Abstract
Summary: The first open source software suite for experimentalists and curators that (i) assists in the annotation and local management of experimental metadata from high-throughput studies employing one or a combination of omics and other technologies; (ii) empowers users to uptake community-defined checklists and ontologies; and (iii) facilitates submission to international public repositories.
Availability and Implementation: Software, documentation, case studies and implementations at http://www.isa-tools.org
Contact: isatools@googlegroups.com
1 HIGH-THROUGHPUT OMICS STUDIES
The development of high-throughput genomic and post-genomic (hereafter, ‘omics’) technologies entails changes in the handling, processing and sharing of data (Schofield et al., 2009). Omics datasets are often complex and rich in context. Studies may run material through several kinds of assay, using both omics and other technologies; for example, studying the effect of a compound on rat liver through transcriptome, proteome and metabolome profiling (using high-throughput sequencing and two kinds of mass spectrometry, respectively) alongside conventional analyses (e.g. histopathology). Such data must be accompanied by enough contextual information (i.e. metadata; sample characteristics, technology and measurement types; instrument parameters and sample-to-data relationships) to make datasets comprehensible and reusable if they are to underpin future investigations.
Many funders and journals require that researchers share data, and encourage the enrichment and standardization of experimental metadata (Field et al., 2009). Consequently, more and richer studies are flowing into public databases. However, two bottlenecks can significantly hamper this process, necessitating urgent solutions. First, international public repositories for ‘omics data such as GEO (Barrett et al., 2009), ArrayExpress (Parkinson et al., 2009), PRIDE (Vizcaíno et al., 2010), ENA, SRA and DRA (Shumway et al., 2010), have their own submission formats, data models and terminologies, created for specific types of assay. This complicates the submission process for researchers producing multi-assay studies (and greatly increases the risk that these datasets become irrevocably fragmented). Secondly, the shortage of curators to check and annotate submissions to public repositories—a situation unlikely to change soon—necessitates better annotation at source (by experimentalists or community-based efforts; Howe et al., 2008). Free software, with automated content validation, is required to facilitate the collection, management and curation of a variety of study inhouse, and to format those data for submission to public repositories. Such software should support community-defined reporting standards, such as the minimum information checklists listed by the MIBBI Portal (Taylor et al., 2007), and ontologies, (Côté et al., 2006; Smith et al., 2007; Noy et al., 2009).
The Investigation/Study/Assay (ISA) infrastructure described here is the first general-purpose format and freely available desktop software suite designed to regularize local management of experimental metadata by enabling curation at source, supporting community-defined reporting standards and preparing studies for submission to public repositories.
2 THE ISA FORMAT AND SOFTWARE SUITE
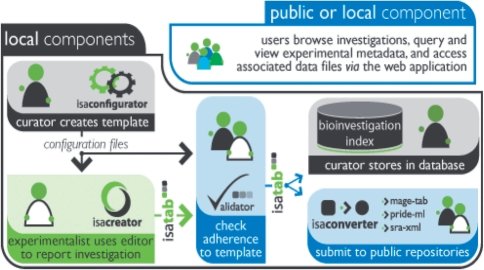
The software suite comprises five platform-independent Java-based software components for local use, including a relational database (Fig. 1), built around the ISA-Tab format. The components work both as stand-alone applications and as a unified system to assist in the local management and storage of experimental metadata, and to facilitate data submission to international public repositories. All components run as ‘desktop’ applications; in addition, the database component features a web-based query interface.
Fig. 1.
The role of each ISA software component, showing their interrelations, target users and the flow of information through the system.
2.1 ISA-Tab: an extensible, cross-domain format
‘Investigation’, ‘Study’ and ‘Assay’ are the three key entities around which the general-purpose ISA-Tab format for structuring and communicating metadata is built (Sansone et al., 2008). Investigation contains all the information needed to understand the overall goals and means used in an experiment; Study is the central unit, containing information on the subject under study, its characteristics and any treatments applied. Each Study has associated Assay(s), producing qualitative or quantitative data, defined by the type of measurement (i.e. gene expression) and the technology employed (i.e. high-throughput sequencing). The hierarchical structure of ISA-Tab enables the representation of studies employing one or a combination of omics and other technologies, overcoming the fragmentation of the existing submission formats built for specific types of assay. To ensure conversion, ISA-Tab has been designed with reference to these existing ‘omics formats (Jones et al., 2007), complementing and extending their work where necessary; for example, it shares both syntax and the use of easily-manipulable tab-delimited text files with ArrayExpress’ MAGE-Tab (Rayner et al., 2006). Additionally, where omics-based technologies are used in clinical or non-clinical studies, ISA-Tab complements existing biomedical formats such as the Study Data Tabulation Model (http://www.cdisc.org/sdtm), endorsed by the US Food and Drug Administration. ISA-Tab also complements the XML formats used by the PRIDE, ENA, SRA and DRA repositories, and consequently offers a way to render their experimental metadata documents in a more user-friendly format. Note though that ISA-Tab is simply a format; the decision on how to regulate its use (i.e. enforcing the filling of required fields, or the use of ontologies) is left to local administrators' use of ISA software components, or the growing number of other systems and groups implementing the format (e.g. Krestyaninova et al., 2009; SysMO-DB http://www.sysmo-db.org/community; XperimentR, http://www.imperial.ac.uk/bioinfsupport/resources/data_management/; more given on the ISA web site).
2.2 ISAcreator: a user-friendly editor
This desktop application enables users (i.e. experimentalists) to compile experimental metadata sets, and to import and edit existing ISA-Tab formatted files. It breaks down overall descriptions into relatively simple parts, uses graphical abstraction to enable visualization of the information described and facilitates time-efficient description of experimental steps by remembering prior behaviour (through user profiles). ISAcreator's aesthetically pleasing interface makes extensive use of Java Swing and external open source libraries (e.g. Prefuse, http://prefuse.org/). The editor uses a style of form- and spreadsheet-based data entry that is likely to be familiar to researchers, augmenting basic functionality such as ‘auto-fill’ and ‘undo’ with advanced features, listed below.
2.2.1 Ontology support
A dedicated ‘widget’ allows ontology terms to be searched for and inserted in real time via the BioPortal (Noy et al., 2009) and the Ontology Lookup Service (Côté et al., 2006). Terms from those sources are imported along with core metadata (identifiers, definitions and ontology version); term selection is facilitated by a search history displaying prior choices (through user profiles).
2.2.2 Design wizard
An alternative way for users to enter information that leverages common patterns to reduce repetitive tasks by guiding users through a series of questions that elicit information about the design of the Study and associated Assay(s).
2.2.3 Spreadsheet import
As a second alternative, this widget enables the mapping and import of information from existing spreadsheets; also the reformatting and reannotation of legacy data.
2.2.4 Data file chooser
This widget appends data files located either local to the operator, or identified by FTP on a remote system, to an experimental metadata sets. Upon completion of a valid investigation report, ISAcreator outputs a compressed ‘ISArchive’ containing the ISA-Tab-formatted metadata and either the actual data files, or a reference to them, if necessary (e.g. because of their large size), consisting of their address and file name.
2.3 ISAconfigurator: standards-compliant templates
This desktop application allows ‘power users’ (i.e. community curators) to customize the fields displayed by ISAcreator, and for example, to meet the requirements of one or more MIBBI minimum information checklists by declaring certain fields mandatory, or by specifying allowed values (e.g. drawn from a set of ontology terms, or formatted in a specific manner). Configuration files from ISAconfigurator are read by ISAcreator, which then generates interface components as required.
2.4 ISAvalidator: adherence to templates
This desktop application also reads configuration files and checks both that completed ISA-Tab files meet specified requirements and that associated data files have been linked. Whether ISA-Tab files are created with ISAcreator or another way (e.g. with spreadsheet software), ISAvalidator checks that the document is syntactically correct and internally consistent, and reports on errors (i.e. missing or incorrect values).
2.5 BioInvestigation Index: local storage
An ISArchive provides a simple way to store and share information in a structured manner, but those tasks are better performed by uploading such a file to an instance of our ‘BioInvestigation Index’ (BII), or another system that implements ISA-Tab import. The BII includes a management tool and relational database (tested with Oracle, MySQL and PostgreSQL). The former enables validation and loading of an ISArchive and provides simple permissions functionality to link users (or groups of users) to studies. The latter manages the storage of experimental metadata, which can be collectively searched and browsed via a query interface or web services; the destination for associated data files, and their protocol for transfer, is custom defined by the local administrator on installation. As an example, a publicly accessible instance of the BII, maintained by the European Bioinformatics Institute (http://www.ebi.ac.uk/bioinvindex), has proven useful as a curation and storage system for multi-assay studies, and as a mechanism for submitting data files to ArrayExpress, PRIDE, ENA and SRA. Installation of the BII system requires some knowledge of database management. However, it is portable enough to be easily installed in individual labs, to maximize the efficiency with which high-throughput studies can be managed and shared among users that have been granted access to them.
2.6 ISAconverter: submission to public repositories
ISAconverter recodes the relevant parts of ISArchives as MAGE-Tab, PRIDE XML or SRA-XML (used by ArrayExpress, PRIDE and ENA, SRA and DRA, respectively), enabling combined submission to public omics repositories. It is readily extensible to support export of other formats, e.g. SOFT required by GEO (Barrett et al., 2009). Mappings for format elements are available in the ISA-Tab specification and documentation on the ISA web site.
3 COLLABORATIONS AND CASE STUDIES
Developed for the European multi-site ‘CarcinoGENOMICS’ project (Vinken et al., 2008), the ISA software suite version one was released in early 2009. The core ISA developers are engaged with an ever-growing number of collaborators: case studies from early implementers already provide evidence of the diverse life science scenarios in which the suite's various components have been successfully tested and are being used with large datasets (details on the ISA web site). The main limitations recorded to date are simply the person hours required to specify the standards and ontologies to be used and to actually curate studies.
Demonstrable acceptance and community engagement has also brought a new funding stream for this project, allowing us to continue the collaborative development of this exemplar system that supports data sharing policies, promotes the uptake of community-defined reporting standards and ontologies and enables curation at source (Field et al., 2009). The ISA components, in particular the BII, have been designed to provide core functionalities. Inevitably, each collaborator has additional in-house requirements that are too specific to be included as core functionality. This may be due to the nature of their studies or their need for one or more ISA software components to be interoperable with existing systems. To support further collaborative development, the core ISA developers are setting up an environment for distributed development, and are augmenting the ISA code base with Application Programming Interfaces (APIs). Ongoing collaborative activities include: a module to enable the analysis of ISA-Tab formatted metadata and any associated data, using R; integration with other data management and analysis systems (e.g. Fang et al., 2009; MetWare, http://metware.org); and giving assistance to the growing number of projects exploring the tools and underlying format (e.g. Sage http://sagecongress.org/WP/workstreams/Standards; Kawaji et al., 2009). Other collaborative activities include an enhanced user authentication system, support for additional formats such as RDF, OWL and SOFT, converters to/from lab equipment-related file formats (e.g. sampling robots and mass spectrometers) and improved packaging and distribution mechanisms to offer a single download bundle to facilitate installation.
ACKNOWLEDGEMENTS
The ISA developers owe debts of gratitude to many collaborators, as listed at: http://isatab.sf.net/people_funding.html.
Funding: CarcinoGENOMICS, NuGO, BBSRC (BB/I000917/1, BB/G000638/1, BB/E025080/1), NERC-NEBC and EMBL.
Conflict of Interest: none declared.
REFERENCES
- Barrett T, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté RG, et al. The ontology lookup service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics. 2006;7:97. doi: 10.1186/1471-2105-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, et al. ArrayTrack: an FDA and public genomic tool. Methods Mol. Biol. 2009;563:379–398. doi: 10.1007/978-1-60761-175-2_20. [DOI] [PubMed] [Google Scholar]
- Field D, et al. ‘Omics Data Sharing. Science. 2009;9:234–236. doi: 10.1126/science.1180598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D, et al. Big data: the future of biocuration. Nature. 2008;4:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, et al. The Functional Genomics Experiment model (FuGE): an extensible framework for standards in functional genomics. Nat. Biotechnol. 2007;25:1127–1133. doi: 10.1038/nbt1347. [DOI] [PubMed] [Google Scholar]
- Kawaji H, et al. The FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Genome Biol. 2009;10:R40. doi: 10.1186/gb-2009-10-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krestyaninova M, et al. A System for Information Management in BioMedical Studies—SIMBioMS. Bioinformatics. 2009;25:2768–2769. doi: 10.1093/bioinformatics/btp420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy NF, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37:W170–W173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, et al. ArrayExpress update-from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:868–872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner TF, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone SA, et al. The first RSBI (ISA-TAB) workshop: “can a simple format work for complex studies?”. OMICS. 2008;12:143–149. doi: 10.1089/omi.2008.0019. [DOI] [PubMed] [Google Scholar]
- Schofield PN, et al. Post-publication sharing of data and tools. Nature. 2009;10:171–173. doi: 10.1038/461171a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumway M, et al. Archiving next generation sequencing data. Nucleic Acids Res. 2010;38:870–871. doi: 10.1093/nar/gkp1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, et al. MIBBI: a minimum information checklist resource. Nat. Biotechnol. 2007;26:889–896. [Google Scholar]
- Vinken M, et al. The carcinoGENOMICS project: critical selection of model compounds for the development of omics-based in vitro carcinogenicity screening assays. Mutat. Res. 2008;659:202–210. doi: 10.1016/j.mrrev.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, et al. The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 2010;38:736–742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]