold. We use peak height to substitute SNR that was used by Morris et al. (2005) and Du et al. (2006), because noise cannot be exactly estimated in either time domain or wavelet domain. Finally, the union results of two quantifying rules are the final detected peaks.
Fig. 2.
GDWavelet Method's Flowchart. Raw MS data is smoothed by bivariate shrinkage estimator in SWT domain to keep true signal and to reduce noise. Without removing baseline, smoothed signal is used to estimate parameters of peaks by zero-crossing lines in multi-scale Gaussian derivative wavelet domain. After removing peaks with frequency response and width less than a threshold, we get all peak candidates. All peak candidates are quantified by PR in envelop analysis and peak height. Union results are final output.
We randomly select 19-th sample of CAMDA, 2006 to illustrate how GDWavelet method detects MS peaks. In Figure 3a, blue signal represents raw signal and red one is signal smoothed by (2). A zoom in subfigure draws the peak which is used to show its zero-crossing lines in Figure 3b. Using one zero-crossing line in multi-scale of the Gaus1 and two zero-crossing lines in multi-scale of the Gaus2, position, height, SD, and frequency response of this peak are estimated. In Figure 3c, we quantify peaks by two rules: peak height and PR (in Envelope analysis). The circles are results from peak height-based quantification. The stars are from PR-based quantification. Union results include all peaks with heights larger than a threshold or PR one. Figure 3d shows receiver operating characteristic (ROC) curves of four related methods. GDWavlet gives the best performance.
3 EXPERIMENTS AND DISCUSSIONS
3.1 Experimental setup
Cruz-Marcelo et al. (2008) and Emanuele and Gurbaxani (2009) presented the extensive studies to compare the performance of state-of-the-art methods for SELDI data preprocessing, including CWT (Du et al., 2006), Cromwell (Coombes et al., 2005; Morris et al., 2005), PROcess (Li et al., 2006), Ciphergen and SpecAlign (Wong et al., 2005). They concluded that CWT (Du et al., 2006) has the best performance. Another method which also works well is Cromwell (Coombes et al., 2005; Morris et al., 2005). In this section, our GDWavelet method will be compared with the Cromwell (Coombes et al., 2005; Morris et al., 2005), the CWT (Du et al., 2006) and our previous method, GaborEnvelop (Nguyen et al., 2009). Cromwell method is implemented by MATLAB which can be downloaded from (UT-MD Anderson Cancer Centre, 2002). The CWT method (Du et al., 2006) was implemented in R (called ‘MassSpecWavelet’) and Version 1.12 can be downloaded from Du et al. (2009). GaborEnvelop (Nguyen et al., 2009) is implemented in MATLAB.
We evaluate the performance of above methods by the ROC curve. Both simulated and real data are used. The first simulated data was proposed by Morris et al. (2005) and Coombes et al. (2005) and is available for download at Simulated Proteomics Spectra (2005). In this data, hundreds of mean spectrum samples with hundreds of proteomics datasets are generated.
Based on the simulation engine developed by Morris et al. (2005) and code (R and MATLAB) to generate simulated data proposed by Cruz-Marcelo et al. (2008) and Zhang et al. (submitted for publication), we also create two new simulated datasets to investigate noise affection on different algorithms. The 100 spectrums with 20 − 30 true peaks are created first, and Gaussian and real noise are added separately to get two datasets. When Gaussian noise is added, each sample includes 20% of protein peaks which are below three time of SNR. Real noise is extracted from real data in which there is no true peaks. There is only noise from 26000 (index) to end in first sample of CAMDA, 2006. Real noise probability density function is built. Using this function, noise with different SD will be created. Based on this configuration, we create about 20–30 true peaks and more small energy peaks in simulated data.
The CAMDA dataset (2006) of all-in-1 Protein Standard II (Ciphergen Cat. # C100–007) is the real dataset. Because we know polypeptide composition and position in this dataset, we can estimate the sensitivity and the FDR. There are seven polypeptides which create seven true peaks at 7034, 12 230, 16 951, 29 023, 46 671, 66 433 and 147 300 of the m/z values.
The sensitivity and FDR of four methods are calculated for 60 real MS signals and three simulated MS datasets with 100 signals each. The SNR thresholding values are increased gradually to calculate the ROC curves of Cromwell and CWT methods. The SNR thresholding values are chosen from 0 to 20 for Cromwell method and from 0 to 120 for CWT method. In our GDWavelet method, the peak height ratio, which is defined as the ratio of current peak height over average height of peaks, is changed from 0 to 10 to build the ROC curve. We plot the average ROC curves in Figures 4 and 5. We should notice that we take average of all ROC points with the same SNR threshold (for Cromwell and CWT) and the same peak height rate (for our GDWavelet method).
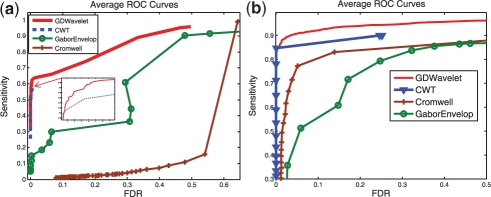
Fig. 4.
ROC Curves—simulated data with Gaussian noise. Average ROC curves of four methods (Cromwell, CWT, GaborEvelop and GDWavelet). (a) Obtained from 100 mean simulated MS signals which can be downloaded from (Simulated Proteomics Spectra, 2005). There are 149 true peaks in this data. (b) Obtained from 100 simulated MS signals in which Gaussian noise is added. There are 20–30 true peaks in this data.
Fig. 5.
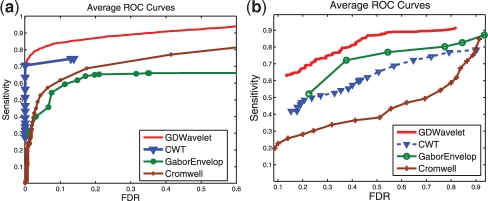
ROC curves—simulated data with real noise and real data. Average ROC curves of four methods (Cromwell, CWT, GaborEvelop and GDWavelet). (a) Obtained from 100 simulated MS signals in which real noise is used. There are 20–30 true peaks in this data. (b) Obtained from 60 MS signals (CAMDA, 2006). There are seven true peaks in this data.
3.2 Experimental results
Three simulated datasets and one real SELDI-TOF dataset are used to create ROC curves in Figures 4 and 5. In all four datasets, the performance of Cromwell is not stable and gets worse than CWT and GDWavelet. Between GaborEnvelop which used Envelope analysis and CWT which used ridge lines and SNR in peak quantification, GaborEnvelop is better than CWT in real data in Figure 4b and CWT is better than GaborEnvelop in simulated data. In all cases, our GDWavelet method has much better performance than GaborEnvelop and CWT methods. At the same FDR, the sensitivity of our method is consistently higher than GaborEnvelops and CWTs. It is clear that utilizing both of Envelope analysis and Gaussian derivative wavelet in peak quantification made significant contributions to detect both high energy and small energy peaks. Bivariate shrinkage estimator in wavelet domain guarantees that denoising step in our method saves more true signal than in Morris et al. (2005). Zero-crossing lines-based peak parameters estimations in our article is more accurate and robust to noise than ridge lines in Du et al. (2006). Envelope analysis is more efficiently used in GDWavelet than in GaborEnvelop. Therefore, the GDWavelet has better peak detection results than Cromwell, GaborEnvelop and CWT. Thus, it is an efficient and accurate method to detect peaks in both real and simulated MS data. In Figures 4 and 5, CWT's ROC curves is limited in small FDR because two thresholds of the length of ridge lines and the scale corresponding to the maximum amplitude on the ridge line are used as default in MassSpecWavelet (Du et al., 2009). Finally the runtime of GDWavelet algorithm is comparable to CWT method, because both methods need more computational time to decompose a signal to many scale using continuous wavelet transform.
4 CONCLUSIONS
In this article, we proposed new zero-crossing line theory in multi-scale of Gaussian derivative wavelet to estimate parameters of peaks in MS which has been assumed as a mixture of Gaussian. A novel GDWavelet method was proposed to efficiently and accurately detect MS peaks by integrating bivariate shrinkage model, Gaussian derivative and Envelope analysis. The bivariate shrinkage estimator in SWT domain was used to reduce noise and still keep true peaks. All parameters of a Gaussian peak, estimated by multi-scale in Gaussian derivative wavelet and Envelope analysis, have been used to remove false peaks. The peak height and PR were introduced as a nice substitution of the previous SNR method to identify true peaks. Both simulated data and real MS data are used to evaluate our GDWavelet method. Simulated data were created with both Gaussian noise and real noise. Our GDWavelet method gave out a much better performance in the ROC curves than three other state-of-the-art peak detection methods. GDWavelet algorithm will be extended and test with other kinds of MS (such as MALDI-TOF) as future work. Based on GDWavelet method, many MS data-related applications will be improved, such as protein identification, biomarker discovery, cancer classification, etc.
Funding: NSF-CCF 0830780; NSF-CCF 0939187; NSF-CCF 0917274; NSF-DMS 0915228; NSF-CNS 0923494; UTA-REP.
Conflict of Interest: none declared.
REFERENCES
- CAMDA. Conference contest. 2006 Available at http://camda.duke.edu/camda06/datasets/index.html (last accessed data July 12, 2010) [Google Scholar]
- Coombes K, et al. Improved peak detection and quantification of mass spectrometry data acquired from surface-enhanced laser desorption and ionization by denoising spectra with the undecimated discrete wavelet transform. Proteomics. 2005;5:4107–4117. doi: 10.1002/pmic.200401261. [DOI] [PubMed] [Google Scholar]
- Cruz-Marcelo A, et al. Comparison of algorithms for pre-processing of seldi-tof mass spectrometry data. Bioinformatics. 2008;24:2129–2136. doi: 10.1093/bioinformatics/btn398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, et al. Mass spectrum processing by wavelet-based algorithms. 2009 Available at http://bioconductor.org/packages/2.5/bioc/html/MassSpecWavelet.html (last accessed date July 12, 2010) [Google Scholar]
- Du P, et al. Improved peak detection in mass spectrum by incorporating continuous wavelet transform-based pattern matching. Bioinformatics. 2006;22:2059. doi: 10.1093/bioinformatics/btl355. [DOI] [PubMed] [Google Scholar]
- Donoho D, et al. De-Noising by soft-thresholding. IEEE Trans. Inf. Theory. 1995;41:613–627. [Google Scholar]
- Emanuele V, Gurbaxani B. Benchmarking currently avaible SELDI-TOF MS preprocessing techniques. Proteomics. 2009;9:1754–1762. doi: 10.1002/pmic.200701171. [DOI] [PubMed] [Google Scholar]
- Huang H, et al. Array CGH data modeling and smoothing in stationary wavelet packet transform domain. BMC Genomics. 2008;9:S2–S17. doi: 10.1186/1471-2164-9-S2-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. SELDI-TOF mass spectrometry protein data. New York: Springer; 2006. pp. 91–109. Ch. 6. [Google Scholar]
- Mallat S. Wavelet Tour of Signal Processing - The Sparse Way. Academic Press; 2009. [Google Scholar]
- Myers C, et al. Accurate detection of aneuploidies in array CGH and gene expression microarray data. Bioinformatics. 2004;20:3533–3543. doi: 10.1093/bioinformatics/bth440. [DOI] [PubMed] [Google Scholar]
- Morris J, et al. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics. 2005;21:1764–1775. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- Nguyen N, et al. Peak detection in mass spectrometry by gabor filters and envelope analysis. JBCB. 2009;7:547–569. doi: 10.1142/s0219720009004229. [DOI] [PubMed] [Google Scholar]
- Nguyen N, et al. Stationary wavelet packet transform and dependent Laplacian bivariate shrinkage estimator for array-CGH data smoothing. J. Comput. Biol. 2010;17:139–152. doi: 10.1089/cmb.2009.0013. [DOI] [PubMed] [Google Scholar]
- Selesnick IW, et al. Hilbert transform pairs of wavelet bases. IEEE Signal Process. Lett. 2001;8:170–173. [Google Scholar]
- Sendur L, et al. Bivariate shinkage function for wavelet-based denoising exploiting interscale dependency. IEEE Trans. Signal Process. 2002;50:2744–2756. [Google Scholar]
- 2005. [(last accessed date July 12, 2010)]. Simulated Proteomics Spectra. Available at http://bioinformatics.mdanderson.org/Supplements/Datasets/Simulations/index.html. [Google Scholar]
- UT-MD Anderson Cancer Center. The new model processor for mass spectrometry data. 2002 Available at http://bioinformatics.mdanderson.org/cromwell.html (last accessed date July 12, 2010) [Google Scholar]
- Vo A, et al. Scaling theorems for zero crossings of bandlimited signals. IEEE Trans. Pattern Anal. Mach. Intell. 1996;18:309–320. [Google Scholar]
- Wong J, et al. Specalign processing and alignment of mass spectra datasets. Bioinformatics. 2005;21:2088–2090. doi: 10.1093/bioinformatics/bti300. [DOI] [PubMed] [Google Scholar]
- Yuille A, et al. Scaling theorems for zero crossings. IEEE Trans. Pattern Anal. Mach. Intell. 1986;8:15–25. doi: 10.1109/tpami.1986.4767748. [DOI] [PubMed] [Google Scholar]