Abstract
Summary: Cytoscape Web is a web-based network visualization tool–modeled after Cytoscape–which is open source, interactive, customizable and easily integrated into web sites. Multiple file exchange formats can be used to load data into Cytoscape Web, including GraphML, XGMML and SIF.
Availability and Implementation: Cytoscape Web is implemented in Flex/ActionScript with a JavaScript API and is freely available at http://cytoscapeweb.cytoscape.org/
Contact: gary.bader@utoronto.ca
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Increasing amounts of high-throughput data are being collected, stored, shared and analyzed on the web, highlighting the need for effective web-based data visualization. Network visualization components are especially valuable to help researchers interpret their data as part of data analysis tools. However, current web-based network visualization components lack many useful features of their desktop counterparts. Medusa (Hooper and Bork, 2005) is a Java applet originally used in the STRING database (Jensen et al., 2009) and by many other web sites for network visualization, but lacks advanced features, such as detailed customization of the network view. jSquid (Klammer et al., 2008) expands Medusa's functionality, but does not provide an easy way for the client web site to change and interact with the network view after it has been rendered. TouchGraph (http://www.touchgraph.com/navigator.html) is another Java applet for network visualization, but provides only one mode of network interaction designed for exploration and is not easily customizable. yFiles Flex (http://www.yworks.com/en/products_yfilesflex_about.html) is a rich Internet application with a feature-rich user interface, an architecture that balances client/server work and supports efficient data communication. This commercial software is customizable within the bounds of the code already written, but is not open source. Cytoscape (http://www.cytoscape.org/) is an open source Java network visualization and analysis tool that provides a large array of useful features (Shannon et al., 2003), but is not specifically designed for use on the web except via Java WebStart or as a library to generate static network images for web display. The field of network visualization is lacking an interactive, easily customizable, open source, web-based visualization component.
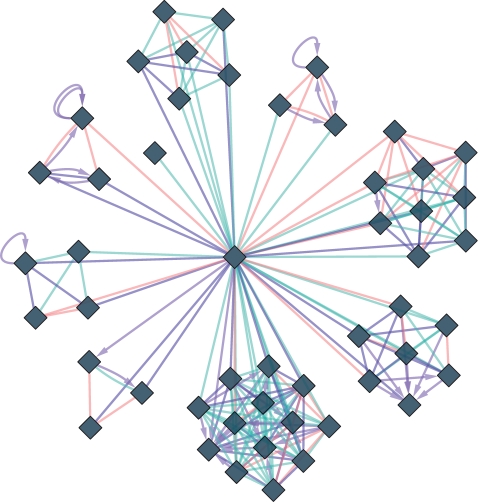
Cytoscape Web is an interactive, web-based network visualization tool, modeled after the popular Cytoscape software (Fig. 1). Using basic programming skills, Cytoscape Web can be customized and incorporated into any web site. Cytoscape Web is not intended as a replacement for the Cytoscape desktop application, for example, it contains none of the plugin architecture functionality of Cytoscape; instead it is intended as a low overhead tool to add network visualization to a web application.
Fig. 1.
A Cytoscape Web network with a customized visual style.
2 IMPLEMENTATION
Cytoscape Web is a client-side component that requires no server-side implementation, and allows developers to choose any server-side technology, if necessary. The main network display component of Cytoscape Web is implemented in Flex/ActionScript, but a JavaScript application programming interface (API) is provided so all the customization and interaction with the network view can be easily built in JavaScript without needing to change and compile the Flash code. This architecture has the advantage of using the Flash platform to implement complex and interactive vector images that behave consistently across major browsers, but without requiring the web site to be entirely built with this technology. In other words, the web site itself can rely on web standards (HTML, CSS and JavaScript) for embedding and interacting with Cytoscape Web. This design also offers the possibility of migrating the implementation to other technologies—such as scalable vector graphics (SVG) and HTML5—in the future without making major API changes. The choice of Flash rather than Java is motivated by the fact that Java applets can be slow to launch, require the download of the large Java runtime and make it difficult to create custom (non-Swing) user interfaces without writing low-level graphics code.
3 AVAILABLE FEATURES
3.1 Features
Similarly to Cytoscape, Cytoscape Web allows the client application to define a network of nodes and edges and customize their attributes. Data can be loaded into Cytoscape Web through one of the supported XML-based exchange formats (GraphML or XGMML) or a simple tab-delimited text (Cytoscape's SIF format). The network data can also be exported to any of the above-mentioned formats. The client can dynamically change node and edge visual styles (e.g. color, size and opacity), using any of the following methods: (i) specifying default visual properties for all elements; (ii) mapping node and edge attributes (e.g. name, interaction type and weight) to visual styles; and (iii) overriding default or mapped styles by setting a bypass style. For instance, different types of interactions can be mapped to edge colors (e.g. protein–protein to blue, protein–DNA to green), and the edge width can be used to represent interaction weight. Then the developer can use the bypass mechanism to create a first neighbors highlight feature, for example, by setting the nodes and edges that belong to the neighbor set to the color red. When the first neighbors' bypass is removed, the colors are automatically restored to their default or mapped values. These three options, combined with more than 20 visual properties for nodes and edges, provide flexibility and enable each Cytoscape Web-based application to define its own semantics, styles and features. For example, iRefWeb (http://wodaklab.org/iRefWeb/), an interface to the interaction Reference Index (iRefIndex) database (Razick et al., 2008), uses a basic implementation of Cytoscape Web to display all interactions in which a single query gene participates. Alternatively, GeneMANIA (http://www.genemania.org; Warde-Farley et al., 2010), a gene function prediction tool, uses a more complex implementation of Cytoscape Web to extend a user's input gene list and display interactions among the genes. Cytoscape Web communicates with the GeneMANIA server, mediated by client-side JavaScript, to display gene or network-specific highlights and associated information interactively.
Cytoscape Web's API can be used to implement the following features: a filter for nodes and edges, which temporarily removes the filtered out elements based on attribute data; functions for adding and deleting nodes and edges at runtime; the ability to export the whole network as an image, either to a PNG (Portable Network Graphics) file or to a publication quality vector format (PDF). Cytoscape Web provides the ability to pan and zoom the network and choose different network layouts, including force directed. The layout parameters can be customized, but if none of the available layouts produces the desired results, the web application can run an external layout algorithm—in JavaScript or on the server-side, for instance—and pass the results, via node positions, back to Cytoscape Web for visualization.
3.2 Performance
Cytoscape Web works best with small- to medium-sized networks, generally with up to a few hundred nodes and edges. Larger networks can be visualized, but the user interaction can become sluggish around 2000 elements (nodes or edges)—800 nodes and 1200 edges, for example (tested on an Apple laptop computer with 2 GHz dual core CPU and 4 GB RAM). Use of the force-directed layout is the major bottleneck in the initial rendering of a typical network. However, faster layouts are available and overall performance is dependent upon the client web site implementation and the end user configuration. Additional performance statistics for Cytoscape Web are available in the Supplementary Material.
3.3 Documentation
Cytoscape Web is actively developed as an open source project and is freely available at http://cytoscapeweb.cytoscape.org/. This web site includes a tutorial, with ready to use sample code, the API documentation and a showcase of major Cytoscape Web features. The online examples can be freely used as a template for building web sites containing Cytoscape Web.
3.4 Future directions
Future plans include the implementation of custom graphics for nodes and edges, additional network layouts and support for importing and closer integration with Cytoscape [e.g. importing/exporting networks in Cytoscape (.cys) file format].
Supplementary Material
ACKNOWLEDGEMENTS
Cytoscape is developed through an ongoing collaboration between the University of California at San Diego, the Institute for Systems Biology, Memorial Sloan-Kettering Cancer Center, Institut Pasteur, Agilent Technologies, Unilever, the University of Michigan, the University of California at San Francisco and the University of Toronto. We gratefully acknowledge the contributions of many Cytoscape developers who developed software that Cytoscape Web was based on. We thank the entire GeneMANIA team for support during the development of Cytoscape Web.
Funding: Genome Canada through the Ontario Genomics Institute (grant number 2007-OGI-TD-05); the U.S. National Institute of General Medical Sciences of the National Institutes of Health (grant number 2R01GM070743-06).
Conflict of Interest: none declared.
REFERENCES
- Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klammer M, et al. jSquid: a Java applet for graphical on-line network exploration. Bioinformatics. 2008;24:1467–1468. doi: 10.1093/bioinformatics/btn213. [DOI] [PubMed] [Google Scholar]
- Razick S, et al. iRefIndex: a consolidated protein interactions database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.